ABSTRACT
Since antimicrobial resistance, especially β-lactam resistance genes were common in clinical Escherichia coli strains, this study had designed and developed multiplex amplification platform for rapid and accurate detection of such resistance genes in 542 clinical E. coli isolates. The obtained specimens were subjected to bacteriological examination, antimicrobial susceptibility testing, and detection of β-lactamase genes and plasmid replicons. The major virulence genes were detected by 7 groups of multiplex PCR and eight groups of multiplex PCR were designed to detect 8 different plasmid replicons including parA-parB, iteron, repA, and RNAI. It was found that most MDR isolates were co-resistant to penicillins (AMP) and fluoroquindones (LVX, CIP) and distribution of LVX and CIP resistance was significantly higher among female than male gender. RNAI (AY234375) showed the highest detection rate, followed by the iteron (J01724) and repA (M26308), indicating the relatively higher carriage rate of corresponding plasmids. BlaOXA acquired the highest carriage rate, followed by group 2 blaCTX-M and blaSHV-1, indicating their prevalence among clinical E. coli. Among the β-lactamase genes, blaOXA acquired the highest carriage rate, followed by group 2 blaCTX-M and blaSHV-1, indicating their prevalence among clinical E. coli. The RNAI (AY234375) showed the highest detection rate, followed by the iteron (J01724) and repA (M26308), indicating the relatively higher carriage rate of the corresponding plasmids by clinical E. coli isolates. It is shown that the developed multiplex amplification methodology is applicable to AMR detection, and such identification of plasmid replicons and β-lactamase genes may aid in the understanding of clinical E. coli isolate epidemiology.
Graphical Abstract
1. Introduction
In recent decades, antimicrobial resistance (AMR) in microorganisms has been considered to be a leading concern for human health, which has posed a significant obstacle for therapeutic treatment on various clinical infections. In 2016, the United Nations High-Level meeting had declared that progress toward achieving several of the Sustainable Development Goals (SDGs) is threatened by AMR [Citation1]. In 2020, the World Health Organization (WHO) had announced the launch of 2 new AMR indicators in the monitoring framework of the SDGs linked to the health target 3, which monitor proportion of bloodstream infections (BSIs) due to 2 typical Super-bugs, Escherichia coli and Staphylococcus aureus [Citation2]. Consequently, in this study, a strategy design based on multiplex amplification and further application for rapid detection on antimicrobial resistance and β-lactam resistance genes had been performed on a large scale of clinical Escherichia coli strains [Citation3].
Bacterial factors are reported to be associated with E. coli pathogenesis and progression [Citation4]. Moreover, pathogenic E. coli strains show great diversity in gene content, virulence factors, genomic Islands, and pathogenicity Islands [Citation5]. Pathogenic Escherichia coli are capable of causing various diseases in humans, including several types of diarrhea, urinary tract infections, sepsis, and meningitis [Citation6,Citation7]. Due to the widely application of antibiotics, the emergence of E. coli isolates with multiple antibiotic-resistant phenotypes, involving coresistance to four or more unrelated families of antibiotics, has been considered a serious health concern [Citation8,Citation9]. A large number of genes are responsible for antibiotic resistance. As the first identified antibiotic, β-lactams have become widely applied. Introduction of β-lactams in clinical settings was quickly followed by the emergence of numerous β-lactamases [Citation10]. Identification of β-lactamase genes is important to understand resistance epidemiology and identify resistant strains [Citation11]. β-lactamase genes has been reported to frequently transferred by plasmids [Citation11–13]. The spread of extended-spectrum β-lactamases (ESBLs) is an emerging global public health problem. Until the late 1990s, TEM and SHV enzymes were the most common extended-spectrum β-lactamases (ESBLs). In the late 1990s, the prevalence of both TEM and SHV decreased, whereas that of CTX-M increased, especially associated with Escherichia coli species. Within a few years, CTX-M ESBL-producing E. coli had spread across the world, involved in both nosocomial outbreaks and community-acquired infections [Citation14].
Plasmids are capable of increasing bacterial genetic diversity, acquiring and losing genes, and can be horizontally exchanged among bacterial populations by conjugation or mobilization [Citation15]. Plasmids contain genes essential for initiation and control of replication and accessory genes [Citation16,Citation17]. Replicons contribute to the replication of plasmid and represent the acquisition of plasmids.
This study collected and analyzed the 10-year antimicrobial resistance data of clinical E. coli isolates and aimed in identifying the distribution of β-lactamase genes and plasmid replicons, which may aid in the understanding of its epidemiology and the development of control strategies.
2. Material and methods
2.1 Bacterial strain
During 2008 to 2018, a total of 542 E. coli isolates were isolated from the First Affiliated Hospital of Guangzhou Medical University (FAHGMU). The large proportion of patients in FAHGMU are from Guangdong and other adjacent provinces, which can represent the epidemiology of E. coli in Southern China. Both inpatients and outpatients were included in this study. For the criteria of bacterial selection, strains were averagely distributed in different months, with an average of 4 to 5 strains isolated in 1 month. Also, strains were selected according to the location of medical settings and wards, as well as their antimicrobial resistance profile. The isolates datas included demographic information such as date, age, gender of patients, department of isolation and infection types.
