ABSTRACT
Colorectal cancer (CRC) is a common malignancy of the gastrointestinal tract. CircRNAs have been reported to play regulatory roles in many cancers, including CRC. This study focuses on the role of circ_0007331 in CRC. Differentially expressed circRNAs in CRC were screened using the GEO database. RT-qPCR was used to analyze mRNA expression. StarBase and TargetScan were used to predict targeting relationships and then verified by the dual luciferase reporter assay along with the RNA pull-down assay. CCK-8 as well and transwell assays were used to measure cell viability, migration, and invasion. Protein levels were determined using western blotting. circ_0007331 is expressed more frequently in patients with CRC. The inhibition of circ_0007331 expression reduced the viability, colony formation, migration, and invasion of CRC cells. However, inhibition of miR-205-5p or elevation of high-mobility group A2 (HMGA2) can reverse the function of inhibited circ_0007331 in tumor cells. This study demonstrated that the circ_0007331/miR-205-5p/HMGA2 axis promotes CRC development. Thus, circ_0007331 may be a potential biomarker for CRC.
Graphical Abstract
1. Introduction
Colorectal cancer (CRC) is a common gastrointestinal malignancy. According to statistics, CRC accounts for approximately 10% of all cancer-related deaths annually [Citation1]. The incidence of CRC in males and females accounts for 9% and 8% of all cancers in men and women, respectively [Citation2]. CRC is either asymptomatic or asymptomatic in its early stages, making it difficult to detect. The 5-year survival rate of patients with early diagnosis and surgery can be 90%, whereas that of patients with advanced CRC with metastasis is lower than 10% [Citation3,Citation4]. Hence, CRC is a serious threat to human health and requires further investigation.
Circular RNA (circRNA) is a type of special non-coding RNA that is transcribed by RNA polymerase II to form a covalently closed single-stranded circular RNA molecule without a 5’ cap and 3’ poly (A) tail [Citation5]. CircRNAs are widely expressed in human tissues and organs, and their expression levels can reach more than 10 times the corresponding mRNA [Citation6]. In addition, its circular structure makes it more stable and conserved than linear non-coding RNAs [Citation7,Citation8]. Moreover, circRNAs exhibit specific expression patterns in different cells [Citation9]. These special features allow circRNAs to modulate gene expression at the transcriptional and post-transcriptional levels [Citation10]. Chen et al. [Citation11] identified 10,245 differentially expressed circRNAs in colorectal cancer expression profiles, among which 6,264 were upregulated. In addition, hsa_circ_001569 was found to increase the proliferation and invasion of tumor cells in different CRC cell lines [Citation12]. circ_BANP can promote the proliferation of colorectal tumor cells through the PI3K/AKT pathway [Citation13], indicating that circRNAs play an important regulatory role in CRC. circ_0007331 is a circular RNA located on chromosome 3, which is reported to be related to colon cancer [Citation14], but its mechanism in CRC remains to be further clarified.
The high-mobility group (HMG) is a class of small non-histone proteins abundant in the nucleus, among which HMGA2 is highly expressed in malignant tumors [Citation15]. The HMGA2 gene, 200 kb in length, is located at 12q13-15 [Citation16] and is widely expressed during embryonic development, but is expressed at low levels in adult tissues [Citation17]. HMGA2 was reported to take part in the regulation of tumor development. For instance, lncRNA HIT000218960 promotes the proliferation and migration of gastric cancer cells by overexpressing HMGA2 [Citation18], and miRNA let-7 affects the progression of esophageal squamous cell carcinoma by negatively regulating HMGA2 [Citation19]. HMGA2 is highly expressed [Citation20]. Silencing HMGA2 can cause tumor cell apoptosis by arresting cell growth in the G2/M phase [Citation21].
Therefore, this study aimed to explore the function and regulation of circ_0007331 in CRC cells. We hypothesized that circ_0007331 regulating the proliferation, migration and invasion via targeting the miR-205-5p/HMGA2 axis.
