ABSTRACT
Ubiquitin-conjugating enzyme E2 B (UBE2B) can form a heterodimer with ubiquitin E3 ligase RAD18. In this study, we aimed to explore new substrates of the UBE2B/RAD18 complex and their regulatory effects in ovarian cancer. Protein physical interactions were predicted using GeneMANIA. Serial sections of commercial ovarian cancer tissue arrays were used to check the protein expression of UBE2B, RAD18, and ZMYM2. Immunofluorescence staining and co-immunoprecipitation assays were performed to check their location and interactions. Cycloheximide chase assay was applied to explore the influence of UBE2B and RAD18 on ZMYM2 degradation. Xenograft tumor models were constructed to assess the influence of the UBE2B-ZMYM2 axis on in vivo tumor growth. A strong positive correlation between UBE2B and ZMYM2 and a moderate positive correlation between RAD18 and ZMYM2 were observed in 23 ovarian cancer cases. In CAOV4 and OVCAR3 cells, myc-ZMYM2 interacted with UBE2B and RAD18. UBE2B and ZMYM2 could be detected in the samples immunoprecipitated by anti-RAD18. UBE2B overexpression or knockdown did not alter ZMYM2 mRNA expression. UBE2B overexpression increased ZMYM2 monoubiquitination but reduced its polyubiquitination. RAD18 knockdown impaired UBE2B-induced ZMYM2 monoubiquitination. UBE2B overexpression significantly enhanced the stability of ZMYM2 protein, the effect of which was weakened by RAD18 knockdown. UBE2B overexpression significantly enhanced the growth of xenograft tumors derived from CAOV4 cells. ZMYM2 knockdown remarkedly suppressed tumor growth and impaired the growth-promoting effect of UBE2B overexpression. In conclusion, this study revealed a novel regulatory effect of the UBE2B/RAD18 complex on ZMYM2 monoubiquitination and stability in ovarian cancer.
Graphic Abstract
1. Introduction
Ubiquitination is an important post-transcriptional modification of proteins for degradation or stabilization, depending on the type of ubiquitin chain formed [Citation1]. Ubiquitin has 76 amino-acid residues with seven internal lysine residues (K6, K11, K27, K29, K33, K48, K63) [Citation1]. Proteins can be modified by monoubiquitin or polyubiquitination (via covalent linking of additional ubiquitin molecules) on these lysine residues [Citation1,Citation2]. The number of ubiquitin and linkage types have distinct physiological properties. K48-linked polyubiquitination can target proteins for proteasomal degradation. In contrast, K63-linked polyubiquitination can stabilize some proteins and play proteasome-independent roles [Citation3,Citation4]. Monoubiquitination is involved in multiple cellular processes, such as chromatin regulation, protein sorting, and trafficking, and protein stability [Citation5]. Ubiquitination requires at least three categories of enzymes: ubiquitin-activating enzymes (E1s), ubiquitin-conjugating enzymes (Es), and ubiquitin-protein ligases (E3s) [Citation6].
Dysregulated genes such as mRNA, lncRNA and microRNAs related to ubiquitination-related enzymes are quite common in cancer initiation and progression, including ovarian cancer [Citation4,Citation7–10]. Ubiquitin-specific peptidase 13 (USP13) is frequently amplified in ovarian cancer. It acts as a master regulator of ATP citrate lyase and oxoglutarate dehydrogenase, which enhance mitochondrial TCA cycle metabolism [Citation7]. Ubiquitin-conjugating enzyme E2 B (UBE2B or RAD6B) overexpression enhances DNA repair and stem cell signaling in ovarian cancer [Citation11,Citation12]. These alterations further lead to acquired chemoresistance [Citation11,Citation12]. Mechanistically, UBE2B promotes monoubiquitination of FA Complementation Group D2 (FANCD2) and DNA-associated Proliferating Cell Nuclear Antigen (PCNA), two proteins that are critical for platinum drugs-induced DNA crosslink repair and DNA damage tolerance [Citation12]. Inhibiting UBE2B using a small molecule inhibitor significantly sensitizes chemoresistant ovarian cancer cells to carboplatin [Citation12]. These findings suggest that UBE2B might be a potential therapeutic target in ovarian cancer. However, the underlying regulatory network in the tumor cells is still not fully understood.
UBE2B can form a heterodimer with RAD18 (an E3 ligase), promoting mono-ubiquitination of proliferating cell nuclear antigen (PCNA) [Citation13] and histone H2A [Citation14], and polyubiquitination of O6-methylguanine-DNA methyltransferase (MGMT) [Citation15]. We hypothesized that there might be some unknown ubiquitination substrates of the UBE2B/RAD18 complex in ovarian cancer. Therefore, we aimed to explore new substrates of the UBE2B/RAD18 complex and their regulatory effects on ovarian cancer growth.
2. Materials and methods
2.1. Bioinformatic data mining
Gene expression of normal fallopian tubes (n = 5) and ovary tissues (n = 88) from the Genotype-Tissue Expression (GTEx), gene expression and survival data of ovarian cancer cases in TCGA pan-cancer data (n = 418) were obtained from the UCSC Xena database (https://xenabrowser.net/datapages/) [Citation16]. Kaplan-Meier (K-M) survival analysis was performed using the log-rank test, with the best cutoff identified in the Receiver operating characteristics (ROC) analysis for progression or death detection. Hazard Ratio (HR) and 95% confidence interval (CI) were calculated.
