ABSTRACT
Endometrial cancer (EC) is one of the most common gynecological tumors with an increasing incidence. CircRNA plays an essential regulatory role in EC. Our objective was to investigate the potential mechanism of circRNAs derived SPOC Domain Containing 1 (SPOCD1) in EC progression. Seven circRNAs from SPOCD1 were analyzed by circBase and their expression was verified by quantitative real-time polymerase chain reaction. Only the expression of hsa_circ_0011324 was significantly increased in cancer tissues. The cell lines Ishikawa and RL95-2 which interfered with or overexpressed hsa_circ_0011324 were constructed and cell functions were tested. Results revealed hsa_circ_0011324 overexpression promoted cell proliferation, migration, and invasion; while silence of hsa_circ_0011324 had opposite effect on cell functions. RNA22 website and Targetscan website were applied to analyze downstream genes regulated by hsa_circ_0011324. Then, the expression of downstream genes was detected in EC tissues. Results indicated hsa-miR-497/16-5p expression were down-regulated, and mechanistic target of rapamycin kinase (mTOR) was up-regulated in EC. Furthermore, hsa_circ_0011324 regulated mTOR expression and cell functions by affecting hsa-miR-497/16-5p. And the potential mechanism was hsa_circ_0011324 competes with mTOR to directly bind to hsa-miR-497/16-5p. In conclusion, hsa_circ_0011324 could sponge hsa-miR-497/16-5p targeted mTOR to participate in EC progress. Our study may provide a new therapeutic target for EC.
GRAPHICAL ABSTRACT
Introduction
Endometrial cancer (EC) is one of the most common gynecological malignancies [Citation1]. In recent years, under aging population, increasing obesity and diabetes influence, EC incidence has gradually increased [Citation2], and the age of onset of EC has gradually become younger [Citation3]. Endometrium changes under a variety of risk factors. Some non-physiological hyperplastic endometrium can be reverted to normal endometrium [Citation4], while some are precancerous lesions and develop into EC of different pathological types [Citation5]. The 5-year survival rate of EC patients decreased with EC stage development. The 5-year survival rate of 80% women diagnosed with EC stage I was >95% [Citation6], while the 5-year survival rate of advanced patients was <20% [Citation7]. Early detection and timely treatment are the key to improve patient’s prognosis. At present, there is no method to detect precancerous lesions or early cancers in time. Therefore, to explore a simple, accurate and economical screening method has become the focus of current research.
Circular RNA (circRNA) is a special class of non-coding RNA molecules (sometimes expressed in vivo), which is also the latest research hotspot in RNA field [Citation8,Citation9]. Different from traditional linear RNA (linear RNA with 5’ and 3’ ends), circRNA molecules have a closed ring structure and are not affected by RNA exonuclease, so their expression is more stable and not easy to degrade [Citation10,Citation11]. In terms of function, recent studies have shown that circRNA molecules are rich in microRNA (miRNA) binding sites and act as miRNA sponges in cells, thereby lifting miRNA inhibition on target genes and increasing target genes expression. This mechanism is known as competing endogenous RNAs (ceRNAs) mechanism [Citation12]. J Jia Y, et al. reported circRNA hsa_circRNA_0001776 down-regulated leucine rich repeats and immunoglobulin like domains 2 and inhibited EC proliferation and promoted apoptosis through sponging miR-182 [Citation13]. Wu B, et al. also reported a novel tumor regulator hsa_circ_0075960 acted as a sponge for miR-361-3p/SH2B adaptor protein 1 in EC cells and regulated EC progression by regulating miR-361-3p [Citation14]. This indicated that circRNA played a vital regulatory role in EC through interactions with EC-associated miRNAs.
Our published papers have shown that SPOC Domain Containing 1 (SPOCD1) accelerated ovarian cancer progression and inhibited apoptosis through PI3K/AKT pathway [Citation15]. We speculated the circRNAs that come from SPOCD1 also participate in EC progression. In this project, 7 circRNAs (hsa_circ_0011324, hsa_circ_0011325, hsa_circ_0011326, hsa_circ_0011327, hsa_circ_0011328, hsa_circ_0011329, hsa_circ_0011,330) from SPOCD1 were further analyzed by circBase. We used quantitative real-time polymerase chain reaction (qRT-PCR) to verify the differential expression of 7 circRNAs from SPOCD1, and found that hsa_circ_0011324 had significant differential expression. However, whether hsa_circ_0011324 participated in EC development and its specific mechanism has not been studied yet. Therefore, based on the above background, in this research, we investigated the mechanism of hsa_circ_0011324 in EC progression. Our goal is to provide a new strategies and targets for EC treatment.
Materials and methods
Collection of clinical samples
Sixty-four samples of cancer (CA) and para-cancerous (Para-CA) tissues came from patients with endometrial cancer in The First Hospital of Lanzhou University. None of the enrolled patients received chemotherapy or radiotherapy before surgery. The diagnosis and identification of EC were based on the standards of the World Health Organization. This study was approved by the institutional ethical board of The First Hospital of Lanzhou University (LDYYLL2021-99), and specimens were collected after each patient’s informed consent was obtained. The main clinical and pathological characteristics of these patients were collected. All patients were followed up for 5 years.
Sanger sequencing and RT-qPCR
Sanger sequencing was performed according to previously reported methods [Citation16,Citation17]. In our study, RNA was extracted from samples of CA and Para-CA using TriQuick Reagent (R1100, Solarbio), which was reversed into complementary DNA (cDNA) using HiScript III RT SuperMix for qPCR (+gDNA WIper) (R323-01, Vazyme). The primers of hsa_circ_0011324 in were used for amplification according to the protocol of 2× Taq Master Mix (P111-03, Vazyme). The amplified products were sequenced in Sangon biotech (Shanghai, China).