2. Materials and methods
2.1 Bioinformatic analysis
A microarray profile GPL19978 related to CRC was downloaded from the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo). All differentially expressed genes (DEGs) of osteoporotic group (n = 3) and non-osteoporotic group (n = 3) were identified and then screened under the standard od P < 0.05 and |log2FC| ≥ 1.5. Additionally, the target miRNA of circ_0007331 was predicated in Starbase online database (http://starbase.sysu.edu.cn/). And the target gene of miR-205-5p was predicated by targetscan online database (http://www.targetscan.org/vert_71/).
2.2 Patients
CRC samples and adjacent non-tumor tissue samples were collected from CRC patients (n = 33) at Yuebei People’s Hospital from 1 January 2019, to 1 November 2020.
The study was approved by the Yuebei People’s Hospital. All the participants signed an informed consent form.
2.3 Cell culture
HCT 8 as well as SW620 cells (ATCC, Manassas, VA) were cultivated in DMEM (Thermo Fisher Scientific, USA). 10% FBS (Gibco, Waltham) and 1% penicillin/streptomycin (Gibco, Waltham) were added to the cell culture medium.
2.4 RT-qPCR
According to a previous study [Citation22], RNA was extracted from HCT 8 and SW620 cells using a commercially available kit (Takara, Japan). Then, cDNA was synthesized, and PCR was performed using a real-time PCR detection system (Bio-Rad, USA). The primer sequences used were as follows.
circ_0007331: F: 5ʹ-GAATGGGATTCGAGACCTG-3ʹ, R: 5ʹ-TTCTTCCAAAGCTGCCTGT-3ʹ
miR-205-5p: F: 5’-TCCTTCATTCCACCGGAGTCTG-3’, R: 5’-GCG AGCACAGAATTAATACGAC-3’;
HMGA2: F: 5’-AGA UUGAGAUUGAAAGUGCCU-3’, R: 5’-GCA CUUUCAAUCUCAAUCUCU-3’
GAPDH: F: 5’-GAGTCCACTGGCGTCTTCAC-3’, R: 5’-ATCTT GAGGCTGTTGTCATACTTCT-3’.
2.5 Cell viability assay
According to a previous study [Citation23], after resuspending the cells, they were inoculated into 96-well plates at 100 μl/well. Added to each well was 10 μl CCK8 reagent (AmyJet Technology Co., Ltd.), and it was cultured for 4 h at 37°C. A microplate reader (Nanjing DeTie Experimental Equipment Co., Ltd.) was used to measure the absorbance at 450 nm.
2.6 Colony formation assay
According to a previous study [Citation24], HCT 8 and SW620 cells were inoculated into 6-well plates and cultured for 7 days with the culture medium refreshed every 2 days. Thereafter, HCT 8 as well as SW620 cells were stained by crystal violet (0.1%) for ten minutes. The colonies were observed under a microscope (Nikon, Tokyo, Japan).
2.7 Transwell assay
According to a previous study [Citation25], 1 × 10 6 of HCT 8 as well as SW620 cells were seeded into a 24-well upper chamber precoated with Matrigel (Corning Inc., USA). After 48 h, the upper chamber cells were removed, and the invading cells were fixed with methanol and stained with crystal violet. The cells were then examined under a microscope.
2.8 Dual luciferase reporter assay
According to a previous study [Citation26], the binding sites of circ_0007331 and miR-205-5p were predicted using StarBase 3.0. After cultivation for 24 h, cells were lysed. A luciferase reporter assay kit (BioVision Tech Co., Ltd.) was used to analyze luciferase activity 48 h after co-transfection with miR-205-5p mimic/control as well as luciferase reporter vectors. Luciferase activity was determined using a commercial kit (Promega).
2.9 RNA pull-down
According to a previous study [Citation27], the MagCapture RNA Pull Down Assay Kit (Whatman Co., Ltd.) was used for the RNA pull-down assay, according to the manufacturer’s protocols. The proteins were collected for mass spectrometry analysis.
2.10 Western blot
According to a previous study [Citation28], protein extracts were loaded onto 10% SDS gel electrophoresis. Then The protein extracts were then transferred to a PVDF membrane (Millipore) and incubated with primary antibodies overnight at 4°C. One day later, the membranes were incubated with secondary antibodies at room temperature for two hours. Finally, the bands were captured using an enhanced chemiluminescence system (Thermo Fisher Scientific, Inc.).