2.2. Cell culture and treatment
Human ovarian clear cell adenocarcinoma cell line OVTOKO was obtained from JCRB Cell Bank (Osaka, Japan). Ovarian serous adenocarcinoma cell line PEO1 was obtained from ECACC (Salisbury, UK), while CAOV4, OVCAR3, and A2780 were obtained from ATCC (Manassas, VA, USA). Primary human normal ovarian surface epithelial (HOSE) cells were purchased from Procell (Wuhan, China). Cells were cultured with the recommended mediums at 37°C in the presence of 5% CO2.
Lentiviral ZMYM2 (NM_003453) overexpression particle with N-terminal myc-tag (myc-ZMYM2), UBE2B (NM_003337) overexpression particle with N-terminal flag-tag (flag-ZMYM2) was generated based on pHBLV-CMVIE-IRES-Puro plasmid from HanBio (Shanghai, China). Lentiviral expression of HA-tagged ubiquitin, pLenti-puro-HA-Ubiquitin (#74218) (HA-Ub) was purchased from Addgene (Watertown, MA, USA). Recombinant lentiviral shRNA particles were generated based on pHBLV-U6-Puro plasmids. The following shRNA sequences were used: shZMYM2#1, 5’-GCCTTGGAATACAGGAGTATT-3’ and #2, 5’-ATACTTCCAGATGGGTCAATA-3’; shUBE2B#1, 5’-GCAGTTATATTTGGACCAGAA-3’ and #2, 5’-CGGGATTTCAAGCGGTTACAA-3’; shRAD18#1, 5’-TGCTTCGAGTATTTCAACATT-3’ and #2, 5’-GCCTGGGAAGCATCACATAAA-3’. Lentiviruses were generated following the standard procedures recommended by HanBio. In brief, HEK293T cells were co-transfected with recombinant lentiviral constructs, packaging plasmid psPAX2, and envelope plasmid pMD2.G, following recommended protocols [Citation17]. Virus-containing supernatant at 72 h post transfection was collected, filtered through a 0.45 μm filter, centrifugated, and stored in −80°C before further use. Cells were infected at the multiplicity of infection (MOI) of 10, with 6 µg/mL polybrene.
MG132, a proteasomal inhibitor, and cycloheximide (CHX, a protein synthesis inhibitor) were purchased from AbMole (Houston, TX, USA).
2.3. Western blotting and co-immunoprecipitation (co-IP) assay
Cells were lysed and protein concentrations were quantified using a BCA kit (Beyotime, Shanghai, China), according to the manufacturer’s instructions. 30 μg proteins were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto nitrocellulose membranes. The membranes were blocked with 5% milk in TBST for 1 h at room temperature and then incubated with primary antibodies in the blocking solution at 4°C overnight. After incubation, the membranes were washed and incubated with a horseradish peroxidase-conjugated secondary antibody in blocking solution at room temperature for 1 h. Then, the target protein signals were developed by an enhanced chemiluminescence reagent (Beyotime), on a ChemiDoc MP Imaging System (BioRad, Irvine, CA, USA). The following primary antibodies and dilutions were used: rabbit anti-UBE2B (1:1000, 10733-1-AP, Proteintech, Wuhan, China), rabbit anti-RAD18 (1:1000, 18333-1-AP), rabbit anti-ZMYM2 (1:1000, GTX31821, GeneTex, Shenzhen, China), anti-β-actin (1:1000, AC-40, GeneTex).
Co-IP assay was performed as described previously [Citation18]. 8 × 106 cells were harvested and lysed in IP lysis buffer containing protease inhibitor (Thermo Fisher Scientific). Cell debris was then removed by centrifugation. The soluble fractions were collected for IP assays. For myc-tag and RAD18 immunoprecipitations, an 800 μl aliquot of the lysate was incubated with mouse Myc monoclonal antibody (3 μg, 60003-2-Ig, Proteintech) or rabbit anti-RAD18 (4 μg, 18333-1-AP, Proteintech) and 50 μl of a 1:1 slurry of protein G-Sepharose for 2 h at 4°C. The Sepharose beads were collected by centrifugation, washed with 1X PBS with 0.2% Tween 20, boiled in 2× SDS loading buffer, and resolved on SDS-PAGE.
2.4. Immunohistochemistry (IHC) staining
Serial ovarian tissue sections were purchased from Taibosi Bio (Xian, China). IHC staining was conducted using the Leica BOND-III platform (Leica, Wetzlar, Germany). Antigen retrieval was performed using BOND Epitope Retrieval Solution for 20 min at 100°C. Specimens were incubated with rabbit anti-UBE2B (1:200, 10733-1-AP), rabbit anti-RAD18 (1:200, DF7314, Affinity Biosciences, Changzhou, China), rabbit anti-ZMYM2 (1:200, GTX31821, GeneTex, Shenzhen, China) for 20 min at room temperature, followed by visualization with the Leica Bond detection kit at room temperature. The specimens were then counterstained with hematoxylin. Protein expression is scored following the methods used in the Human Protein Atlas (HPA) [Citation19]. In brief, the IHC images were manually scored by two independent pathologists without authorships in this study. Staining intensity (negative, weak, moderate or strong) and the fraction of stained cells (<25%, 25–75% or >75%) were combined to give a final score as follows: negative (0), low (1), medium (2) and high (3).