2.2 Bacterial Isolation and Identification
Bacterial identification to the species level on all tested strains had been performed by standard procedures: colony morphology, Gram staining, the API commercial kit and the Vitek 2 automated system [Citation18]. Isolation of E. coli was performed using standard bacteriological methods. The clinical samples were incubated in Violet Red Bile Agar (VRBA, Huankai Microbial, China) for 24 h at 37°C and those pink colored presumptive E. coli were sub-cultured in Eosin Methylene Blue (EMB, Huankai Microbial, China) agar to get a pure colony. Colonies with metallic green sheen were characterized micro-scopically using Gram stain and other different biochemical tests such as indole production, methyl-red test, Voges-Proskauer test, citrate utilization (IMViC) test, oxidase test, sugar fermatation, triple sugar iron [Citation19]. Furthermore, the identified isolates were confirmed by using a species-specific set of primers targeting 16S rRNA gene of E. coli (27FYM: 5′- AGAGTTTGATYMTGGCTCAG-3′; 1492 R: 5′- GGTTACCTTGTTACGACTT-3′) [Citation20].
2.3 Antimicrobial susceptibility testing
Antimicrobial susceptibility testing was performed by disk diffusion according to the Clinical and Laboratory Standard Institute (CLSI), and minimum inhibitory concentrations (MICs) determined through Vitek 2 automated system (Vitek AMS; bioMerieux Vitek Systems Inc., Hazelwood, MO) [Citation17–19]. The 13 tested antimicrobial agents classified into 7 antibiotic groups: penicillins (AMP: Ampcillins), β-lactam (TZP: piperacillin-tazobactam; AMC: Amoxicillin-clavulanate), cephems (CAZ: ceftazidime; FEP: cefepime), monobactams (ATM: aztreonam), carbapenems (IPM: imipenem; MEM: meropenem), aminoglycosides (GEN: gentamicin; TOB: tobramycin; AMK: amikacin) and fluoroquinolones (LVX: levofloxacin; CIP: ciprofloxacin). In addition, the susceptibility analysis was interpreted based on the criteria on E. coli of CLSI (2019).
Multidrug-resistant (MDR) was defined as acquired non-susceptibility to at least one agent in more than three antimicrobial categories, and extensively drug-resistant (XDR) was defined as non-susceptibility to at least one agent in all but two or fewer antimicrobial categories [Citation21]. Pandrug-resistant (PDR) was defined as non-susceptibility to in all antimicrobial categories listed.
2.4 Detection of β-lactamase genes
The major β-lactamase genes were detected by seven groups of multiplex PCR [Citation22,Citation23]. Three sets of primers were used in group 1 for the amplication of blaTEM (blaTEM-1 and blaTEM-2), blaSHV-1, and blaOXA (blaOXA-1, blaOXA-4 and blaOXA-30) genes, respectively. Group 2 and group 3 mainly amplified blaCTX-M, including group 1 blaCTX-M (blaCTX-M-1, blaCTX-M-3 and blaCTX-M-15), group 2 blaCTX-M (blaCTX-M-2) and group 9 blaCTX-M (blaCTX-M9- and blaCTX-M-14), and group 8/25 blaCTX-M (blaCTX-M-8, blaCTX-M-25, blaCTX-M-26 and blaCTX-M-39 to blaCTX-M-41). Five sets of primers were used in group 4 to detect blaAAC (blaAAC-1 and blaAAC-2), blaFOX (blaFOX-1 to blaFOX-5), blaMOX (blaMOX-1, blaMOX-2, blaCMY-1, blaCMY-8 to blaCMY-11 and blaCMY-19), blaDHA (blaDHA-1 and blaDHA-2), blaCIT (blaLAT-1 to blaLAT-3, blaBIL-1, blaCMY-2 to blaCMY-7, blaCMY-12 to blaCMY-18 and blaCMY-21 to blaCMY-23) and blaEBC (blaATC-1 and blaMIR-1). BlaVEB (blaVEB-1 to blaVEB-6), blaPER (blaPER-1 and blaPER-3) and blaGES (blaGES-1 to blaGES-9 and blaGES-11) genes were detected by group 5. BlaGES (blaGES-1 to blaGES-9 and blaGES-11) and blaOXA-18 were amplified by group 6, while blaIMP, blaVIM (blaVIM-1 and blaVIM-2) and blaKPC (blaKPC-1 to blaKPC-5) were detected by group 7. Annealing temperature at 60°C was used for group 1–5, while 57°C and 55°C were selected for group 6 and 7, respectively. One min was chosen for extention time since all the PCR products were shorter than 1 kb. All PCR products were run on 2% agarose gel. To further avoid false-positive results from multiplex PCR, all positive samples were further subjected to separate PCR confirmation using single pairs of primers from the corresponding multiplex PCR. For samples yielding positive results from single PCR reaction, Sanger sequencing was further performed to ensure accuracy. All experiments were performed in triplicate.
2.5 Detection of plasmid replicons
Eight groups of multiplex PCR were designed to detect 8 different plasmid replicons [Citation22,Citation23]. Group 1 targeted on parA-parB (AF250878), iteron (BX664015), and RNAI (M20413). The target genes in group 2 were oriγ (Y00768), repABC (U27345), and repA (NC_003292). One iteron (J01724) and two repA genes (M26308 and U12441) were selected to be detected in group 3. Similarly, iteron (M20134), repA (K02380), and repA2 (AH003523) were detected in group 4. Three repA genes (X73674, K00053, and AE006471) were amplified in group 5. In group 6–8, three different RNAI (AY234375, M93063, and M28718) were detected, respectively. For the multiplex PCR groups except for group 6, annealing temperature at 60°C and extention for 1 min were used. In group 6, 52°C was selected for annealing temperature. All PCR products were run on 2% agarose gel.