2.11 Xenograft model assay
According to a previous study [Citation29], twelve BALB/c nude mice (male, 4-week-old) were provided by Guangdong Medical Laboratory Animal Center (Guangdong, China). The animal experiment was under the approval of Yuebei People’s Hospital and met the demands of laboratory animal welfare and ethics. SW620 cells transfected with sh-nc or sh-circ_0007331 (2 × 106) were injected into the back of mice and allowed to grow for 42 days to establish the Xenograft model. Finally, Xenograft volume and weight were examined.
2.12 Statistical analysis
Statistical analyses were performed using GraphPad Prism 7.0 (Graph-Pad Software, USA). Data are presented as mean ± standard deviation (SD). Two groups were analyzed using the Student’s t-test. The contrast between the two groups was analyzed by analysis of variance. Statistical significance was set at p < 0.05.
3. Results
Circ_0007331 expression was increased in CRC. The inhibition of circ_0007331 decreased the tumor cell viability, clone formation, migration, and invasion through the miR-205-5p/HMGA2 axis. Additionally, the inhibition of circ_0007331 suppressed the development of CRC in vivo.
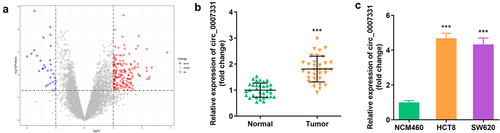
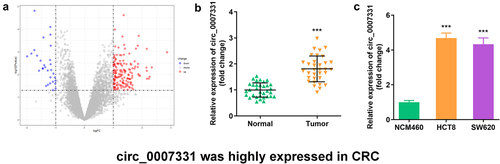
3.1. circ_0007331 was highly expressed in CRC
We analyzed the microarray data of GPL19978 from the GEO database to select circRNAs related to CRC, and 156 circRNAs were significantly upregulated in CRC tissues, while 30 circRNAs were notably downregulated (). In addition, circ_0007331 was expressed more in CRC tissues, HCT8 cells, and SW620 cells ().
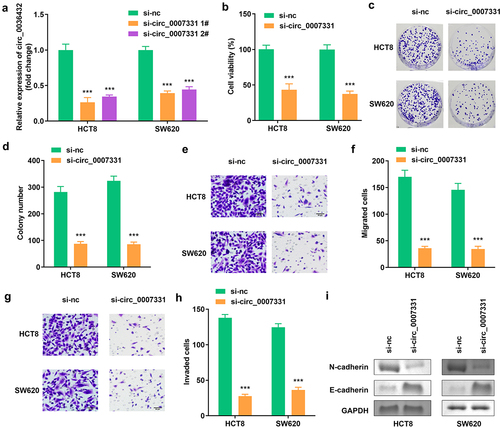
3.2 Knockdown of circ_0007331 reduced cell viability, colony formation, migration along with invasion of HCT8 and SW620 cells
The expression of circ_0007331 was prominently decreased compared to the blank vector, indicating the success of transfection, and 1# was used in the following experiments (). Knockdown of circ_0007331 observably reduced the viability and colony formation of HCT8 and SW620 cells (). Moreover, the migration and invasion of HCT8 and SW620 cells were reduced after circ_0007331 knockdown (), while the protein expression of N-cadherin decreased, whereas that of E-cadherin increased. ().
Figure 2. Circ_0007331 knockdown declined aggressiveness of CRC cells. (a) Expression of circ_0007331 in HCT8 as well as SW620 cells. (b) Cell viability of HCT8 as well as SW620 cells. (c, d) Clone formation of HCT8 as well as SW620 cells. (e, f) The migration of CRC cells. (g, h) The invasion of CRC cells. (i) The protein expression of N-cadherin and E-cadherin. *** P < 0.001 versus si-nc.
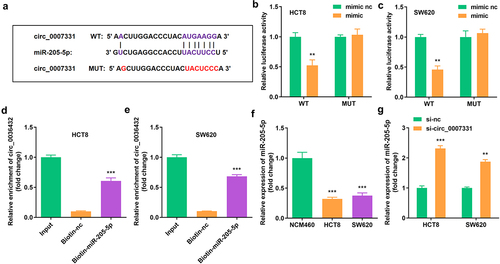
3.3. circ_0007331 directly targeted miR-205-5p
The binding region of miR-205-5p and circ_0007331 was obtained in Starbase online database (http://starbase.sysu.edu.cn/) (), which was verified using a dual luciferase reporter assay along with an RNA pull-down assay (). miR-205-5p expression was markedly suppressed in HCT8 and SW620 cells (), whereas circ_0007331 knockdown notably upregulated the expression of miR-205-5p in HCT8 and SW620 cells ().
