ABSTRACT
As an endocrine and metabolic disorder, polycystic ovarian syndrome (PCOS) is common in females at childbearing age. Our work was intended to uncover the underlying role of LINC00173 and its potential regulatory mechanism in PCOS based on two cell lines (PCOS granulosa cells and KGN cells) and an in vivo model established from Sprague Dawley rats. It was revealed that LINC00173 and JAG1 expressions were upregulated, while miR-124-3p was poorly expressed in PCOS patients and PCOS rats. Functional assays showed that LINC00173 overexpression repressed proliferation and stimulated apoptosis in granulosa cells and KGN cells, while LINC00173 downregulation exhibited the opposite effects. Besides, it was verified that LINC00173 upregulated JAG1 expression in KGN cells via competitively binding to miR-124-3p. Similarly, miR-124-3p abundance was inversely related to LINC00173 and JAG1 level in PCOS. Subsequently, rescue assays elucidated that miR-124-3p upregulation or downregulation eliminated the effects on KGN cell proliferation and apoptosis mediated by LINC00173 overexpression or knockdown. In addition, it was found that the JAG1 level in KGN cells was adversely modulated by miR-124-3p and positively modulated by LINC00173. Moreover, it was further demonstrated that the reduced cell vitality and increased apoptosis of KGN cells induced by overexpressing LINC00173 could be relieved by JAG1 deletion. These findings suggested that LINC00173 could be a latent regulating factor for PCOS progression via modulating the miR-124-3p/JAG1 cascade.
GRAPHICAL ABSTRACT
Introduction
As a common endocrine and metabolic disorder in women at childbearing age [Citation1], polycystic ovarian syndrome (PCOS) is characterized by irregular menstruation, chronic anovulation, and hyperandrogenism [Citation2] and may result in hirsutism, infertility, and even menstrual disturbances [Citation3]. Given the increasing morbidity of PCOS in recent years, PCOS has become a threat to reproductive health worldwide [Citation4]. As PCOS is of complex pathological physiology, obscure etiology, and varied clinical manifestations, PCOS diagnosis remains challenging in clinical endocrinology [Citation5]. Regardless of achievements made in PCOS treatment, most PCOS patients still need life-long treatment [Citation6]. Therefore, the molecular mechanism of PCOS pathogenesis should be studied in depth for the development of novel diagnostic and therapeutic methods for PCOS.
The granulosa cell layer, as an important component of the ovarian follicle, surrounds the oocyte [Citation7]. During the oocyte developmental process, the granulosa cell layer offers nutrients for the oocyte and transfers gene regulators to it [Citation8]. Follicular atresia is a vital process in ovary in follicle degeneration [Citation7]. Besides, granulosa cell (GC) apoptosis is a major cause of follicular atresia [Citation9]. In PCOS cases, abnormal GC proliferation and apoptosis were observed in folliculogenesis [Citation10], indicating GC dysfunction could be an important indicator of PCOS. Moreover, PCOS symptoms could be mitigated by increasing GC proliferation and reducing GC apoptosis [Citation11,Citation12]. Therefore, PCOS onset is closely correlated to the dysregulation of GC proliferation and apoptosis. Since human granulosa-like tumor cell line (KGN) retains the physiologic features of GCs and exhibits long-time and stable proliferative capability [Citation13], it is widely utilized in the studies of GC cellular functions and molecular regulatory mechanisms in PCOS.
Recently, emerging studies have demonstrated that long non-coding RNAs (lncRNAs), non-protein-coding transcripts with more than 200 nucleotides [Citation14], can act as vital regulators in various human diseases via participating in basic biological processes, including cell proliferation, differentiation, and apoptosis [Citation15]. In PCOS, lncRNAs have been proven to regulate metabolism, endocrine function, as well as GC proliferation and apoptosis [Citation16]. For example, Zhu et al. revealed that lncRNA ZFAS1 aggravated PCOS progression by inhibiting proliferation and accelerating apoptosis in GCs via the miRNA-129/HMGB1 axis [Citation17]. As demonstrated by Liu et al., lncRNA PVT1 accelerated PCOS development through regulating GC proliferation and apoptosis via miRNA-17-5p/PTEN cascade [Citation18]. Besides, Yang et al. uncovered that lncRNA BANCR induced GC apoptosis in PCOS, thereby promoting PCOS progression [Citation19]. In a previous report, Wang et al. discovered that LINC00173 expression was upregulated in follicle fluid from PCOS patients [Citation20]. Nevertheless, the specific regulatory mechanism of LINC00173 in PCOS has not yet been explored.
In this work, we focused on the analysis of the potential function and downstream mechanism of LINC00173 in PCOS. It was found that LINC00173 was elevated in PCOS patients and promoted GC apoptosis PCOS progression in rats via the miR-124-3p/JAG1 pathway. Therefore, our findings could provide new biomarkers concerning PCOS pathogenesis.