2.6 Statistical analysis
In this study, the susceptibility of E. coli was defined as resistant (resistant and intermediate resistant) and susceptible in this study. The antimicrobial susceptibility test results were collected and managed in WHONET (version 5.6). Chi-square test or Fisher’s exact test were used to analyze the difference of proportions, if appropriate. A value of p < 0.05 was defined as statistically significant.
3. Results
The obtained E. coli isolates were subjected to bacteriological examination, antimicrobial susceptibility testing, detection of β-lactamase genes and plasmid replicons. The identification of plasmid replicons and β-lactamase genes, especially MDR, XDR, and PDR strains, may aid in the understanding of clinical E. coli isolates epidemiology.
3.1 Phenotypic characteristics of the recovered isolates
The colonies of the recoverd E. coli isolates showed bright pink colonies on VRBA agar plates and showed a distinctive metallic green sheen on EMB agar. All E. coli isolates were lactose fermenting colonies, methyl red, pidole-positive, catalase, oxidase-negative, urea hydrolysisi, critrate utilization, Voges-Proskauer, and did produce H2S in biochemical tests. Moreover, all isolates PCR amplicons were compared in the GenBank database using the BLAST program and the result indicated that these strains belonged to Escherichia coli.
3.2 Characteristics of antimicrobial resistance of E. coli
E. coli isolates collected from 7 different specimen sources during 10-year period were compared antimicrobial resistance in this study (). Females were the most affected group of patients (286 samples, 58.4%) as compared to males (181 samples, 37.0%). Maximum number of E. coli were isolated in the age group of 41–70 (271 samples, 55.3%). Urine was the most predominant specimen source of E. coli (46.9%), followed by blood (24.7%) and sputum (12.2%). Other remaining sources included secretion, pleural fluid, pus and wound, only making up a small proportion.
Table 1. Characteristics of specimen and drug resistance profiles of E. coli.
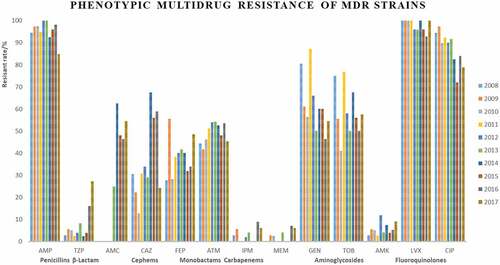
The resistance rates of penicillins (AMP) and fluoroquindones (LVX, CIP) remained high during the study period (vary between 58.8% and 91.2%, 79.7% and 90.2%, 54.9% and 81.3% over the study period, respectively; ). Monobactams (ATM) and aminoglycosides (GEN, TOB) kept fluctant (between 31.9% and 74.5%). Meanwhile, increased rates in β-lactam use for clinical treatment accompanied these increasing resistance rates, the resistance rate of β-lactam (TZP, AMC, 3.5% to 40.7%) and cephems (CAZ, FEP, 16.7% to 40.3%) increased during 10-year period.
3.3 MDR, XDR and PDR profiles in E. coli
Approximately 92.9% of isolates were resistant to at least one antimicrobial agent (). Among them, 76.9% 27.9%, 1.4% were MDR, XDR, and PDR, respectively (). The results showed that many MDR isolates were co-resistant to penicillins (AMP) and fluoroquindones (LVX, CIP) and distribution of LVX and CIP resistance was significantly higher among female than male gender. These results were not exactly consistent with previous research [Citation24,Citation25], its reasons may be due to the geographical difference of E. coli.
Table 2. Characteristics of antimicrobial susceptibility testing profiles of E. coli.
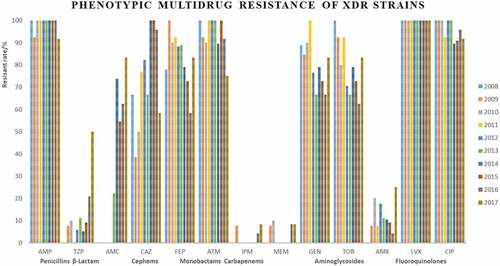
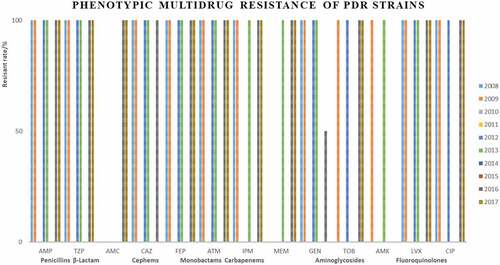
Strains Most of XDR strains were resistant to penicillins, cephems, monobactams, and fluoroquinolones (). Only 7 strains were identified as PDR strains, isolated in 2008, 2009, 2012, 2013, 2016, and 2017 (, ).