2.5. Immunofluorescent (IF) staining
CAOV4 and OVCAR3 cells with myc-ZMYM2 overexpression were grown on coverslips. When reached about 50% of confluence, cells were washed and fixed with 4% paraformaldehyde in Tris-buffered saline (TBS), treated with 0.1% Triton X-100 in TBS, and blocked with 3% BSA at room temperature for 30 min. Then, the cells were then incubated with mouse anti-myc tag (1:100, 60003-2-Ig) and rabbit anti-UBE2B (1:100, 10733-1-AP) or rabbit anti-ZMYM2 (1:50, GTX31821) at 4°C overnight. After being washed with TBS, the cells were incubated with goat anti-rabbit IgG (Dylight 488-conjugated) and goat anti-mouse IgG (Dylight 549-conjugated) secondary antibodies (Thermo Fisher Scientific, Carlsbad, CA, USA). Nuclei were stained with DAPI. Immunofluorescent images were obtained using a fluorescence microscope (IX73, Olympus, Tokyo, Japan) [Citation20].
2.6. Quantitative reverse transcription PCR (RT-qPCR)
Total RNA was extracted from cells using the Qiagen RNeasy mini kits (Qiagen, Valencia, CA, USA). Then, Purified RNA was reversely transcribed to cDNA by using SuperScript III First-Strand Synthesis System for RT-PCR (Thermo Fisher Scientific). qPCR was performed on a 7500 Real-Time PCR System (Thermo Fisher Scientific), using the Power SYBR Green PCR Master Mix. GAPDH mRNA was amplified and used as an internal control for normalization [Citation21]. The following primers were used: GAPDH, forward, 5’-CGACCACTTTGTCAAGCTCA-3’, and reverse, 5’-TTACTCCTTGGAGGCCATGT-3’; UBE2B, forward, 5’- TCCTTCAGAATCGATGGAGTCCA-3’, and reverse, 5’- GCTGGACTGTTAGGATTCGGTTC-3’; ZMYM2, forward, 5’- TGTTCCAGTGCCTATCCCTGTG-3’, and reverse, 5’- TCTCACTGCTGTCCAATGGAGC-3’; RAD18, forward, 5’- GGATTGTCCTGTTTGCGGGGTT-3’, and reverse, 5’-GTTTTGGGCAGCGGCTTCCTTT-3’.
2.7. Colony formation assays
Cells (200/well) were seeded in 24-well plates and incubated at 37°C with 5% CO2 for 10 days. Colonies were visualized by staining with 0.5% crystal violet in 10% neutral buffered formalin for 20 min at room temperature. The plates were then scanned. The numbers of colonies were counted manually using a bright-field microscopy [Citation7].
2.8. Xenograft tumor models
Animal studies were approved by the ethics committee of West China Second University Hospital, China and conducted in Jinruijie Biotechnology Service Center, Chengdu, China. Female athymic nude mice (BALB/c-nu/nu), approximately 6–8 weeks old and weighing 18–20 g were purchased from Vital River Laboratory Animal Technology (Beijing, China) and were housed in specific-pathogen-free (SPF) conditions. Cells (2 × 106) with lentiviral mediated UBE2B overexpression alone, ZMYM2 knockdown alone or in combination were prepared as suspensions in a total volume of 100 μL of 1:1 (v:v) PBS/Matrigel. Then, the cell suspension was injected subcutaneously into the flank of the mice (n = 6/group). Subcutaneous tumor caliper measurements were taken twice weekly. Mice were sacrificed on day 42 after tumor cell inoculation. Then, the tumors were removed, weighed, and sectioned for IHC staining of Ki-67 [Citation7].
2.9. Statistical analysis
All experiments were performed in triplicate with at least three technical repeats. All data were analyzed by GraphPad Prism 8.10 software [Citation22]. Data were presented as the mean ± standard deviation (SD). Statistical analyses were done using Welch’s unpaired t-test or one-way ANOVA with post-hoc Sidak’s multiple comparisons test to check the statistical significance. Significance was reflected by a p-values: * and # p < 0.05; ** and ## p < 0.01; *** and ### p < 0.001.
3. Results
In this study, we explored new substrates of the UBE2B/RAD18 complex and their regulatory effects on ovarian cancer growth. We hypothesized that ZMYM2 might be a potential substrate. Via molecular and cellular studies, we demonstrated that the UBE2B could promote RAD18 mediated ZMYM2 monoubiquitination and increase its stability in ovarian cancer, thereby enhancing ovarian cancer cell proliferation.
3.1. Higher UBE2B expression was linked to a poorer prognosis of ovarian cancer
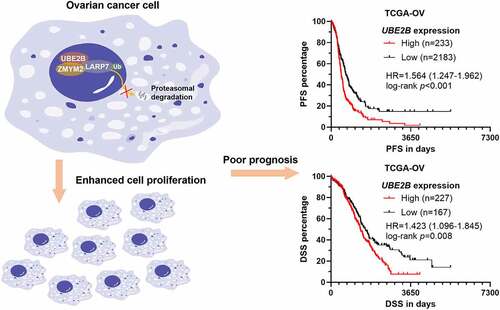
Since UBE2B has been characterized as a prognostic biomarker in multiple tumors [Citation23,Citation24], we further investigated its expression and prognostic significance in ovarian cancer. RNA-seq from the GTEx and TCGA projects were extracted for re-reanalysis. Interestingly, UBE2B expression was significantly lower in ovarian cancer tissues (n = 418) compared to normal fallopian tubes (n = 5) and ovary (n = 88) (). Then, survival difference was compared between patients with high and low UBE2B expression, using the best cutoff value. Results showed that higher UBE2B expression was linked to poorer PFS (HR = 1.564, 95%CI: 1.247–1.962, p < 0.001) () and DSS (HR = 1.423, 95%CI: 1.096–1.845, p = 0.008) ().