Table 1. The primers used in this study
In our study, we evaluated hsa_circ_0011324, hsa_circ_0011325, hsa_circ_0011326, hsa_circ_0011327, hsa_circ_0011328, hsa_circ_0011329, hsa_circ_0011330, hsa-miR-497-5p, hsa-miR-16-5p, and mTOR expression using qRT-PCR according to previously reported methods [Citation16]. 2−ΔΔCt was used to detect gene expression. U6 or GAPDH as internal reference. showed the primers used in this study.
Fluorescence in situ hybridization (FISH)
We performed FISH according to previous research [Citation18]. In brief, precooled 4% paraformaldehyde (PFA) was placed and fixed at 4°C for 40 min. PFA was aspirated at room temperature and washed twice with a film breaker (0.1% Triton X-100 in phosphate-buffered saline (PBS)), 5 min each time. The probe was dropped onto the slipper, the slipper was sealed, desaturated at 85°C for 5 min, then taken out and hybridized overnight in an oven at 37°C and stained with DAPI. Mounting Medium sealed the film and wait until the Mounting Medium was solidified before taking pictures. The probe sequences used was as follows: hsa_circ_0011324: AAAGGCCCAATTTTACCAAAT-CY5, NC: ATCAGTCATCGACAATTAAAC-CY5. They were synthesized by Generay Biotech (Shanghai, China).
Cell culture and treatment
The EC cells Ishikawa (#CC1103) and RL95-2 (#CC1106) were derived from CellCook. Ishikawa culture conditions were RPMI 1640 (#11875, Gibco) supplemented with 15% fetal bovine serum (FBS). RL95-2 was cultured with DMEM/F12 (#11330, Gibco) supplemented with 10% FBS containing 5 μg/mL insulin (#HY-P0035, MCE).
Ishikawa and RL95-2 cell lines with stable interference or overexpression of hsa_circ_0011324 were constructed and cultured, which were divided into Ishikawa-shNC, Ishikawa-shCIRC, Ishikawa-LVNC and Ishikawa-LVCIRC groups; RL95-2-shNC, RL95-2-shCIRC, RL95-2-LVNC, RL95-2-LVCIRC groups according to requirements. Interference and overexpression of hsa_circ_0011324 and its control virus were prepared by Hanbio Biotechnology (Shanghai, China).
To investigate whether hsa_circ_0011324 regulate EC progression through hsa-miR-497-5p and hsa-miR-16-5p, Ishikawa and RL95-2 cell lines with stable overexpression of hsa_circ_0011324 were transfected with hsa-miR-497-5p mimics and hsa-miR-16-5p mimics. The mimics of hsa-miR-497-5p or hsa-miR16-5p and their controls were provided by GenePharma (Shanghai, China). The above cell transfection steps were followed by Lipofectamine™ 2000 (BL623B, Biosharp) protocol.
Cell counting Kit-8 (CCK-8) assay
Cell proliferation was determined using CCK-8 assay kit (G4103, Servicebio) as previous study [Citation19]. In simple terms, cells in the logarithmic growth phase were taken, rinsed once with PBS, and digested by 0.25% trypsin into a single cell suspension. After digestion was terminated in complete medium, the cells were suspended in complete medium. Cells were counted, and the cell concentration was adjusted to 3 × 104 cells/mL, and then inoculated into 96 well plates with 100 μL per well that was 3 × 103 cells/well. Each cell line was tested for three multiple wells per day. 100 μL of complete medium was added to each well, and culture was continued at 37°C in a 5%CO2 incubator. After 12, 24, 48, 72 and 96 h, CCK-8 reagent was added in the ratio of 1:10 that was 100 μL culture solution was added into 10 μL detection solution. After incubation at 37°C for 2 h, the absorbance value at 450 nm was measured by a microplate reader.
Clone formation assay
Clone formation assay was conducted as previous study method [Citation20]. After cell counting, the cells were diluted to 1 × 104cell/mL, and then diluted to 1 × 103cell/mL and inoculated into 6-well plates. After inoculation, we added medium to 2 mL/well for each well, shook the culture plate from left to right in horizontal position to make the cells in the hole as evenly as possible. After one to two weeks of culture, the medium was removed from each well, rinsed with PBS or normal saline twice, fixed with 4% PFA for 15 min, rinsed with PBS twice, and 200 μL crystal violet staining solution was added to each well. After 20 min, the 6-well plate was rinsed under running water. After rinsing, the number of colonies was calculated.
Transwell migration assay
As previous study method, we performed Transwell migration assay [Citation20]. Cells in logarithmic growth phase were taken and rinsed once with PBS, then digested by 0.25% trypsin into single cell suspension. Appropriate amount of cell suspension was taken and centrifuged at 800 rpm for 5 min. After supernatant was aspirated and cells were suspended with basal medium, cell concentration was adjusted to 1 × 106/mL with basal medium, 100 μL was added to upper chamber of Transwell chamber, and 600 μL of complete medium was added to lower chamber. Cells in upper chamber were wiped with cotton swabs, fixed with 4% PFA for 15 min, stained with 1% crystal violet for 10 min, and observed whether the cells passed through the pores under a microscope.
Transwell invasion assay
As previous study method, we performed Transwell invasion assay [Citation20]. Matrigel was dissolved overnight at 4°C, diluted with Matrigel: medium (1:3) in precooled basal medium, 40 μL was added to precooled Transwell chamber, the action should be slow to avoid bubbles. Matrigel was solidified after incubation at 37°C for 2 h. We added 100 μL and 600 μL basal medium to the upper and lower chambers, hydrated overnight at 37°C. The next day the medium was aspirated. After trypsin digestion, an appropriate amount of cell suspension was taken and centrifuged at 800 rpm for 5 min. After supernatant was aspirated and cells were suspended with basal medium, cell concentration was adjusted to 1 × 106/mL with basal medium, 100 μL was added to upper chamber of Transwell chamber, and 600 μL of complete medium was added to lower chamber. The cells in upper chamber were wiped with cotton swabs, fixed with 4% paraformaldehyde for 15 minutes, stained with 1% crystal violet for 10 minutes, observed under a microscope and photographed.