3.4 Distribution of β-lactamase genes
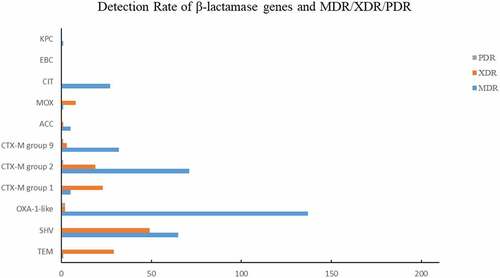
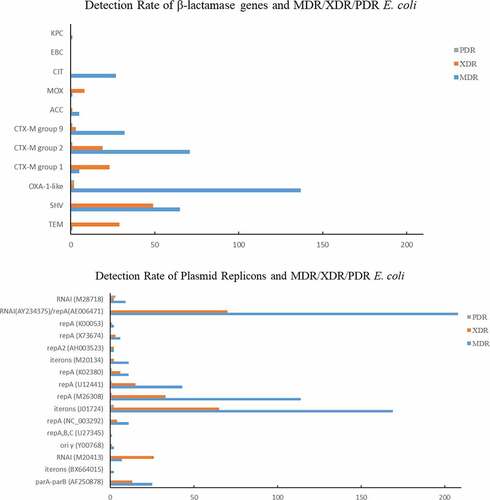
According to characteristics of antimicrobial resistance to β-lactam (TZP, AMC), the resistance rate increased during the study period (from 3.5% to 40.8%). Among all β-lactamase genes, blaOXA-1 acquired the highest carriage rate (195/542, 35.98%), followed by group 2 blaCTX-M (90/542, 16.61%), blaSHV-1 (78/542, 14.39%) and group 9 blaCTX-M (39/542, 7.20%), indicating their prevalence among clinical E. coli in South China (). The carriage rate of blaOXA-1 was high (40.82–62.50%) from 2008 to 2012, 2015, and 2018, but decreased (4.08–5.56%) in 2013–2014 and 2016–2017. The carriage rate of group 2 blaCTX-M and blaSHV-1 was fluctuant.
Table 3. Distribution of β-lactamase genes and plasmid replicons of E. coli.
Among the 378 MDR strains, blaOXA-1 acquired the highest carriage rate (137/378, 36.24%), followed by group 2 blaCTX-M (71/378, 18.78%), blaSHV-1 (65/378, 17.20%), group 9 blaCTX-M (32/378, 8.47%), and blaCIT (27/378, 7.14%) (). Among the 137 XDR strains, blaOXA-1 acquired the highest carriage rate (49/137, 35.77%), followed by blaSHV-1 (29/137, 21.17%), group 2 blaCTX-M (23/137, 16.79%), and group 9 blaCTX-M (19/137, 13.87%) (). Among the 7 PDR strains, 2 strains carried blaOXA-1, 1 strains carried group 2 blaCTX-M (23/137, 18.78%), and another one carried group 9 blaCTX-M, respectively.
3.5 Distribution of plasmid replicons
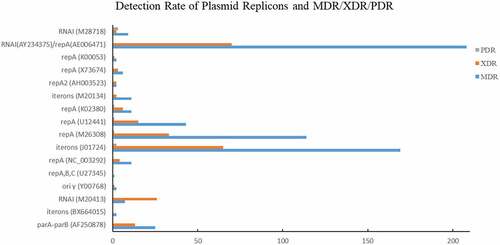
Eight groups of plasmid replicons were detected among the 542 E. coli isolates (). The RNAI (AY234375) in group 6 showed the highest detection rate (285/542, 52.58%), followed by the iteron (J01724, 233/542, 42.99%) and repA (M26308, 135/542, 24.91%) in group 3, indicating the relatively higher carriage rate of corresponding plasmids by clinical E. coli isolates. In group 1, RNAI (M20413) showed relatively higher detection rate (61/542, 11.25%), followed by parA-parB (AF250878, 31/542, 5.72%) and iteron (BX664015, 6/542, 1.11%). The identification rate of RNAI (M20413) was relatively stable among strains isolated in different years, except for in 2013 (none detected). In group 2, the identification rates of oriγ (Y00768), repABC (U27345), and repA (NC_003292) were 2.40%, 0.92%, and 4.43%, respectively. The oriγ (Y00768) was only detected in 2012 and 2018, while the repABC (U27345) was only detcted in 2009, 2011, and 2018. In group 3, except for iteron (J01724) and repA (M26308), repA (U12441) was carried by 135 (8.49%) E. coli strains isolated in 2012 and 2014–2016. The detection rate of the three genes in group 4 was relatively low, but the lowest identification rate was acquired by group 5. One of the repA genes (K00053) in group 5 was not detected. Only 1 strain isolated in 2010 carried the RNAI from group 7 (M93063).
Among those MDR strains, the identification rates of RNAI (AY234375)/repA (AE006471), iterons (J01724), repA (M26308) were 55.03% (208/378), 44.71% (169/378), and 30.16% (114/378), respectively (). Approximately 51.10% (70/137) and 47.45% (65/137) XDR strains carried RNAI (AY234375)/repA (AE006471) or iterons (J01724).
4. Discussion
In this study, females were more susceptible to E. coli infection, which is consistent with global trends that higher prevalence of urinary tract infections in female patients than in males [Citation26–28]. Our result showed that urine was the main source of E. coli. Urinary tract infections (UTIs) caused by uropathogenic Escherichia coli (UPEC) are one of the most common outpatient bacterial infections [Citation29]. The emergence of high rates of antibiotic resistance and multidrug-resistant MDR-phenotype from urinary tract infections related bacteria becomes a public health concern worldwide [Citation30]. Moreover, the high resistance rates of penicillins (AMP) and fluoroquindones (LVX, CIP) during this period was in agreement with previous study [Citation27,Citation31].