Figure 3. Circ_0007331 directly targeted miR-205-5p (a) The binding sites between circ_0007331 and miR-205-5p. (b, c) The luciferase activity of HCT8 and SW620 cells. (d, e) The interaction between circ_0007331 and miR-205-5p. (f) miR-205-5p expression in HCT8 and SW620 cells. (g) miR-205-5p expression in cells with circ_0007331 knockdown. **P < 0.01, *** P < 0.001 versus control.
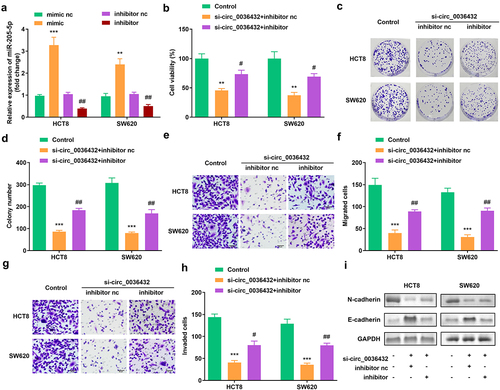
3.4 Inhibition of miR-205-5p abrogated the effects of depletion of circ_0007331 in HCT8 and SW620 cells
shows that, compared with the blank vector, miR-205-5p expression was increased by the mimic and downregulated by the inhibitor. miR-205-5p inhibition remarkably improved the viability and colony formation of HCT8 and SW620 cells, which reversed the decrease caused by circ_0007331 knockdown (). In addition, the depletion of miR-205-5p enhanced the migration and invasion of HCT8 and SW620 cells (). In addition, the protein expression of N-cadherin and E-cadherin was reversed after the inhibition of miR-205-5p().
Figure 4. miR-205-5p inhibition reversed the effects of circ_0007331 knockdown. (a) Expression of circ_0007331 in HCT8 as well as SW620 cells. (b) Cell viability of HCT8 as well as SW620 cells. (c, d) Clone formation of HCT8 as well as SW620 cells. (e, f) The migration of CRC cells. (g, h) The invasion of CRC cells. (i) The protein expression of N-cadherin and E-cadherin. #P < 0.05, ##P < 0.01, **P < 0.01, *** P < 0.001 versus control.
3.5. miR-205-5p directly targeted HMGA2
To determine the regulatory axis, including circ_0007331 and miR-205-5p, TargetScan (http://www.targetscan.org/vert_71/) was used to predict miR-205-5p’s target gene; HMGA2 was found to be the regulatory axis (). Dual luciferase reporter assay and RNA pull-down assay were performed to confirm the targeting interaction of HMGA2 and miR-205-5p (). showed that HMGA2 was expressed highly in HCT8 and SW620 cells. Moreover, high miR-205-5p expression decreased HMGA2 expression in HCT8 and SW620 cells ().
Figure 5. miR-205-5p directly targeted HMGA2 (a) The binding sites between HMGA2 and miR-205-5p. (b, c) The luciferase activity of HCT8 and SW620 cells. (d, e) The interaction between HMGA2 as well as miR-205-5p. (f) Expression of HMGA2 in HCT8 and SW620 cells. (g) Expression of HMGA2 in cells with miR-205-5p overexpression. **P < 0.01, *** P < 0.001 versus control.