Figure 1. Higher UBE2B expression was linked to a poorer prognosis of ovarian cancer. (a) The expression of UBE2B in the normal fallopian tube (GTEx-FT, n = 5) and ovary tissues (n = 88) (GTEx-OV) and ovarian cancer tissues in TCGA-OV (n = 418). (b,c) K-M survival curves were generated to detect the difference in PFS (b) and DSS (c) between patients with high and low UBE2B expression, using the best cutoff value (Youden Index) identified in ROC analysis.
3.2. UBE2B and RAD18 expression are positively correlated with ZMYM2 protein levels in ovarian tumor tissues
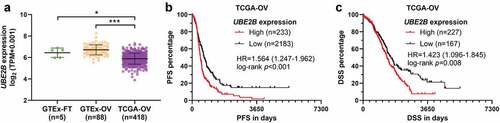
Using GeneMANIA [Citation25], we predicted the interactome of UBE2B. Results showed that UBE2B might interact with both RAD18 and ZYYM2. The mutual interactions between RAD18 and ZMYM2 were also predicted (). The complex formed by UBE2B and RAD18 has been validated by multiple previous studies [Citation26,Citation27]. Using RNA-seq data from TCGA, we found that RAD18 expression was significantly higher in the tumor group than normal fallopian and ovary tissues (). Then, we examined the protein expression of UBE2B, RAD18, and ZMYM2 in multiple ovarian cancer cell lines and normal HOSE cells. Western blot assay showed that UBE2B expression varied significantly in different cell lines. A2780 and OVCAR3 cells had significantly higher UBE2B expression than other cell lines (). Interestingly, these two cell lines also had the highest expression of RAD18 and ZMYM2 ().
Figure 2. UBE2B and RAD18 expression are positively correlated with ZMYM2 protein levels in ovarian tumor tissues. (a) Proteins with potential interactions with UBE2B. The prediction was conducted using GeneMANIA. (b) The expression of RAD18 in the normal fallopian tube (GTEx-FT, n = 5) and ovary tissues (n = 88) (GTEx-OV) and ovarian cancer tissues in TCGA-OV (n = 418). (c) The expression of UBE2B, RAD18, and ZMYM2 in multiple ovarian cancer cell lines (OVTOKO, CAOV4, PEO1, A2780, and OVCAR3) and normal HOSE cells. (d-f) IHC staining images of UBE2B (d), RAD18 (e), and ZMYM2 (f) in 24 ovarian cancer tissues. Representative images of high (red frames) and low (blue frames) protein expression were provided in the right panels. **, p < 0.01.
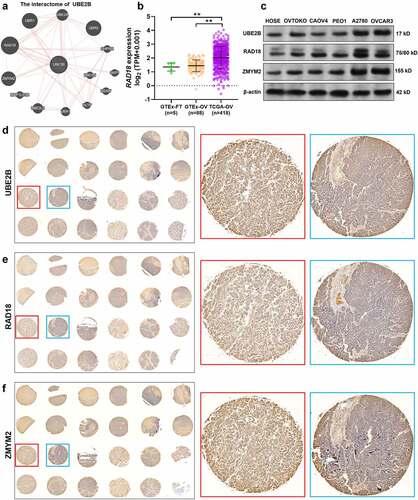
Then, using serial tissue sections of primary ovarian cancer tissues (n = 24), we assessed the expression UBE2B, RAD18, and ZMYM2 at the protein levels by IHC staining ()). IHC staining scores were assessed among 23 cases (1 case was deleted due to tissue loss). Positive protein expression was observed in all 23 cases ()). In some samples with strong UBE2B and RAD18 staining, ZMYM2 expression was strong (, red frames). In some tissues with middle UBE2B and RAD18 staining, ZMYM2 expression was weaker (, blue frames). Therefore, we calculated the correlations between the staining scores of UBE2B and ZMYM2 and between RAD18 and ZMYM2 (). Spearman’s correlation analysis confirmed a strong positive correlation between UBE2B and ZMYM2 (Spearman’s r = 0.68, ) and a moderate positive correlation between RAD18 and ZMYM2 (Spearman’s r = 0.54, ). These findings imply that there might be some unrevealed regulations among them.
Table 1. The correlation between UBE2B and ZMYM2 IHC staining scores
Table 2. The correlation between RAD18 and ZMYM2 IHC staining scores
3.3. UBE2B interacts with both RAD18 and ZMYM2 in ovarian cancer cells
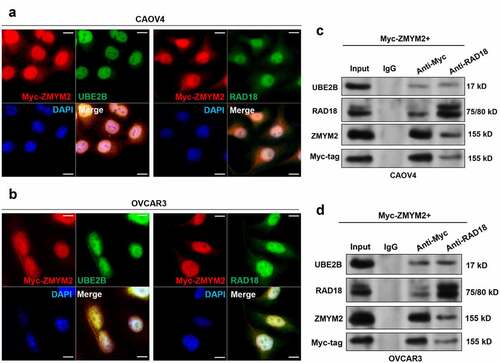
CAOV4 and OVCAR3 cells with relatively lower and higher UBE2B expression were selected as representative cell lines for further studies. IF staining results confirmed the co-localization of UBE2B and RAD18 with ZMYM2 in the two cell lines (). Co-IP assays showed that myc-ZMYM2 interacted with UBE2B and RAD18 (). UBE2B and ZMYM2 were also detected in the samples immunoprecipitated by anti-RAD18 ().