Multidrug resistance has been increased all over the world that is considered a public health threat gave a warning to the potential and proper use of antibiotics [Citation32,Citation33]. Several recent investigations reported the emergence of multidrug-resistant bacterial pathogens from different origins including humans, birds, cattle, and fish that increase the need for routine application of the antimicrobial susceptibility testing to detect the antibiotic of choice as well as the screening of the emerging MDR strains [Citation34–39]. The rate of MDR was higher than in other study from China, which reported only 47.9% of MDR strains [Citation24]. It is known that susceptibility patterns may vary in diferent geographical regions and can be change over time. It is worth noting that MDR E. coli usually implies a signifcant increase in resistance and pathogenic potential and indicates complicated treatment and bad prognosis of infections [Citation40]. Interestingly, resistance rates to antimicrobial drugs were higher in isolates from females than in those from male patients. Thus, it may be more difficult to eradicaten female infections and susceptibility analysis of isolates to antibiotics prior to treatment choice is recommended. For example, German national guideines agent as a first choice for the treatment of uncomplicated cystitis [Citation30].
Over the last decade, blaCTX-M has become increasingly common worldwide, to the point that their prevalence easily surpassing those of blaSHV and blaTEM ESBL genes [Citation14]. Based on CTX-M amino acid sequences, these enzymes have been classified into five major groups, groups 1, 2, 8, 9 and 25. Previous study revealed that the incidence of these blaCTX-M genotypes varies geographically. Our results showed that group 2 blaCTX-M may be the most widely disseminated genotype in southern China, followed by group 9 and group 1.
Strains carrying blaCIT were mainly isolated in 2010 and 2011. Six each group 1 blaCTX-M and blaAAC were detected, with group 1 blaCTX-M identfied in strains isolated in 2008 and 2009, and blaAAC carried by the strains isolated from 2010 to 2012. Two strains isolated in 2008 and 2011, respectively, carried blaMOX. Only one each strain isolated in 2011, 2018, and 2010 was identified to carry blaTEM, blaEBC, and blaKPC, respectively. In contrast, no group 8/25 blaCTX-M, blaAAC, blaFOX, blaDHA, blaGES, blaPER, blaVEB, blaOXA-48, blaIMP, and blaVIM genes were identfied.
Among the 94 isolates that were resistant to β-Lactam, surprisingly 64 did not have any of the 21 β-lactam resistance genes. These results suggested that there may be no relation between the presence of blaTEM, blaSHV, blaOXA-1-like, blaCTX-M, blaACC, blaMOX, blaCIT, blaEBC, and blaKPC with β-lactam production and other types of β-lactam resistant genes may be responsible for the phenotype. More importantly, the expression of β-lactam genes depends upon the environmental conditions such as the presence of antibiotics and genes presence shown by PCR does not necessarily indicate its expression.
Plasmids are extra-chromosomal circular fragments of DNA that replicate autonomously in E. coli. Plasmids appear to increase bacterial genetic diversity, containing genes essential for initiation and control of replication and accessory genes that may be useful to their bacterial host such as antimicrobial resistance or virulence genes [Citation41]. Since the origin of replication is a constant and conserved part of a plasmid, replicon typing focused on this portion of the plasmid is a more sensitive and specific method for identifying phylogenetically related plasmids than restriction-based analysis of the entire plasmid. The classfication of plasmid is usually based on PCR-based replicon typing (PBRT) method which relied on the phenomenon of incompatibility that closely related plasmids cannot coexist stably in the same cell [Citation42].
Plasmids are significantly correlated with bacterial pathogenicity. IncFI (pEM4) and IncI (pEM6) are found to determine virulence and colicine I production and resistance to tetracycline [Citation43]. RNAI, a small countertranscript RNA which can inhibit translation of repA mRNA, expressed the incompatibility of IncI plasmids in the PBRT scheme. IncI plasmids generate both a thick pilus (tra genes) for DNA transfer and athin pilus (pil genes) that appears to stabilize the mating apparatus in liquid media but not on solid surfaces, which is reported to result in different conjugation efficiencies and biofilm and/or adherence properties [Citation44,Citation45]. ESBL and plasmid-mediated (p)AmpC genes have been described on IncI plasmids in E. coli. blaCTX-M-1 is the most often identified gene on IncI plasmid, followed by blaCMY-2 and blaCTX-M-15 [Citation46]. RepA, broadly distributed throughout the low G + C Gram-positive bacteria, is the target site of IncA/C, IncN, IncF and IncT plasmid In the PBRT (PCR-based replicon typing) scheme. Incompatibility group IncA/C plasmids are large, low copy plasmids that associated with enteric pathogens of humans and animals [Citation47,Citation48] IncA/C plasmids are associated with MDR and can encode ESBLs (blaTEM, blaSHV, but rarely blaCTX-M), AmpC (blaCMY, blaDHA), carbapenemases (blaOXA, blaNDM, blaIMP) and enzymes modifying groups of antibiotics: sulfonamides (sul1, sul2), aminoglycosides (aphA1, aadA, aadB, strA, strB, aacC), tetracyclines, chloramphenicol (floR, catA1) and trimethoprim (dfrA) [Citation49,Citation50]. IncN plasmids carry a great variety of resistance determinants against ESBL, sulfonamides, quinolones, aminoglycosides, tetracyclines and streptomycin [Citation51]. The most frequently described resistance genes on IncF plasmids are ESBL genes (genes encoding carbapenemases, aminoglycoside-modifying enzymes and plasmid-mediated quinolone resistance (PMQR) genes). IncF plasmids also drive the spread of blaNDM and the rmtB gene (mostly reported in China) [Citation52]. IncT plasmids usually carry kanamycin- (Rst1) or sulfonamide resistance genes [Citation53]. ParA-parB is the target site for typing IncHI plasmids in the PBRT scheme, which has been reported to contain large virulence plasmid pWR100 [Citation54]. IncH plasmids are reported to be associated with multidrug resistance because, besides ESBL genes, they often carry genes encoding for resistance to sulfonamides, aminoglycosides, tetracyclines and streptomycin [Citation55]. IncP is a group of broad-host-range, low-copy-number plasmids, the copy number of which is controlled by iterons [Citation56]. A recurring theme in the duplication of prokaryotic replicons is the recognition of the replication origin by cis-encoded initiators that bind to repeated nucleotide sequences called iterons. They were reported to carry genes conferring resistance to ESBL, sulfonamides, aminoglycosides and tetracyclines [Citation57,Citation58]. Oriγ is the target site of IncX, a group of narrow-host-range plasmids encoding primarily AMR determinants against ESBL and quinolones. These plasmids encode primarily AMR determinants against ESBL and quinolones [Citation59,Citation60]. Its six known subtypes (X1-X6) were releveant to tetracycline and trimethoprim resistance determinants. IncX plasmids were able to form cointegrants with Salmonella serotype-specific plasmid-carrying virulence genes which resulted in a broadening of the host range of the new plasmid [Citation61].