Immunohistochemistry (IHC)
We performed IHC following with previous study method [Citation20]. First, xylene and ethanol were used for dewaxing and hydration. Then, fresh 3% H2O2 was prepared with distilled water or PBS and sealed at room temperature for 5–10 min. After that, it was heated by boiling and heated by electric furnace or water bath. Five percent BSA was used and sealed at room temperature for 15 minutes. We discarded the excess liquid and dropped mTOR Monoclonal Antibody (66,888-1-IG, Proteintech) overnight at 4°C. Polymerized HRP labeled anti-rabbit/mouse IgG secondary antibody was incubated. DAB was used for color rendering. Hematoxylin was redyed and washed twice with distilled water for 2 min each. Finally, after dehydration, transparent neutral resin was sealed and stored at room temperature.
Western blot analysis
We detected protein expression via western blot analysis following with previous study [Citation20]. Total protein was extracted with RIPA lysate. Protein was quantified according to BCA protein determination Kit (BL521A, Biosharp). SDS-PAGE loading buffer was mixed, and the mixture was heated in a boiling water bath at 100°C for 5 minutes. The protein was adsorbed on PVDF membrane by gel electrophoresis and sealed with 5% skim milk solution at room temperature for 90 min. mTOR Monoclonal Antibody (66,888-1-IG, Proteintech) and GAPDH (AB9485, ABCAM) were incubated overnight. The second antibody was incubated. Exposure was performed using an ultra-sensitive ECL chemiluminescence substrate (BL520A, Biosharp). GAPDH was acted as an internal reference to detect expression levels.
RNA immunoprecipitation (RIP) assay
In our study, we used AGO2-RIP assay to confirm whether hsa_circ_0011324 can compete with mTOR to bind to miRNA [Citation21]. Ishikawa and RL95-2 cell lines with stable overexpression of hsa_circ_0011324 were cultured and divided into Ishikawa-LVNC, Ishikawa-LVCIRC, RL95-2-LVNC and RL95-2-LVCIRC groups according to requirements. The cells were collected for RIP test. After RNA was obtained, hsa_circ_0011324 and mTOR mRNA expressions were evaluated by qRT-PCR. Using the Magna RIP™ RNA-Binding Protein Immunoprecipitation Kit (#17-700) provided by Millipore, RIP was performed according to the instructions. The antibody used was Anti-Argonaute-2 antibody [EPR10411] (ab186733, ABCAM).
Bioinformatics prediction and dual-luciferase assay
RNA22 website and Targetscan website were used to analyze downstream genes regulated by hsa_circ_0011324. Then, dual-luciferase assay was conducted following with the method in previous study [Citation20]. 293 T cells were purchased from CellCook (#CC4003) and cultured in DMEM high Glucose (#11995, Gibco) with 10% FBS. According to the TransDetect® Double-Luciferase Reporter Assay Kit (FR201-01, TransGen Biotech), the binding effect of hsa_circ_0011324 with hsa-miR-497/16-5p and the binding effect of hsa-miR-497/16-5p with mTOR mRNA 3ʹUTR (untranslated regions) were detected. Fluorescence reporter plasmid was synthesized by Genecfps (Jiangsu, China).
Statistical analysis
Graphpad Prism 8.0 software was used for statistical analysis. Measurement data were expressed as mean ± standard deviation (SD). Student’s t-test was used between the two groups, and one-way analysis of variance (ANOVA) was performed for inter-group comparison. The disease-free survival (DFS) and overall survival (OS) rates were calculated through Kaplan-Meier, and differences were compared using a log-rank test. chi-square (and Fisher’s exact) test detected the relationship between hsa_circ_0011324 and clinicopathological parameters. Cox regression (Univariate and multivariate) analyzed OS in EC patients. Pearson Correlation Coefficient studied the correlation between hsa_circ_0011324 and hsa-miR-497-5p, hsa_circ_0011324 and hsa-miR-16-5p. P < 0.05 indicated the difference was statistically significant.
Results
Our published papers have shown that SPOCD1 accelerated ovarian cancer progression. We speculated the circRNAs that come from SPOCD1 also participate in EC progression. In this project, 7 circRNAs (hsa_circ_0011324, hsa_circ_0011325, hsa_circ_0011326, hsa_circ_0011327, hsa_circ_0011328, hsa_circ_0011329, hsa_circ_0011330) from SPOCD1 were found in circBase. We found that only hsa_circ_0011324 had significant differential expression in EC via qRT-PCR detection. Therefore, based on the above background, in this research, we investigated the mechanism of hsa_circ_0011324 in EC progression. Results showed that hsa_circ_0011324 could sponge hsa-miR-497/16-5p targeted mTOR to participate in EC progress.