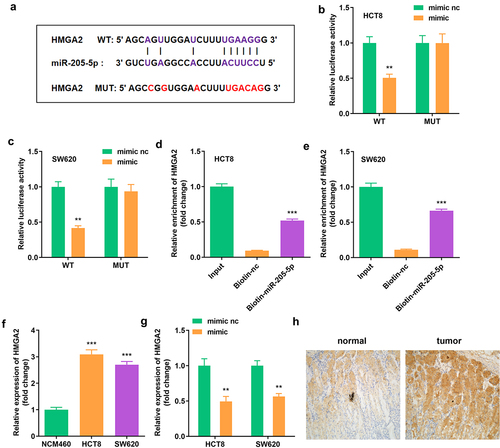
Figure 6. Elevated HMGA2 neutralized the function of miR-205-5p on cell viability, colony formation, migration as well as invasion (a) HMGA2 expression in HCT8 as well as SW620 cells. (b) Cell viability of HCT8 as well as SW620 cells. (c, d) Clone formation of HCT8 as well as SW620 cells. (e, f) The migration of CRC cells. (g, h) The invasion of CRC cells. (i) The protein expression of N-cadherin and E-cadherin. #P < 0.05, ##P < 0.01, **P < 0.01, *** P < 0.001 versus control.
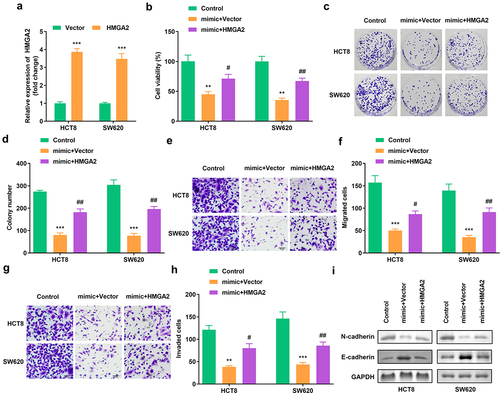
3.6 Elevated HMGA2 neutralized the function of miR-205-5p on cell viability, colony formation, migration as well as invasion
A notable increase in HMGA2 expression resulted in a successful transfection (). HMGA2 overexpression rescued cell viability and colony formation (). The migration and invasion of CRC cells was also enhanced after HMGA2 overexpression (). Furthermore, the protein expression of N-cadherin was increased, whereas that of E-cadherin was decreased. ().
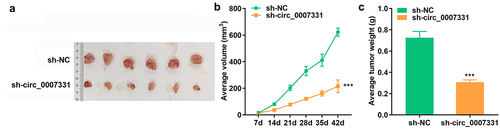
3.7 Depletion of circ_0007331 suppressed tumor growth in vivo
To further confirm the role of circ_0007331 in CRC, the xenograft tumor model was established and the tumor volume and weight were analyzed. The results showed that the volume () and weight () of tumors in sh-circ_0007331 group were significantly decreased compared with control group.
4. Discussion
CRC is a common malignant digestive tract tumor with high morbidity as well as mortality [Citation30], and its incidence has increased rapidly in recent years [Citation31,Citation32]. This study reveals that the dysregulated circ_0007331 is connected with the occurrence and development of CRC. circ_0007331 knockdown reduced the activity and proliferation of tumor cells as well as migration and invasion through the circ_0007331/miR-205-5p/HMGA2 axis. To our knowledge, this is the first study to investigate the regulatory role of circ_0007331 in CRC development.
Circular RNAs are involved in various malignant tumors by inducing angiogenesis, and promoting tumor immune escape and the formation of an inflammatory microenvironment [Citation33]. For example, circ_0078710 regulates miR-31 to accelerate the proliferation and migration of liver cancer cells [Citation34], while circ_0014717 affects the tumor cell cycle and suppresses the development of CRC by upregulating the expression of P16 protein [Citation35]. In this study, circ_0007331 levels increased in CRC. The activity, proliferation, migration, and invasion abilities of tumor cells were weakened after the knockdown of circ_0007331, suggesting that circ_0007331 plays a regulatory role in CRC.
Given that circRNAs can act as ‘ sponges ‘ to regulate miRNAs, we predicted that circ_0007331 targets miR-250-5p. It has been reported that miR-250-5p is a regulatory factor in various cancers, such as targeting ZEB1 to inhibit the aggressiveness of prostate cancer cells [Citation36], promoting the development of colorectal cancer by targeting VEGFA [Citation37], and suppressing the vitality and invasion of ovarian cancer cells by promoting apoptosis [Citation38]. Here, miR-250-5p was less expressed in CRC but increased in tumor cells with circ_0007331 knockdown. In addition, inhibition of miR-250-5p can rescue the effect of circ_0007331 knockdown on cells, indicating that miR-250-5p is an important part of the circ_0007331 regulatory axis.