Figure 3. UBE2B interacts with both RAD18 and ZMYM2 in ovarian cancer cells. (a,b) IF staining of myc-ZMYM2 (red) and UBE2B (green) in CAOV4 (a) and OVCAR3 (b) cells. (c,d). Co-IP assays were performed using whole-cell lysates from CAOV4 and OVCAR3 cells with indicating myc-ZMYM2 overexpression. IP was performed using mouse anti-myc tag or rabbit anti-RAD18.
3.4. UBE2B promotes RAD18 mediated ZMYM2 monoubiquitination and stabilization
Since UBE2B is a ubiquitin-conjugating enzyme and RAD18 is an E3 ubiquitin-protein ligase, we then detect whether these proteins can modulate the stability of ZMYM2 protein. UBE2B overexpression in CAOV4 and cells or UBE2B knockdown in OVCAR3 cells did not alter ZMYM2 mRNA expression (). However, UBE2B overexpression increased ZMYM2 protein level (), while UBE2B knockdown decreased ZMYM2 protein level (). MG132 treatment abrogated UBE2B knockdown induced ZMYM2 downregulation (). RAD18 knockdown decreased ZMYM2 protein level and canceled UBE2B overexpression increased ZMYM2 increment (). RAD18 overexpression also increased ZMYM2 protein level, but this effect was hampered when UBE2B was knocked down (). MG132 treatment abrogated these alterations ()). These findings imply that UBE2B might be a positive regulator of ZMYM2 protein level, possibly via the ubiquitin-proteasomal pathway.
Figure 4. UBE2B promotes RAD18 mediated ZMYM2 monoubiquitination and stabilization. (a and c). QRT-PCR (a) and western blotting (b) results showing the expression of ZMYM2 at the mRNA and protein levels in CAOV4 cells 48 h after lentiviral mediated flag-UBE2B overexpression. (b and d). QRT-PCR (b) and western blotting (d) assays to check the expression of ZMYM2 at the mRNA and protein levels 48 h after lentiviral mediated UBE2B knockdown in OVCAR3 cells, with or without the treatment of MG132 (10 μM, 10 h). (e and g) Western blotting assays to detect the expression of UBE2B, RAD18, and ZMYM2 in CAOV4 cells 48 h after lentiviral mediated flag-UBE2B overexpression alone or combined with RAD18 knockdown. Additional MG132 treatment (10 μM, 10 h) was administrated in panel G. (f and g)Western blotting assays to detect the expression of UBE2B, RAD18, and ZMYM2 in OVCAR3 cells 48 h after lentiviral mediated UBE2B knockdown alone or combined with RAD18 overexpression in OVCAR3 cells, with (h) or without (f) the treatment of MG132 (10 μM, 10 h). (i) Co-IP assays to detect the ubiquitination status of ZMYM2 in CAOV4 cells with indicating selective combinations of HA-Ub overexpression, flag-UBE2B overexpression, ZMYM2 overexpression, and RAD18 knockdown. Immunoprecipitation was performed using rabbit anti-myc tag (1:50, 16286-1-AP). Ubiquitinated ZMYM2 was detected using mouse anti-HA tag (66006-2-Ig). (j–l) Representative images (j) and quantitative analysis (k-l) of CHX chase assay in CAOV4 cells with RAD18 knockdown alone, UBE2B overexpression alone, or in combination. 24 h after lentiviral infection, cells were treated with 50 μg/mL of CHX for indicated times to inhibit protein synthesis. Quantitative results were generated by using the ImageJ software. ZMYM2 expression was normalized to that of β-actin. The normalized ZMYM2 expression at 0 h was defined as 1.0 for each panel. n = 3 per group. *, comparison between shNC and shRAD18 or between vector and UBE2B groups. #, comparison between UBE2B and shRAD18+ UBE2B groups. n.s., not significant; ** and ##, p < 0.01; *** and ###, p < 0.001. Scale bar: 20 μm.
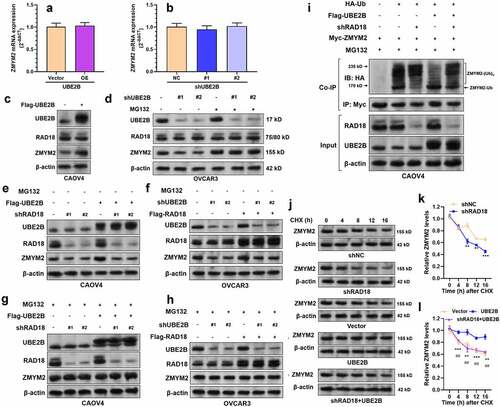
To validate this hypothesis, we performed co-IP assays to detect the ubiquitination of ZMYM2. Results showed that RAD18 knockdown increased ZMYM2 polyubiquitination but reduced its monoubiquitination (). UBE2B overexpression increased ZMYM2 monoubiquitination but reduced its polyubiquitination (). RAD18 knockdown impaired UBE2B overexpression induced ZMYM2 monoubiquitination increment (). These findings suggest that both UBE2B and RAD18 act as positive modulators of ZMYM2 monoubiquitination. Then, we conducted CHX chase assays to check the stability of ZMYM2 protein. Knockdown of RAD18 significantly shortened the half-life of ZMYM2 protein ()). UBE2B overexpression significantly enhanced the stability of ZMYM2 protein ()). RAD18 knockdown weakened the effect of UBE2B overexpression on prolonging the half-life of ZMYM2 protein ()). Collectively, these findings suggested that UBE2B and RAD18 cooperatively enhance ZMYM2 stability by increasing its monoubiquitination.