1. Hsa_circ_0011324 expression in EC and its correlation with prognosis
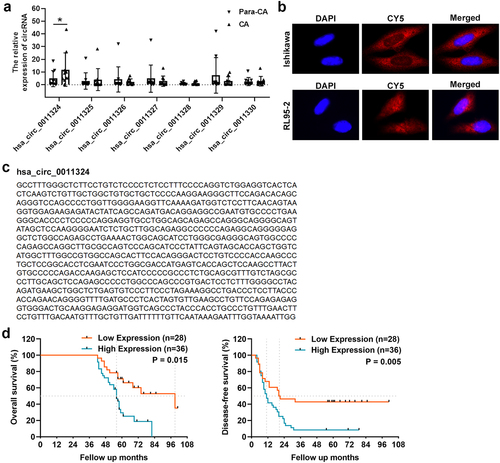
First, we used CA and Para-CA samples to examine SPOCD1-derived circRNAs (hsa_circ_0011324, hsa_circ_0011325, hsa_circ_0011326, hsa_circ_0011327, hsa_circ_0011328, hsa_circ_0011329, and hsa_circ_0011330) expression using qRT-PCR. Only hsa_circ_0011324, which originated from exon 16 of the SPOCD1 gene, was significantly increased in CA (). Therefore, we selected hsa_circ_0011324 for further study. Firstly, we searched circBase to find the sequence of hsa_circ_0011324 ( left). Then Sanger sequencing was used to confirm the loop structure of hsa_circ_0011324, and results indicated the head-to-tail splice site was consistent with the sequence from circBase ( right). Next, FISH experiment was used to evaluate cellular localization of hsa_circ_0011324 in EC cell lines (Ishikawa and RL95-2). And results demonstrated hsa_circ_0011324 was mainly localized in cytoplasm (), indicated that hsa_circ_0011324 may participate in EC progression through ceRNAs mechanism. Afterward, we verified the hsa_circ_0011324 expression and its correlation with prognosis in EC tissues. Kaplan–Meier survival curves showed the high expression of hsa_circ_0011324 in EC tissues was closely related to poor OS and DFS (). showed the relationship between hsa_circ_0011324 and clinicopathological parameters. Among them, hsa_circ_0011324 was closely related to tumor staging. showed the univariate and multivariate analysis of OS in EC patients. Among them, circRNA expression and Clinical Stage FIGO had significant differences.
Table 2. Relationship between hsa_circ_0011324 expression and clinicopathological parameters
Table 3. Univariate and multivariate analysis of Overall Survival (OS) in EC patients (n = 64)
Figure 1. Hsa_circ_0011324 expression in endometrial cancer and its correlation with prognosis. (a). Quantitative real-time polymerase chain reaction analysis of SPOCD1-derived circRNAs (hsa_circ_0011324, hsa_circ_0011325, hsa_circ_0011326, hsa_circ_0011327, hsa_circ_0011328, hsa_circ_0011329, and hsa_circ_0011330) expression in CA (cancer tissue) and Para-CA (para-cancerous) came from patients with endometrial cancer. (b). Left is the sequence of hsa_circ_0011324 from circBase, right is the Sanger sequencing analysis to confirm the loop structure of hsa_circ_0011324. (c). Fluorescence in situ hybridization experiment detected the localization of hsa_circ_0011324 in endometrial cancer cell lines (Ishikawa and RL95-2). (d). Kaplan-Meier survival curves for endometrial cancer patients according to hsa_circ_0011324 expression level. * P < 0.05.
2. Hsa_circ_0011324 promoted EC progression
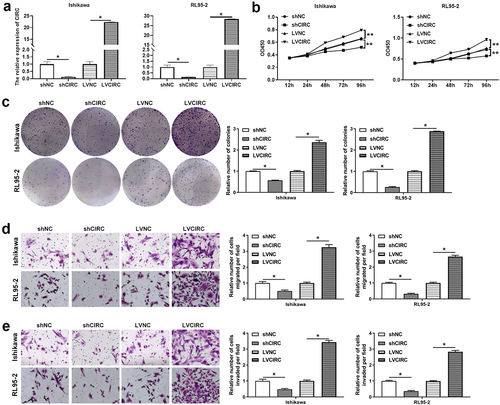
indicated that hsa_circ_0011324 maybe participate in the EC progression. Therefore, we further studied the hsa_circ_0011324 function in EC cell lines (Ishikawa and RL95-2). Firstly, Ishikawa and RL95-2 cell lines that interfere with or overexpress hsa_circ_0011324 were constructed. showed the cell lines Ishikawa and RL95-2 that interfered with or overexpressed hsa_circ_0011324 were successfully constructed. After that, we performed cell function tests on Ishikawa and RL95-2 cells. Hsa_circ_0011324 overexpression promoted cell proliferation, migration, and invasion (). Silence of hsa_circ_0011324 inhibited cell proliferation, migration, and invasion (). Base on the above results, we found that hsa_circ_0011324 overexpression promoted EC progression, while knockdown of hsa_circ_0011324 inhibited EC progression.
Figure 2. Hsa_circ_0011324 promoted endometrial cancer progression. (a). Quantitative real-time polymerase chain reaction determined hsa_circ_0011324 expression changes in Ishikawa and RL95-2 cells after stable overexpression or interference with hsa_circ_0011324. After stable overexpression or interference with hsa_circ_0011324 in Ishikawa and RL95-2 cells, (b) Cell Counting Kit-8 evaluated cell proliferation changes. (c). Clone formation detected the number of cell clones. (d) and (e). Transwell measured cell migration (d) and invasion (e) changes. CIRC indicates hsa_circ_0011324; shNC indicates the negative control of interference with hsa_circ_0011324; shCIRC indicates interference with hsa_circ_0011324; LVNC indicates the negative control of hsa_circ_0011324 overexpression; LVCIRC indicates hsa_circ_0011324 overexpression. * P < 0.05, ** P < 0.01.
3. Hsa-miR-497/16-5p were down-regulated while mTOR was up-regulated in EC tissues
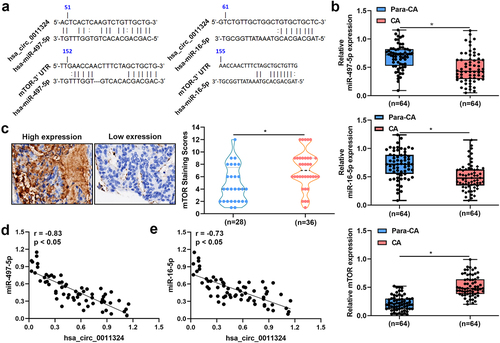
To study whether hsa_circ_0011324 regulated downstream genes to participate in EC progression, RNA22 website and Targetscan website were used to analyze the potential downstream genes of hsa_circ_0011324. Results indicated that hsa_circ_0011324 might inhibit the targeted inhibitory effect of hsa-miR-497-5p and hsa-miR-16-5p on mTOR by sponging hsa-miR-497-5p and hsa-miR-16-5p (). showed the expression of hsa-miR-497-5p and hsa-miR-16-5p was low, while mTOR expression was high in CA compared to Para-CA. Pearson Correlation Coefficient revealed hsa-miR-497-5p and hsa-miR-16-5p expressions in EC was negatively correlated with hsa_circ_0011324 (). These results suggested that the downstream genes (hsa-miR-497/16-5p and mTOR) involved in EC progression.