5. Conclusions
This study had designed and developed multiplex amplification platform for rapid and accurate detection of such resistance genes in 542 clinical E. coli isolates. It was found that most MDR isolates were co-resistant to penicillins (AMP) and fluoroquindones (LVX, CIP) and distribution of LVX and CIP resistance was significantly higher among female than male gender. Among the β-lactamase genes, blaOXA acquired the highest carriage rate, followed by group 2 blaCTX-M and blaSHV-1, indicating their prevalence among clinical E. coli in South China. The RNAI (AY234375) showed the highest detection rate, followed by the iteron (J01724) and repA (M26308), indicating the relatively higher carriage rate of corresponding plasmids by clinical E. coli isolates. A high percentage of the isolated E. coli strains were multidrug resistant (MDR) to penicillins: ampicillin, aminoglycosides, and fluoroquinolones; and are harboring the blaOXA-1, group 2 blaCTX-M, and blaSHV-1 genes, as well as RNAI(AY234375)/repA(AE006471), iterons (J01724) genes. It is shown that such identification of plasmid replicons and β-lactamase genes may aid in the understanding of clinical E. coli isolates epidemiology. A strategy design based on multiplex amplification and further application for rapid detection on antimicrobial resistance and β-lactam resistance genes in clinical Escherichia coli strains.
Data availability
All data generated or analyzed during this study are included in this article.
The authors confirm that the data supporting the findings of this study are available within the article
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- World Health Organization. Executive Board 140th Session: EB140/11 Antimicrobial resistance. 2017.
- Kotwani A, Joshi J, Kaloni D. Pharmaceutical effluent: a critical link in the interconnected ecosystem promoting antimicrobial resistance. Environ Sci Pollut Res Int. 2021;28(25):32111–32124.
- Song B, Yu J, Sun Y, et al. Microfluidics for the rapid detection of Escherichia coli O157:H7 using antibody-coated microspheres. Bioengineered. 2021;12(1):392–401.
- Lin WH, Wang MC, Liu PY, et al. Escherichia coli urinary tract infections: host age-related differences in bacterial virulence factors and antimicrobial susceptibility. J Microbiol Immunol Infect. 2021;S1684-1182(21):00091.
- Reitzer L, Zimmern P. Rapid growth and metabolism of uro-pathogenic Escherichia coli in relation to urine composition. Clin Microbiol Rev. 2019;33(1):e00101e19.
- Liu S, Dai J, Kang Z, et al. Production of novel NaN3-resistant creatine amidinohydrolase in recombinant Escherichia coli. Bioengineered. 2015;6(4):248–250.
- Singh AP, Aijaz S. Generation of a MDCK cell line with constitutive expression of the Enteropathogenic E. coli effector protein Map as an in vitro model of pathogenesis. Bioengineered. 2015;6(6):335–341.
- Cohen SP, McMurry LM, Hooper DC, et al. Cross-resistance to fluoroquinolones in multiple-antibiotic-resistant (Mar) Escherichia coli selected by tetracycline or chloramphenicol: decreased drug accumulation associated with membrane changes in addition to OmpF reduction. Antimicrob Agents Chemother. 1989;33(8):1318–1325.
- Maynard C, Fairbrother JM, Bekal S, et al. Antimicrobial resistance genes in enterotoxigenic Escherichia coli O149:K91 isolates obtained over a 23-year period from pigs. Antimicrob Agents Chemother. 2003;47(10):3214–3221.
- Amábile-Cuevas CF, Chicurel ME. Bacterial plasmids and gene flux. Cell. 1992;70(2):189–199.
- Bergström S, Normark S. Beta-lactam resistance in clinical isolates of Escherichia coli caused by elevated production of the ampC-mediated chromosomal beta-lactamase. Antimicrob Agents Chemother. 1979;16(4):427–433.
- Bergstrom CT, Lipsitch M, Levin BR. Natural selection, infectious transfer and the existence conditions for bacterial plasmids. Genetics. 2000;155(4):1505–1519.
- Shannon K, Williams H, King A, et al. Hyperproduction of TEM-1 beta-lactamase in clinical isolates of Escherichia coli serotype O15. FEMS Microbiol Lett. 1990;55(3):319–323.