HMGA2 is a target of miR-250-5p. Studies have shown that high HMGA2 expression is linked to the occurrence, development, and prognosis of malignant tumors of the digestive tract [Citation39], such as enhancing the invasion of gastric cancer by promoting epithelial-mesenchymal transition [Citation40] and increasing the proliferation and differentiation of esophageal cancer cells [Citation41]. This study also verified that increased HMGA2 expression enhances the aggressive behavior of CRC cells by modulating circ_0007331.
5. Conclusion
In conclusion, circ_0007331 expression is increased in CRC. The inhibition of circ_0007331 increased tumor cell viability, clone formation, migration, and invasion through the miR-205-5p/HMGA2 axis. These findings may provide potential therapeutic targets for CRC treatment.
Ethical approval
This study protocol was approved by the Ethics Committee of Yue bei People’s Hospital 201,900,220 on February 20th in 2019.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
Not applicable.
Additional information
Funding
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424.
- Siegel RL, Miller KD, Fuchs HE, et al. Cancer statistics, 2021. CA Cancer J Clin. 2021;71(1):7–33.
- Dahan L, Norguet E, Etienne-Grimaldi MC, et al. Pharmacogenetic profiling and cetuximab outcome in patients with advanced colorectal cancer. BMC Cancer. 2011;11(1):496.
- Courtney RJ, Paul CL, Carey ML, et al. A population-based cross-sectional study of colorectal cancer screening practices of first-degree relatives of colorectal cancer patients. BMC Cancer. 2013;13(1):13.
- Ashwal-Fluss R, Meyer M, Pamudurti NR, et al. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56(1):55–66.
- Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19(2):141–157.
- Li X, Yang L, Chen LL. The biogenesis, functions, and challenges of circular RNAs. Mol Cell. 2018;71(3):428–442.
- Dong R, Ma XK, Chen LL, et al. Increased complexity of circRNA expression during species evolution. RNA Biol. 2017;14(8):1064–1074.
- Nicolet BP, Engels S, Aglialoro F, et al. Circular RNA expression in human hematopoietic cells is widespread and cell-type specific. Nucleic Acids Res. 2018;46(16):8168–8180.
- Xu T, Wang M, Jiang L, et al. CircRNAs in anticancer drug resistance: recent advances and future potential. Mol Cancer. 2020;19(1):127.
- Chen S, Zhang L, Su Y, et al. Screening potential biomarkers for colorectal cancer based on circular RNA chips. Oncol Rep. 2018;39(6):2499–2512.
- Xie H, Ren X, Xin S, et al. Emerging roles of circRNA_001569 targeting miR-145 in the proliferation and invasion of colorectal cancer. Oncotarget. 2016;7(18):26680–26691.
- Zhu M, Xu Y, Chen Y, et al. Circular BANP, an upregulated circular RNA that modulates cell proliferation in colorectal cancer. Biomed Pharmacother. 2017;88:138–144.
- Han Z, Chen H, Guo Z, et al. Bioinformatics analysis: the regulatory network of hsa_circ_0007843 and hsa_circ_0007331 in colon cancer. Biomed Res Int. 2021;2021:6662897.
- Sgarra R, Lee J, Tessari MA, et al. The AT-hook of the chromatin architectural transcription factor high mobility group A1a is arginine-methylated by protein arginine methyltransferase 6. J Biol Chem. 2006;281(7):3764–3772.
- Ozturk N, Singh I, Mehta A, et al. HMGA proteins as modulators of chromatin structure during transcriptional activation. Front Cell Dev Biol. 2014;2:5.
- Ashar HR, Chouinard RA Jr., Dokur M, et al. In vivo modulation of HMGA2 expression. Biochim Biophys Acta. 2010;1799(1–2):55–61.
- Sun L, Yu J, Wang P, et al. HIT000218960 promotes gastric cancer cell proliferation and migration through upregulation of HMGA2 expression. Oncol Lett. 2019;17(6):4957–4963.
- Liu Q, Lv GD, Qin X, et al. Role of microRNA let-7 and effect to HMGA2 in esophageal squamous cell carcinoma. Mol Biol Rep. 2012;39(2):1239–1246.