Figure 3. Hsa-miR-497/16-5p were down-regulated while mTOR was up-regulated in endometrial cancer tissues. (a). RNA22 website combined with targetscan website were used to analyze the potential downstream genes of hsa_circ_0011324. (b). Quantitative real-time polymerase chain reaction detected hsa-miR-497-5p, hsa-miR-16-5p, and mTOR mRNA expressions in CA (cancer tissue) and Para-CA (para-cancerous) came from patients with endometrial cancer. (c). Immunohistochemistry evaluated mTOR expression. (d). Pearson Correlation Coefficient studied the correlation between hsa_circ_0011324 and hsa-miR-497-5p. (e). Pearson correlation coefficient studied the correlation between hsa_circ_0011324 and hsa-miR-16-5p. mTOR, mechanistic target of rapamycin kinase. * P < 0.05.
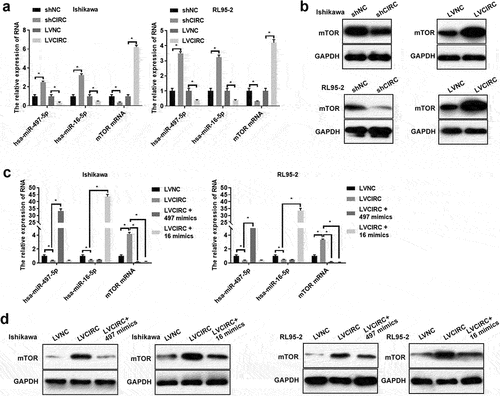
4. Hsa_circ_0011324 regulated mTOR expression by affecting hsa-miR-497/16-5p
Previous researches confirmed hsa-miR-497-5p mimics and hsa-miR-16-5p mimics decreased mTOR expression [Citation22–24]. Therefore, we further studied whether hsa_circ_0011324 regulated mTOR expression via hsa-miR-497/16-5p in EC progression. We firstly detected hsa-miR-497/16-5p and mTOR expression in cell lines Ishikawa and RL95-2 that stably overexpress or interfere with hsa_circ_0011324. showed overexpression of hsa_circ_0011324 inhibited hsa-miR-497/16-5p expression and promoted mTOR expression; while interference with hsa_circ_0011324 promoted hsa-miR-497/16-5p expression and inhibited mTOR expression. Next, we overexpressed hsa-miR-497/16-5p in cell lines Ishikawa and RL95-2 that stably overexpress hsa_circ_0011324. showed that the overexpression of hsa-miR-497/16-5p was successful. In addition, overexpression of hsa_circ_0011324 and then overexpression of hsa-miR-497/16-5p could inhibit the increase of mTOR expression caused by overexpression of hsa_circ_0011324 (). These results demonstrated that hsa_circ_0011324 regulated mTOR expression by affecting hsa-miR-497/16-5p.
Figure 4. Hsa_circ_0011324 regulated mTOR expression by affecting hsa-miR-497/16-5p. After stably overexpression or interference with hsa_circ_0011324 in Ishikawa and RL95-2 cells, (a). Quantitative real-time polymerase chain reaction determined hsa-miR-497/16-5p and mTOR mRNA expression. (b). Western blotting detected mTOR protein expression. After overexpression of hsa_circ_0011324 and then overexpression of hsa-miR-497/16-5p in Ishikawa and RL95-2 cells, (c). Quantitative real-time polymerase chain reaction evaluated hsa_circ_0011324, hsa-miR-497/16-5p, and mTOR mRNA expression. (d) Western blotting assessed mTOR protein expression. mTOR indicates mechanistic target of rapamycin kinase; shNC indicates the negative control of interference with hsa_circ_0011324; shCIRC indicates interference with hsa_circ_0011324; LVNC indicates the negative control of hsa_circ_0011324 overexpression; LVCIRC indicates hsa_circ_0011324 overexpression; LVCIRC+497 mimics indicates overexpression of hsa_circ_0011324 and hsa-miR-497-5p; LVCIRC+16 mimics indicates overexpression of hsa_circ_0011324 and hsa-miR-16-5p. * P < 0.05.
5. Hsa_circ_0011324 was involved in EC progression by affecting hsa-miR-497/16-5p
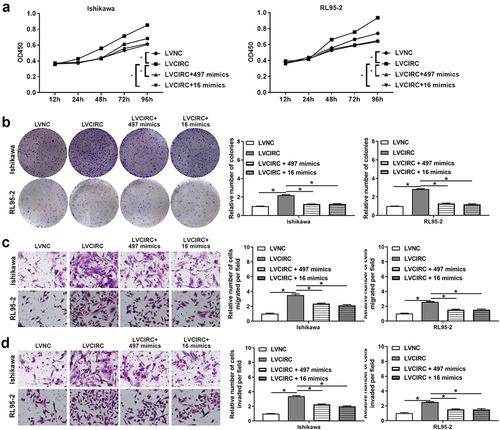
Previous studies showed hsa-miR-497-5p and hsa-miR-16-5p expression were down-regulated in EC [Citation25–27] . Therefore, we further investigated whether hsa_circ_0011324 was involved in EC progression by affecting hsa-miR-497/16-5p. After overexpression of hsa-miR-497/16-5p in Ishikawa and RL95-2 that stably overexpress hsa_circ_0011324, we performed cell function tests. showed hsa_circ_0011324 overexpression and then overexpression of hsa-miR-497/16-5p could inhibit the increase in cell proliferation and the number of cell clones caused by overexpression of hsa_circ_0011324. Besides, overexpression of hsa_circ_0011324 and then overexpression of hsa-miR-497/16-5p could inhibit hsa_circ_0011324 overexpression inducing enhance of cell migration and invasion (). These results showed hsa_circ_0011324 participated in EC progression by affecting hsa-miR-497/16-5p.