- Memariani M, Najar Peerayeh S, Zahraei Salehi T, et al. Occurrence of SHV, TEM and CTX-M β-lactamase genes among enteropathogenic Escherichia coli strains isolated from children with diarrhea. Jundishapur J Microbiol. 2015;8(4):e15620.
- Francia MV, Varsaki A, Garcillán-Barcia MP, et al. A classification scheme for mobilization regions of bacterial plasmids. FEMS Microbiol Rev. 2004;28(1):79–100.
- Zhou XY, Bordon F, Sirot D, et al. Emergence of clinical isolates of Escherichia coli producing TEM-1 derivatives or an OXA-1 beta-lactamase conferring resistance to beta-lactamase inhibitors. Antimicrob Agents Chemother. 1994;38(5):1085–1089.
- Prinarakis EE, Miriagou V, Tzelepi E, et al. Emergence of an inhibitor-resistant beta-lactamase (SHV-10) derived from an SHV-5 variant. Antimicrob Agents Chemother. 1997;41(4):838–840.
- Deng Y, Liu J, Peters BM, et al. Antimicrobial resistance investigation on Staphylococcus strains in a local hospital in Guangzhou, China, 2001-2010. Microb Drug Resist. 2015;21(1):102–104.
- Quinn PJ, Markey BK, and Carter ME, et al. Veterinary microbiology and microbial disease: pathogenic bacteria blackwell scientific publications. London: Oxford: Wiley-Blackwell; 2002. p. 113–115.
- Alizadeh Behbahani B, Noshad M, Falah F. Inhibition of Escherichia coli adhesion to human intestinal Caco-2 cells by probiotic candidate Lactobacillus plantarum strain L15. Microb Pathog. 2019;136:103677.
- Magiorakos AP, Srinivasan A, Carey RB, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18(3):268–281.
- Poirel L, Walsh TR, Cuvillier V, et al. Multiplex PCR for detection of acquired carbapenemase genes. Diagn Microbiol Infect Dis. 2011;70(1):119–123.
- Ellington MJ, Kistler J, Livermore DM, et al. Multiplex PCR for rapid detection of genes encoding acquired metallo-beta-lactamases. J Antimicrob Chemother. 2007;59(2):321–322.
- Xu Y, Sun H, Bai X, et al. Occurrence of multidrug-resistant and ESBL-producing atypical enteropathogenic Escherichia coli in China. Gut Pathog. 2018;10:8.
- Chen Y, Liu Z, Zhang Y, et al. Increasing prevalence of ESBL-producing multidrug resistance Escherichia coli from diseased pets in Beijing, China From 2012 to 2017. Front Microbiol. 2019;10:2852.
- Dehbanipour R, Rastaghi S, Sedighi M, et al. High prevalence of multidrug-resistance uropathogenic Escherichia coli strains, Isfahan, Iran. J Nat Sci Biol Med. 2016;7(1):22–26.
- Bouchillon S, Hoban DJ, Badal R, et al. Fluoroquinolone resistance among gram-negative urinary tract pathogens: global smart program results, 2009-2010. Open Microbiol J. 2012;6:74–78.
- Ali I, Rafaque S, Ahmed S, et al. Prevalence of multi-drug resistant uropathogenic Escherichia coli in Potohar region of Pakistan. Asian Pac J Trop Biomed. 2016;6(1):60–66.
- Lin WH, Wang MC, Liu PY, et al. Escherichia coli urinary tract infections: host age-related differences in bacterial virulence factors and antimicrobial susceptibility. J Microbiol Immunol Infect. 2021;S1684-1182(21):00091.
- Wagenlehner FM, Schmiemann G, Hoyme U, et al. Nationale S3-Leitlinie “Unkomplizierte Harnwegsinfektionen”: empfehlungen zu Therapie und Management unkomplizierter bakterieller ambulant erworbener Harnwegsinfektionen bei erwachsenen Patienten. Urologe A. 2011;50(2):153–169.
- Ramírez-Castillo FY, Moreno-Flores AC, Avelar-González FJ, et al. An evaluation of multidrug-resistant Escherichia coli isolates in urinary tract infections from Aguascalientes, Mexico: cross-sectional study. Ann Clin Microbiol Antimicrob. 2018;17(1):34.
- Algammal AM, Mabrok M, Sivaramasamy E, et al. Emerging MDR-Pseudomonas aeruginosa in fish commonly harbor oprL and toxA virulence genes and blaTEM, blaCTX-M, and tetA antibiotic-resistance genes. Sci Rep. 2020;10(1):15961.
- Algammal AM, Hashem HR, Alfifi KJ, et al. atpD gene sequencing, multidrug resistance traits, virulence-determinants, and antimicrobial resistance genes of emerging XDR and MDR-Proteus mirabilis. Sci Rep. 2021;11(1):9476.
- Makharita RR, El-Kholy I, Hetta HF, et al. Antibiogram and genetic characterization of carbapenem-resistant gram-negative pathogens incriminated in healthcare-associated infections. Infect Drug Resist. 2020;13:3991–4002.
- Enany ME, Algammal AM, Shagar GI, et al. Molecular typing and evaluation of Sidr honey inhibitory effect on virulence genes of MRSA strains isolated from catfish in Egypt. Pak J Pharm Sci. 2018;31(5):1865–1870.
- Abolghait SK, Fathi AG, Youssef FM, et al. Methicillin-resistant Staphylococcus aureus (MRSA) isolated from chicken meat and giblets often produces staphylococcal enterotoxin B (SEB) in non-refrigerated raw chicken livers. Int J Food Microbiol. 2020;328:108669.