3.5. UBE2B overexpression-induced ovarian cancer growth is hampered by ZMYM2 knockdown
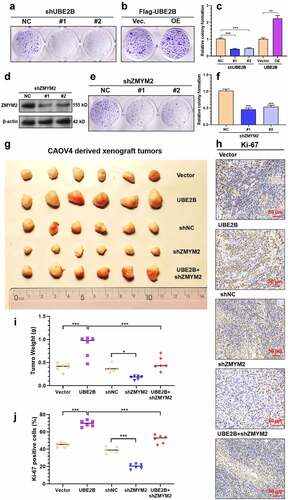
Since we confirmed that UBE2B and RAD18 cooperatively enhance ZMYM2 stability by increasing its monoubiquitination, we further explored the regulation of the UBE2B-ZMYM2 axis on ovarian cancer growth. Knockdown of UBE2B or ZMYM2 () significantly suppressed the colony formation of CAOV4 cells ()). UBE2B overexpression significantly enhanced colony formation (). To further validate the regulatory effects in vivo, we generated xenograft tumor models in nude mice, using CAOV4 cells with UBE2B and ZMYM2 manipulated. UBE2B overexpression substantially enhanced tumor growth (). ZMYM2 knockdown significantly suppressed tumor growth and impaired the growth-promoting effect of UBE2B overexpression (). Ki-67 staining of the tumors confirmed that UBE2B overexpression had the highest level of Ki-67 positive cells (), which was hampered by ZMYM2 knockdown ().
Figure 5. UBE2B overexpression-induced ovarian cancer growth is hampered by ZMYM2 knockdown. (a-f) Representative images (a, b, and e) and quantitation (c and f) of colonies formed by CAOV4 cells with UBE2B knockdown (a and c) or overexpression (b-c), or ZMYM2 knockdown (d-f). n = 3 per group. (g,i) Representative images (g) and quantitation of tumor weight (i) of xenograft tumors derived from CAOV4 cells with UBE2B overexpression alone, ZMYM2 knockdown alone, or in combination. n = 6 per group. (h,j) Representative images (h) and quantitation of the Ki-67 positive cells (j) of xenograft tumors derived from CAOV4 cells with UBE2B overexpression alone, ZMYM2 knockdown alone, or in combination. n = 6 per group. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
4. Discussion
In this study, using data from TCGA, we further confirmed that UBE2B upregulation was associated with poor survival of ovarian cancer patients. As an E2 ubiquitin-conjugating enzyme, UBE2B interacts with different E3 ligases and exerts different modifications [Citation28]. It associates with ring finger protein 20/40 (RNF20/40) and monoubiquitylates Histone H2B, leading to a localized open conformation that supports gene transcription [Citation29]. UBE2B can interact with RAD18 that catalyzes the monoubiquitination of PCNA, which is a DNA-polymerase sliding clamp that participates in DNA synthesis and repair [Citation30].
By performing bioinformatic prediction, we found that ZMYM2 might interact with UBE2B and RAD18. The upregulation of RAD18 in ovarian cancer was confirmed based on RNA-seq data in TCGA database. The tumor-promoting effect of RAD18 upregulation was observed in multiple cancers. It promotes the migration and invasion of esophageal squamous cell cancer by amplifying the JNK-MMPs signaling pathway [Citation31]. High RAD18 expression might predict poor responses to neoadjuvant chemoradiotherapy in patients with locally advanced rectal cancer [Citation32]. In patients with gastric cancer, elevated RAD18 expression is associated with substantially shorter overall survival and recurrence-free survival durations [Citation33]. Knockdown of RAD18 significantly reduces the growth, migration, and invasion of ovarian cancer cells [Citation34]. ZMYM2 encodes a zinc finger protein involved in a histone deacetylase complex [Citation35], stabilizing a transcriptional co-repressor complex containing LSD1–CoREST–HDAC1 [Citation36]. This complex is activated in a series of cancers and suppresses the expression of multiple tumor suppressor genes. Targeting this complex using HDAC inhibitor has been considered a potential therapeutic strategy to overcome the development of cancer stem cells in ovarian cancer [Citation37]. Another recent study reported that ZMYM2 could bind to B-Myb and promote the S-phase entry of HepG2 cells [Citation38]. Knockdown of ZMYM2 could significantly impair G1/S-phase progression [Citation38].
Therefore, we decided to validate the predicted interactions. IHC staining of serial ovarian cancer tissues confirmed a positive correlation between UBE2B and ZMYM2, and between RAD18 and ZMYM2. IF staining and co-IP assays in CAOV4 and OVCAR3 cells confirmed their mutual interaction. One recent study found that UBE2B is intrinsically capable of catalyzing polyubiquitin chain formation. However, this activity is largely abrogated when interacting with RAD18 due to RAD18ʹs competing with ubiquitin for a noncovalent ‘backside’ binding site on UBE2B [Citation26]. This mechanism helps explain why the UBE2B/RAD18 complex is prone to catalyze monoubiquitination of the substrates. Some recent studies suggest that monoubiquitination can increase the half-life of the substrates. For example, Mdm2 catalyzes HDAC3 monoubiquitination and enhances its stability [Citation39]. FBXL10 can stabilize ERRα protein by reducing polyubiquitylation and increasing monoubiquitination in breast cancer cells [Citation40]. In this study, we revealed that UBE2B and RAD18 cooperatively increase ZMYM2 monoubiquitination and reduce its polyubiquitination, thereby enhancing its stability. By using CAOV4 cells in in vitro and in vivo models of tumor growth, we confirmed that UBE2B overexpression significantly enhanced tumor cell growth. ZMYM2 knockdown remarkedly suppressed tumor growth and impaired the growth-promoting effect of UBE2B overexpression.