Figure 5. Hsa_circ_0011324 was involved in endometrial cancer progression by affecting hsa-miR-497/16-5p. After overexpression of hsa_circ_0011324 and then overexpression of hsa-miR-497/16-5p in Ishikawa and RL95-2 cells, (a). Cell Counting Kit-8 measured cell proliferation changes. (b). Clone formation assessed the number of cell clones. C and D. Transwell detected cell migration (c) and invasion (d) changes. LVNC indicates the negative control of hsa_circ_0011324 overexpression; LVCIRC indicates hsa_circ_0011324 overexpression; LVCIRC+497 mimics indicates overexpression of hsa_circ_0011324 and hsa-miR-497-5p; LVCIRC+16 mimics indicates overexpression of hsa_circ_0011324 and hsa-miR-16-5p. * P < 0.05.
6. Hsa_circ_0011324 competes with mTOR to directly bind to hsa-miR-497/16-5p
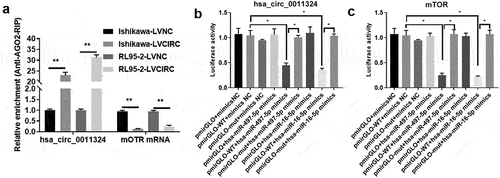
To confirm the relationship between hsa_circ_0011324, hsa-miR-497/16-5p, and mTOR, we performed AGO-RIP and Dual-luciferase assay. AGO2 can enrich hsa_circ_0011324 and mTOR, and overexpression of hsa_circ_0011324 decreased enrichment of mTOR on AGO2 (). Dual-luciferase results further proved hsa_circ_0011324 could directly sponge hsa-miR-497/16-5p (). At the same time, hsa-miR-497/16-5p could directly target mTOR mRNA (). These results illuminated that hsa_circ_0011324 competes with mTOR to directly bind to hsa-miR-497/16-5p.
Figure 6. Hsa_circ_0011324 competes with mTOR to directly bind to hsa-miR-497/16-5p. (a). Overexpression of hsa_circ_0011324 in Ishikawa and RL95-2 cells, then conducted AGO2-RIP (RNA immunoprecipitation) followed with quantitative real-time polymerase chain reaction detection of hsa_circ_0011324 and mTOR. (b) Dual-luciferase assessed the binding effect of hsa_circ_0011324 and hsa-miR-497/16-5p. (c). Dual-luciferase tested the binding effect of hsa-miR-497/16-5p and mTOR mRNA 3ʹUTR (untranslated regions). LVNC indicates the negative control of hsa_circ_0011324 overexpression; LVCIRC indicates hsa_circ_0011324 overexpression; WT indicates wild type; mut indicates mutant type; mimics NC indicates the negative control of mimics. * P < 0.05, ** P < 0.01.
Discussion
EC often occurs in postmenopausal women, and endometrium is a well-known site of cancer affecting women [Citation28]. Currently, EC treatment remains a challenge for gynecological and radiation oncologists [Citation29]. CircRNA plays an essential regulatory role in EC. This project used circBase to analyze the differential expression of 7 circRNAs from SPOCD1, and found hsa_circ_0011324 had significant differential expression. Further, we investigated the mechanism of hsa_circ_0011324 and its regulated downstream genes in EC. Our study illuminated hsa_circ_0011324 could sponge hsa-miR-497/16-5p targeted mTOR to participate in EC progress. This is the first report on the progress and mechanism of hsa_circ_0011324ʹs participation in EC.
With the enhancement of RNA sequencing methods, more and more circRNAs have been identified and their functions have been gradually revealed [Citation30]. In addition, several clinical trials are investigating the biomarkers and therapeutic effects of circRNAs. These efforts might provide a new perspective for gynecological tumors diagnosis and treatment. In addition, ceRNAs mechanism suggested circRNAs can regulate mRNA by competing with same pool miRNAs [Citation31]. In recent years, researchers have reported circRNA-ceRNA mechanism in EC. For example, Wang Y, et al. found circRNA hsa_circ_002577 acted as a miR-625-5p sponge to up-regulate insulin like growth factor 1 receptor and activate PI3K/Akt pathway, thereby accelerating EC progression [Citation32]. Zong ZH, et al. reported circ_PUM1 could compete with miR-136, leading to Notch receptor 3 upregulation, thereby promoting EC development [Citation33]. Liu Y, et al. also found circ_0067835 could compete with miR-324-5p, leading to high mobility group AT-Hook 1 upregulation and thus inducing EC [Citation34]. However, the molecular mechanism of hsa_circ_0011324 involved in EC is still unclear. In this study, we found that hsa_circ_0011324 was high expression in EC tissues, and the high expression of hsa_circ_0011324 was closely associated with poor OS/DFS. In addition, hsa_circ_0011324 overexpression promoted EC cell proliferation, migration, and invasion; while knockdown of hsa_circ_0011324 inhibited EC cell proliferation, migration, and invasion. These results indicated that hsa_circ_0011324 involved in EC progression.
Previous studies have shown that changes in hsa-miR-497-5p expression affected EC occurrence, and the low expression of miR-497-5p was related to high-risk EC [Citation25,Citation35]. In addition, hsa-miR-16-5p was down-regulated in EC and inhibited EC [Citation27]. These findings suggested that hsa-miR-497-5p and hsa-miR-16-5p might be used as good predictive and distinguishing factors for EC, and may be used as potential biomarkers. In addition, miR-497-5p can as a sponge of circSLC6A6 to regulate tumor-related signaling pathway PI4KB/hedgehog and participate EC progress [Citation36]. And long non-coding RNA SNHG25 sequestered miR-497-5p acted as ceRNA in EC, thereby positively regulating fatty acid synthase expression and promoting EC deterioration [Citation26]. In this study, we found hsa-miR-497-5p and hsa-miR-16-5p were low expression in EC, and their expression in EC tissues were negatively correlated with hsa_circ_0011324. These results were consistent with the previously reportion that low expression trend of hsa-miR-497-5p and hsa-miR-16-5p in EC tissues. Moreover, our study suggested hsa_circ_0011324 overexpression promoted cell functions (proliferation, migration, and invasion) via affected the expression of hsa-miR-497/16-5p.