- Algammal AM, Hetta HF, Elkelish A, et al. Methicillin-resistant Staphylococcus aureus (MRSA): one health perspective approach to the bacterium epidemiology, virulence factors, antibiotic-resistance, and zoonotic impact. Infect Drug Resist. 2020;13:3255–3265.
- Algammal AM, Hashem HR, Al-Otaibi AS, et al. Emerging MDR-Mycobacterium avium subsp. avium in house-reared domestic birds as the first report in Egypt. BMC Microbiol. 2021;21(1):237.
- Algammal AM, Mabrok M, Ezzat M, et al. Prevalence, antimicrobial resistance (AMR) pattern, virulence determinant and AMR genes of emerging multi-drug resistant Edwardsiella tarda in Nile tilapia and African catfish. Aquaculture. 2022;548:737643.
- Algammal AM, Hetta HF, Batiha GE, et al. Virulence-determinants and antibiotic-resistance genes of MDR-E. coli isolated from secondary infections following FMD-outbreak in cattle. Sci Rep. 2020;10(1):19779.
- Carattoli A, Bertini A, Villa L, et al. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods. 2005;63(3):219–228.
- Partridge SR, Kwong SM, Firth N, et al. Mobile genetic elements associated with antimicrobial resistance. Clin Microbiol Rev. 2018;31(4):e00088–17.
- Morzejko E, Panek E, Cebrat S. Conjugational cotransfer of IncFI and IncI conjugative plasmids forming aggregate in a pathogenic Citrobacter freundii strain. FEMS Microbiol Lett. 1990;56(1–2):7–11.
- Bradley DE. Characteristics and function of thick and thin conjugative pili determined by transfer-derepressed plasmids of incompatibility groups I1, I2, I5, B, K and Z. J Gen Microbiol. 1984;130(6):1489–1502.
- Komano T. Shufflons: multiple inversion systems and integrons. Annu Rev Genet. 1999;33:171–191.
- Johnson TJ, Shepard SM, Rivet B, et al. Comparative genomics and phylogeny of the IncI1 plasmids: a common plasmid type among porcine enterotoxigenic Escherichia coli. Plasmid. 2011;66(3):144–151.
- Johnson TJ, Lang KS. IncA/C plasmids: an emerging threat to human and animal health? Mob Genet Elements. 2012;2(1):55–58.
- Hancock SJ, Phan MD, Peters KM, et al. Identification of IncA/C plasmid replication and maintenance genes and development of a plasmid multilocus sequence typing scheme. Antimicrob Agents Chemother. 2017;61(2):e01740–16.
- Harmer CJ, Hall RM. pRMH760, a precursor of A/C₂ plasmids carrying blaCMY and blaNDM genes. Microb Drug Resist. 2014;20(5):416–423.
- Call DR, Singer RS, Meng D, et al. blaCMY-2-positive IncA/C plasmids from Escherichia coli and Salmonella enterica are a distinct component of a larger lineage of plasmids. Antimicrob Agents Chemother. 2010;54(2):590–596.
- Carattoli A. Plasmids and the spread of resistance. Int J Med Microbiol. 2013;303(6–7):298–304.
- Rahman M, Shukla SK, Prasad KN, et al. Prevalence and molecular characterisation of New Delhi metallo-β-lactamases NDM-1, NDM-5, NDM-6 and NDM-7 in multidrug-resistant Enterobacteriaceae from India. Int J Antimicrob Agents. 2014;44(1):30–37.
- Carattoli A, Zankari E, García-Fernández A, et al. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother. 2014;58(7):3895–3903.
- Sergueev K, Dabrazhynetskaya A, Austin S. Plasmid partition system of the P1par family from the pWR100 virulence plasmid of Shigella flexneri. J Bacteriol. 2005;187(10):3369–3373.
- Holt KE, Phan MD, Baker S, et al. Emergence of a globally dominant IncHI1 plasmid type associated with multiple drug resistant typhoid. PLoS Negl Trop Dis. 2011;5(7):e1245.
- Norberg P, Bergström M, Jethava V, et al. The IncP-1 plasmid backbone adapts to different host bacterial species and evolves through homologous recombination. Nat Commun. 2011;2:268.
- Poirel L, Villa L, Bertini A, et al. Expanded-spectrum beta-lactamase and plasmid-mediated quinolone resistance. Emerg Infect Dis. 2007;13(5):803–805.
- Novais A, Cantón R, Valverde A, et al. Dissemination and persistence of blaCTX-M-9 are linked to class 1 integrons containing CR1 associated with defective transposon derivatives from Tn402 located in early antibiotic resistance plasmids of IncHI2, IncP1-alpha, and IncFI groups. Antimicrob Agents Chemother. 2006;50(8):2741–2750.
- Venditti C, Fortini D, Villa L, et al. Circulation of blaKPC-3-carrying IncX3 plasmids among Citrobacter freundii isolates in an Italian hospital. Antimicrob Agents Chemother. 2017;61(8):e00505–17.
- Espinal P, Miró E, Segura C, et al. First description of blaNDM-7 carried on an IncX4 plasmid in Escherichia coli ST679 isolated in Spain. Microb Drug Resist. 2018;24(2):113–119.
- Rozwandowicz M, Brouwer MSM, Fischer J, et al. Plasmids carrying antimicrobial resistance genes in Enterobacteriaceae. J Antimicrob Chemother. 2018;73(5):1121–1137.