One point worth noticing is that UBE2B expression at the mRNA level was significantly higher in the normal ovary than in the tumor tissues. However, findings of this study largely supported the oncogenic role of UBE2B in ovarian cancer. This inconsistency might be explained by the following reasons. Firstly, high-grade serous carcinoma mainly originates from fallopian tube secretory cells [Citation41]. Therefore, normal ovary might not serve as the real normal control. Besides, although we found a difference in UBE2B expression between GTEx-normal fallopian tube and primary tumors in TCGA-OV, the number of the GTEx-normal fallopian tube group is small (n = 5), which limits its statistical power. Based on the findings of this study, we infer that aberrant UBE2B expression might be linked to malignant behaviors and have oncogenic properties. However, the detailed mechanisms underlying its dysregulation need to be carefully studied in the future.
5. Conclusion
This study revealed a novel regulatory effect of the UBE2B/RAD18 complex on ZMYM2 monoubiquitination and stability in ovarian cancer. UBE2B promotes RAD18-mediated ZMYM2 monoubiquitination and increases its stability in ovarian cancer, thereby enhancing ovarian cancer cell proliferation.
Highlights
(1) Higher UBE2B expression was linked to a poorer prognosis of ovarian cancer.
(2) UBE2B and RAD18 expression are positively correlated with ZMYM2 protein levels in ovarian tumor tissues.
(3) UBE2B interacts with RAD18 and ZMYM2 in ovarian cancer cells.
(4) UBE2B promotes RAD18 mediated ZMYM2 monoubiquitination and stabilization.
(5) UBE2B promotes ovarian cancer growth partially via maintaining ZMYM2 stabilization.
Data availability
The data and material used to support the findings of this study are available from the corresponding author upon request.
Acknowledgements
The authors have no conflict of interest.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Abu Ahmad Y, Oknin-Vaisman A, Bitman-Lotan E, et al. From the evasion of degradation to ubiquitin-dependent protein stabilization. Cells. 2021;10(9):2374.
- Swatek KN, Komander D. Ubiquitin modifications. Cell Res. 2016;26(4):399–422.
- Choi YB, Harhaj EW. HTLV-1 tax stabilizes MCL-1 via TRAF6-dependent K63-linked polyubiquitination to promote cell survival and transformation. PLoS Pathog. 2014;10(10):e1004458.
- Sun T, Liu Z, Yang Q. The role of ubiquitination and deubiquitination in cancer metabolism. Mol Cancer. 2020;19(1):146.
- Deng L, Meng T, Chen L, et al. The role of ubiquitination in tumorigenesis and targeted drug discovery. Signal Transduct Target Ther. 2020;5(1):11.
- Morreale FE, Walden H. Types of ubiquitin ligases. Cell. 2016;165(1):248–e1.
- Han C, Yang L, Choi HH, et al. Amplification of USP13 drives ovarian cancer metabolism. Nat Commun. 2016;7:13525.
- Li M, Tang Y, Zuo X, et al. Loss of Ras GTPase-activating protein SH3 domain-binding protein 1 (G3BP1) inhibits the progression of ovarian cancer in coordination with ubiquitin-specific protease 10 (USP10). Bioengineered. 2022;13(1):721–734.
- Low SS, Ji D, Chai WS, et al. Recent progress in nanomaterials modified electrochemical biosensors for the detection of microRNA. Micromachines (Basel). 2021;12(11). DOI:10.3390/mi12111409.
- Shin Low S, Pan Y, Ji D, et al. Smartphone-based portable electrochemical biosensing system for detection of circulating microRNA-21 in saliva as a proof-of-concept. Sens Actuators B Chem. 2020;308:127718.
- Somasagara RR, Tripathi K, Spencer SM, et al. Rad6 upregulation promotes stem cell-like characteristics and platinum resistance in ovarian cancer. Biochem Biophys Res Commun. 2016;469(3):449–455.
- Somasagara RR, Spencer SM, Tripathi K, et al. RAD6 promotes DNA repair and stem cell signaling in ovarian cancer and is a promising therapeutic target to prevent and treat acquired chemoresistance. Oncogene. 2017;36(48):6680–6690.
- Bailly V, Lamb J, Sung P, et al. Specific complex formation between yeast RAD6 and RAD18 proteins: a potential mechanism for targeting RAD6 ubiquitin-conjugating activity to DNA damage sites. Genes Dev. 1994;8(7):811–820.
- Mustofa MK, Tanoue Y, Chirifu M, et al. RAD18 mediates DNA double-strand break-induced ubiquitination of chromatin protein. J Biochem. 2021;170(1):33–40.
- Hsu SH, Chen SH, Kuo CC, et al. Ubiquitin-conjugating enzyme E2 B regulates the ubiquitination of O(6)-methylguanine-DNA methyltransferase and BCNU sensitivity in human nasopharyngeal carcinoma cells. Biochem Pharmacol. 2018;158:327–338.