PI3K/AKT/mTOR pathway is a classical pathway in cancer, and this pathway inhibitors are effective and safe in patients with locally advanced, metastatic or recurrent EC [Citation37,Citation38]. Such as Barra F, et al. reported PI3K/AKT/mTOR signaling inhibitors had great clinical value in EC [Citation39]. In this research, we found mTOR was highly expressed in EC. And hsa_circ_0011324 overexpression promoted mTOR expression by affected the expression of hsa-miR-497/16-5p. Furthermore, AGO2-RIP and dual-luciferase verified hsa_circ_0011324 competes with mTOR to directly bind to hsa-miR-497/16-5p. This is the first time we reported hsa_circ_0011324-hsa-miR-497/16-5p-mTOR axis participate in EC
However, the above mechanism was confirmed only in vivo cell experiments. More studies are needed in vitro to verify our conclusion. In the future studies, we will conduct tumorigenesis in nude mice to confirm whether hsa_circ_0011324 involve in EC progression by sponging with hsa-miR-497/16-5p to regulate mTOR; and more EC tissues will be collected to confirm whether hsa_circ_0011324 can be a predictor for EC diagnosis and treatment.
Conclusion
Hsa_circ_0011324 was high expression in EC, which indicated poor OS and DFS. Overexpression of hsa_circ_0011324 promoted the ability of proliferation, migration, and invasion in EC cells; while silence of hsa_circ_0011324 had opposite effect on cell functions. Furthermore, we reported for the first time that hsa_circ_0011324 could sponge hsa-miR-497/16-5p targeted mTOR to participate in EC progress.
Highlights
Hsa_circ_0011324 promoted EC progression.
Hsa-miR-497/16-5p were low expression, while mTOR was high expression in EC.
Hsa_circ_0011324 was involved in EC progression by affecting hsa-miR-497/16-5p.
4. Hsa_circ_0011324 competes with mTOR to directly bind to hsa-miR-497/16-5p.
Ethics statement
The studies involving human participants were reviewed and approved by this present study was approved by the Ethics Committee of The First Hospital of Lanzhou University (LDYYLL2021-99). The patients/participants provided their written informed consent to participate in this study.
Author contributions
YY: design of this study; grammar and language modification. LD and XB: acquisition of data; data analysis; drafting the article. All authors contributed to the article and approved the submitted version.
Supplemental Material
Download TIFF Image (3.2 MB)Acknowledgments
This work was supported by The Foundation of the First Hospital of Lanzhou University (no.2019-44)
Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary material
Supplemental data for this article can be accessed here
Additional information
Funding
References
- Braun MM, Overbeek-Wager EA, and Grumbo RJ. Diagnosis and management of endometrial cancer. Am Fam Physician. 2016;93(6):468–474.
- Siegel RL, Miller KD, Jemal A. Cancer statistics. CA Cancer J Clin. 2018;68(1):7–30.
- Chlebowski RT, Anderson GL, Sarto GE, et al. Continuous combined estrogen plus progestin and endometrial cancer: the women’s health initiative randomized trial. J Natl Cancer Inst. 2016;108(3):djv350.
- Zhang C, Wang EY, Liu F, et al. Routine histologic features in complex atypical hyperplasia can predict the presence of endometrial carcinoma: a clinicopathological study of 222 cases. Hum Pathol. 2018;80(40–46):40–46.
- Chapel DB, Patil SA, Plagov A, et al. Quantitative next-generation sequencing-based analysis indicates progressive accumulation of microsatellite instability between atypical hyperplasia/endometrial intraepithelial neoplasia and paired endometrioid endometrial carcinoma. Mod Pathol. 2019;32(1508–1520):1508–1520.
- Han L, Du J, Zhao L, et al. An efficacious endometrial sampler for screening endometrial cancer. Front Oncol. 2019;9(67). DOI:10.3389/fonc.2019.00067
- Siegel RL, Miller KD, Jemal A. Cancer statistics. CA Cancer J Clin. 2016;66(1):7–30.
- Zhou WY, Cai Z-R, Liu J, et al. Circular RNA: metabolism, functions and interactions with proteins. Mol Cancer. 2020;19(172). DOI:10.1186/s12943-020-01286-3
- Shi Y, Jia X, Xu J. The new function of circRNA: translation. Clin Transl Oncol. 2020;22(2162–2169):2162–2169.
- Qu S, Yang X, Li X, et al. Circular RNA: a new star of noncoding RNAs. Cancer Lett. 2015;365(141–148):141–148.
- Salzman J. Circular RNA expression: its potential regulation and function. Trends Genet. 2016;32(309–316):309–316.
- Huang A, Zheng H, Wu Z, et al. Circular RNA-protein interactions: functions, mechanisms, and identification. Theranostics. 2020;10(3503–3517):3503–3517.
- Jia Y, Liu M, Wang S. CircRNA hsa_circRNA_0001776 inhibits proliferation and promotes apoptosis in endometrial cancer via downregulating LRIG2 by sponging miR-182. Cancer Cell Int. 2020;20(412). DOI:10.1186/s12935-020-01437-y
- Wu B, Ren A, Tian Y, et al. Hsa_circ_0075960 serves as a sponge for miR-361-3p/SH2B1 in endometrial carcinoma. Technol Cancer Res Treat. 2020;19(1533033820983079):153303382098307.