- Sahengbieke S, Wang J, Li X, et al. Circulating cell-free high mobility group AT-hook 2 mRNA as a detection marker in the serum of colorectal cancer patients. J Clin Lab Anal. 2018;32(4):e22332.
- Esmailzadeh S, Mansoori B, Mohammadi A, et al. siRNA-mediated silencing of HMGA2 induces apoptosis and cell cycle arrest in human colorectal carcinoma. J Gastrointest Cancer. 2017;48(2):156–163.
- Chen S, Wei Y, Liu H, et al. Analysis of Collagen type X alpha 1 (COL10A1) expression and prognostic significance in gastric cancer based on bioinformatics. Bioengineered. 2021;12(1):127–137.
- Zhang J, Zhang Y, Li L, et al. Pregnancy-associated plasma protein-A (PAPPA) promotes breast cancer progression. Bioengineered. 2022;13(1):291–307.
- Geng X, Sun Y, Fu J, et al. MicroRNA-17-5p inhibits thyroid cancer progression by suppressing early growth response 2 (EGR2). Bioengineered. 2021;12(1):2713–2722.
- Shan Y, Li Y, Han H, et al. Insulin reverses choriocarcinoma 5- fluorouracil resistance. Bioengineered. 2021;12(1):2087–2094.
- Wang L, Hou S, Li J, et al. Circular RNA circ-LARP1B contributes to cutaneous squamous cell carcinoma progression by targeting microRNA-515-5p/TPX2 microtubule nucleation factor axis. Bioengineered. 2022;13(1):1209–1223.
- Cheng C, Zhang H, Dai Z, et al. Circular RNA circVRK1 suppresses the proliferation, migration and invasion of osteosarcoma cells by regulating zinc finger protein ZNF652 expression via microRNA miR-337-3p. Bioengineered. 2021;12(1):5411–5427.
- Taylor SC, Posch A. The design of a quantitative western blot experiment. Biomed Res Int. 2014;2014:361590.
- Porru M, Pompili L, Caruso C, et al. Xenograft as in vivo experimental model. Methods Mol Biol. 2018;1692:97–105.
- Cancer Genome Atlas N. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337.
- Zhang YL, Zhang ZS, Wu BP, et al. Early diagnosis for colorectal cancer in China. World J Gastroenterol. 2002;8(1):21–25.
- Jemal A, Bray F, Center MM, et al. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90.
- Bach DH, Lee SK, Sood AK. Circular RNAs in cancer. Mol Ther Nucleic Acids. 2019;16:118–129.
- Xie B, Zhao Z, Liu Q, et al. CircRNA has_circ_0078710 acts as the sponge of microRNA-31 involved in hepatocellular carcinoma progression. Gene. 2019;683:253–261.
- Wang F, Wang J, Cao X, et al. Hsa_circ_0014717 is downregulated in colorectal cancer and inhibits tumor growth by promoting p16 expression. Biomed Pharmacother. 2018;98:775–782.
- Li L, Li S. miR-205-5p inhibits cell migration and invasion in prostatic carcinoma by targeting ZEB1. Oncol Lett. 2018;16(2):1715–1721.
- Liu H, Li A, Sun Z, et al. Long non-coding RNA NEAT1 promotes colorectal cancer progression by regulating miR-205-5p/VEGFA axis. Hum Cell. 2020;33(2):386–396.
- Tao P, Yang B, Zhang H, et al. The overexpression of lncRNA MEG3 inhibits cell viability and invasion and promotes apoptosis in ovarian cancer by sponging miR-205-5p. Int J Clin Exp Pathol. 2020;13(5):869–879.
- Wu J, Liu Z, Shao C, et al. HMGA2 overexpression-induced ovarian surface epithelial transformation is mediated through regulation of EMT genes. Cancer Res. 2011;71(2):349–359.
- Sun J, Sun B, Zhu D, et al. HMGA2 regulates CD44 expression to promote gastric cancer cell motility and sphere formation. Am J Cancer Res. 2017;7(2):260–274.
- Mei LL, Wang WJ, Qiu YT, et al. miR-125b-5p functions as a tumor suppressor gene partially by regulating HMGA2 in esophageal squamous cell carcinoma. PLoS One. 2017;12(10):e0185636.