- Goldman MJ, Craft B, Hastie M, et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat Biotechnol. 2020;38(6):675–678.
- Elegheert J, Behiels E, Bishop B, et al. Lentiviral transduction of mammalian cells for fast, scalable and high-level production of soluble and membrane proteins. Nat Protoc. 2018;13(12):2991–3017.
- Dong L, Huang J, Zu P, et al. Transcription factor 3 (TCF3) combined with histone deacetylase 3 (HDAC3) down-regulates microRNA-101 to promote Burkitt lymphoma cell proliferation and inhibit apoptosis. Bioengineered. 2021;12(1):7995–8005.
- Uhlen M, Oksvold P, Fagerberg L, et al. Towards a knowledge-based Human Protein Atlas. Nat Biotechnol. 2010;28(12):1248–1250.
- Olympus Life Science Europa Gmb H. Flexible applications for Olympus digital microscope camera. Biotechnol J. 2008;3(1):31.
- Ayakannu T, Taylor AH, Willets JM, et al. Validation of endogenous control reference genes for normalizing gene expression studies in endometrial carcinoma. Mol Hum Reprod. 2015;21(9):723–735.
- Mitteer DR, Greer BD, Fisher WW, et al. Teaching behavior technicians to create publication-quality, single-case design graphs in graphpad prism 7. J Appl Behav Anal. 2018;51(4):998–1010.
- Huang WL, Luo CW, Chou CL, et al. High expression of UBE2B as a poor prognosis factor in patients with rectal cancer following chemoradiotherapy. Anticancer Res. 2020;40(11):6305–6317.
- Zou R, Xu H, Li F, et al. Increased expression of UBE2T predicting poor survival of epithelial ovarian cancer: based on comprehensive analysis of UBE2s, clinical samples, and the GEO database. DNA Cell Biol. 2021;40(1):36–60.
- Montojo J, Zuberi K, Rodriguez H, et al. GeneMANIA: fast gene network construction and function prediction for Cytoscape. F1000Res. 2014;3:153.
- Hibbert RG, Huang A, Boelens R, et al. E3 ligase Rad18 promotes monoubiquitination rather than ubiquitin chain formation by E2 enzyme Rad6. Proc Natl Acad Sci U S A. 2011;108(14):5590–5595.
- Tsuji Y, Watanabe K, Araki K, et al. Recognition of forked and single-stranded DNA structures by human RAD18 complexed with RAD6B protein triggers its recruitment to stalled replication forks. Genes Cells. 2008;13(4):343–354.
- Game JC, Chernikova SB. The role of RAD6 in recombinational repair, checkpoints and meiosis via histone modification. DNA Repair (Amst). 2009;8(4):470–482.
- Foglizzo M, Middleton AJ, Day CL. Structure and function of the RING domains of RNF20 and RNF40, dimeric E3 ligases that monoubiquitylate histone H2B. J Mol Biol. 2016;428(20):4073–4086.
- Hoege C, Pfander B, Moldovan GL, et al. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419(6903):135–141.
- Zou S, Yang J, Guo J, et al. RAD18 promotes the migration and invasion of esophageal squamous cell cancer via the JNK-MMPs pathway. Cancer Lett. 2018;417:65–74.
- Yan X, Chen J, Meng Y, et al. RAD18 may function as a predictor of response to preoperative concurrent chemoradiotherapy in patients with locally advanced rectal cancer through caspase-9-caspase-3-dependent apoptotic pathway. Cancer Med. 2019;8(6):3094–3104.
- Baatar S, Bai T, Yokobori T, et al. High RAD18 expression is associated with disease progression and poor prognosis in patients with gastric cancer. Ann Surg Oncol. 2020;27(11):4360–4368.
- Xue H, Wu Z, Rao D, et al. Long non-coding RNA LINC00858 aggravates the oncogenic phenotypes of ovarian cancer cells through miR-134-5p/RAD18 signaling. Arch Gynecol Obstet. 2020;302(5):1243–1254.
- Bantscheff M, Hopf C, Savitski MM, et al. Chemoproteomics profiling of HDAC inhibitors reveals selective targeting of HDAC complexes. Nat Biotechnol. 2011;29(3):255–265.
- Gocke CB, Yu H. ZNF198 stabilizes the LSD1-CoREST-HDAC1 complex on chromatin through its MYM-type zinc fingers. PLoS One. 2008;3(9):e3255.
- Wang D, Li W, Zhao R, et al. Stabilized peptide HDAC inhibitors derived from HDAC1 substrate H3K56 for the treatment of cancer stem-like cells in vivo. Cancer Res. 2019;79(8):1769–1783.
- Cibis H, Biyanee A, Dorner W, et al. Characterization of the zinc finger proteins ZMYM2 and ZMYM4 as novel B-MYB binding proteins. Sci Rep. 2020;10(1):8390.
- Choi YM, An S, Bae S, et al. Mdm2 is required for HDAC3 monoubiquitination and stability. Biochem Biophys Res Commun. 2019;517(2):353–358.
- Yang Y, Li S, Li B, et al. FBXL10 promotes ERRalpha protein stability and proliferation of breast cancer cells by enhancing the mono-ubiquitylation of ERRalpha. Cancer Lett. 2021;502:108–119.
- Erickson BK, Conner MG, Landen CN Jr. The role of the fallopian tube in the origin of ovarian cancer. Am J Obstet Gynecol. 2013;209(5):409–414.