- Liu D, Yang Y, Yan A, et al. SPOCD1 accelerates ovarian cancer progression and inhibits cell apoptosis via the PI3K/AKT pathway. Onco Targets Ther. 2020;13(351–359). DOI:10.2147/OTT.S200317
- Long MY, Chen, JW, and Zhu, Y, et al. Comprehensive circular RNA profiling reveals the regulatory role of circRNA_0007694 in papillary thyroid carcinoma. Am J Transl Res. 2020;12(4):1362–1378.
- Xu L, Feng X, Hao X, et al. CircSETD3 (Hsa_circ_0000567) acts as a sponge for microRNA-421 inhibiting hepatocellular carcinoma growth. J Exp Clin Cancer Res. 2019;38(98). DOI:10.1186/s13046-019-1041-2
- Young AP, Jackson DJ, Wyeth RC. A technical review and guide to RNA fluorescence in situ hybridization. PeerJ. 2020;8:e8806.
- Lu H, Xie X, Wang K, et al. Circular RNA hsa_circ_0096157 contributes to cisplatin resistance by proliferation, cell cycle progression, and suppressing apoptosis of non-small-cell lung carcinoma cells. Molecular and Cellular Biochemistry. 2020;475(1–2):63–77.
- Wang H, Wang N, Zheng X, et al. Circular RNA hsa_circ_0009172 suppresses gastric cancer by regulation of microRNA-485-3p-mediated NTRK3. Cancer Gene Therapy. 2021;28(12):1312–1324.
- Zhang D, Yi, S, and Cai, B, et al. Involvement of ferroptosis in the granulosa cells proliferation of PCOS through the circRHBG/miR-515/SLC7A11 axis. Ann Transl Med. 2021;9(16):1348. DOI:10.21037/atm-21-4174
- Xu S, Fu G-B, Tao Z, et al. MiR-497 decreases cisplatin resistance in ovarian cancer cells by targeting mTOR/P70S6K1. Oncotarget. 2015;6(26457–26471):26457–26471.
- Cheng H, Xue, J, and Yang, S, et al. Co-targeting of IGF1R/mTOR pathway by miR-497 and miR-99a impairs hepatocellular carcinoma development. Oncotarget. 2017;8(29):47984–47997. DOI:10.18632/oncotarget.18207
- Lee DE, Brown, JL, and Rosa, ME, et al. microRNA-16 is downregulated during insulin resistance and controls skeletal muscle protein accretion. J Cell Biochem. 2016;117(8):1775–1787.
- Kalinkova L, Kajo K, Karhanek M, et al. Discriminating miRNA profiles between endometrioid well- and poorly-differentiated tumours and endometrioid and serous subtypes of endometrial cancers. Int J Mol Sci. 2020;21(17):6071.
- He Y, Xu S, Qi Y, et al. Long noncoding RNA SNHG25 promotes the malignancy of endometrial cancer by sponging microRNA-497-5p and increasing FASN expression. J Ovarian Res. 2021;14(163). DOI:10.1186/s13048-021-00906-w
- Kumari P, Sharma I, Saha SC, et al. Diagnostic potential of differentially regulated microRNAs among endometriosis, endometrioid ovarian cancer, and endometrial cancer. J Cancer Res Ther. 2021;17(1003–1011):1003–1011.
- Aoki Y, Kanao H, Wang X, et al. Adjuvant treatment of endometrial cancer today. Jpn J Clin Oncol. 2020;50(753–765):753–765.
- Barcellini A, Roccio M, Laliscia C, et al. Endometrial cancer: when upfront surgery is not an option. Oncology. 2021;99(65–71):65–71.
- Shi Y, He, R, and Yang, Y, et al. Circular RNAs: novel biomarkers for cervical, ovarian and endometrial cancer (Review). Oncol Rep. 2020;44(5):1787–1798. DOI:10.3892/or.2020.7780
- Cheng DL, Xiang YY, Ji LJ, et al. Competing endogenous RNA interplay in cancer: mechanism, methodology, and perspectives. Tumour Biol. 2015;36(2):479–488.
- Wang Y, Yin L, Sun X. CircRNA hsa_circ_0002577 accelerates endometrial cancer progression through activating IGF1R/PI3K/Akt pathway. J Exp Clin Cancer Res. 2020;39(169). DOI:10.1186/s13046-020-01679-8
- Zong ZH, Liu Y, Chen S, et al. Circ_PUM1 promotes the development of endometrial cancer by targeting the miR-136/NOTCH3 pathway. J Cell Mol Med. 2020;24(4127–4135):4127–4135.
- Liu Y, Chang Y, Cai Y. Circ_0067835 sponges miR-324-5p to induce HMGA1 expression in endometrial carcinoma cells. J Cell Mol Med. 2020;24(13927–13937):13927–13937.
- Fridrichova I, Kalinkova L, Karhanek M, et al. miR-497-5p decreased expression associated with high-risk endometrial cancer. Int J Mol Sci. 2020;22(1):127.
- Hu J, Peng X, Du W, et al. circSLC6A6 sponges miR-497-5p to promote endometrial cancer progression via the PI4KB/Hedgehog axis. J Immunol Res. 2021;2021(5512391):1–13.
- Roncolato F, Lindemann K, Willson ML, et al. PI3K/AKT/mTOR inhibitors for advanced or recurrent endometrial cancer. Cochrane Database Syst Rev. 2019;10(CD012160). DOI:10.1002/14651858.CD012160.pub2
- Zhou X, Chen Z, Pei L, et al. MicroRNA miR-106a-5p targets forkhead box transcription factor FOXC1 to suppress the cell proliferation, migration, and invasion of ectopic endometrial stromal cells via the PI3K/Akt/mTOR signaling pathway. Bioengineered. 2021;12(2203–2213). DOI:10.1080/21655979.2021.1933679
- Barra F, Evangelisti, G, and Ferro Desideri, L, et al. Investigational PI3K/AKT/mTOR inhibitors in development for endometrial cancer. Expert Opin Investig Drugs. 2019;28(2):131–142.