Materials and methods
Clinical samples
This study has gained ethical approval from Changzhou Hospital of Traditional Chinese Medicine (approval no. 2018-LL-01(F)). Thirty-two PCOS patients (PCOS group) and 32 age-matched non-PCOS patients (Control group) at Changzhou Hospital of Traditional Chinese Medicine were enrolled in this study and signed the written informed consent. PCOS cases were defined as patients meeting two of the following features: 1. oligo-ovulation or anovulation; 2. polycystic ovarian morphology according to ultrasonographic results; 3. clinical and/or biochemical signs of hyperandrogenism [Citation21]. Exclusion criteria for Control group: ovarian abnormalities, irregular menstruation, history of reproductive system disease or appendicitis. No participants took any medications (hormonal medications or oral contraceptives) in the past 3 months or had intrauterine device. Fasting venous blood sample was collected from each participant in the follicular or preovulatory phase of the cycle.
Isolation of ovarian GCs
After ovulation induction was performed properly as per a standard protocol [Citation22], PCOS and non-PCOS patients received oocyte retrieval. For PCOS patients, the remaining follicular fluid samples containing GCs were collected from each patient. Then, GCs were collected from follicular fluid by means of Percoll differential centrifugation and purified with red blood cell lysis buffer. Finally, purified GCs were preserved at −80°C before subsequent experiments.
Cell culture
KGN cell line obtained from the BeNa Culture Collection (Beijing, China) was cultured in the culture medium containing 90% DMEM and 10% FBS in a humidified atmosphere (37°C; 5% CO2).
Cell transfection
LINC00173 overexpression plasmid (oe-LINC00173), empty pcDNA3.1 vector (Vector), short hairpin RNA (shRNA) against LINC00173 (sh-LINC00173), JAG1 (sh-JAG1), and negative control (sh-NC), miR-124-3p mimic, miR-124-3p inhibitor, and corresponding negative controls (NC mimics and NC inhibitor) were synthesized by GenePharma (Shanghai, China) and transfected into KGN cells via Lipofectamine 2000 (Invitrogen, USA).
RT-qPCR assay
TRIzol reagent (Invitrogen) was applied to extract total RNA from blood samples, GCs, or KGN cells. To detect LINC00173 and JAG1 expressions, SYBR Premix Ex Taq II and TaqMan™ Gene Expression Assay (FAM) were used. Meanwhile, TaqMan MicroRNA Reverse Transcription kit was applied to measure miR-124-3p expression on TaqMan Universal Master Mix II (Applied Biosystems) [Citation23]. The relative expression was calculated by 2−ΔΔCt, with GAPDH or U6 as internal control. The primers used were listed below: miR-124-3p (Forward (F)): 5’-TAAGGCACGCGGTGAATGCC-3′ and (Reverse (R)): 5’-GGCGCCTCTCTTGGCATTC-3′; U6 (F): 5′-CTCGCTTCGGCAGCACAT-3′ and (R): 5′-TTTGCGTGTCATCCTTGCG-3′; LINC00173 (F): 5′-GGAATGTTGCGATCCTCTGG-3′ and (R): 5′-CAGCCATGTCTCAGAGGTGA-3′; JAG1 (F): 5′-TGCTACAACCGTGCCAGTGACT-3′ and (R): 5′- TCAGGTGTGTCGTTGGAAGCCA-3′; GAPDH (F): 5′- GTCTCCTCTGACTTCAACAGCG-3′ and (R): 5′-ACCACCCTGTTGCTGTAGCCAA-3′.
CCK-8 assay
For GC and KGN cell viability analysis, GCs and KGN cells were cultivated in 96-well plates. After CCK-8 solution was added (10 μl/well) at the indicated time, KGN cells were cultured for another 2 h. The absorbance values at 450 nm were measured with a microplate reader (EnSpire 2300, USA).
Flow cytometry
To detect GC and KGN cell apoptosis, annexin V-fluorescein isothiocyanate/propidium iodide (Annexin V-FITC/PI) Apoptosis Detection kit was applied to stain cells according to standard protocol. The apoptotic cells were calculated with a flow cytometer (BD Biosciences, USA) [Citation24].
Western blotting
The total protein was extracted by RIPA buffer from GCs and KGN cells, isolated by SDS-PAGE, and blotted onto PVDF membranes, which were sealed with 5% skimmed milk. Subsequently, the membranes were cultured with primary antibody against Bax, c-caspase-3, JAG1, or GAPDH at 4°C for 24 hours and continuously cultivated with the secondary antibody. The immunoblots were visualized with enhanced chemiluminescence and analyzed with ImageJ2X software.
Luciferase activity assay
The 3ʹ-UTR sequences of LINC00173 or JAG1 with wildtype (WT) or mutant (MUT) binding sites for miR-124-3p were amplified and cloned into pmirGLO vectors (Promega, USA) to generate luciferase reporter vectors (LINC00173-WT, LINC00173-MUT, JAG1-WT, or JAG1-MUT). KGN cells were transfected with LINC00173-WT, LINC00173-MUT, JAG1-WT, or JAG1-MUT, together with miR-124-3p mimics or NC mimics via Lipofectamine 2000. Finally, luciferase activity was measured with luciferase assay kit (Promega) after 48 hours’ transfection.
Establishment of PCOS model in rats
Forty female Sprague Dawley (SD) rats (21 days old) bought from SIPPR-BK Lab Animal Ltd. (Shanghai, China) were raised in an environment-controlled animal room (12-h light/dark cycle; 25°C) with adequate water and food supply. All these rats were randomly divided to four groups: Control group (n = 10), PCOS group (n = 10), PCOS+sh-NC (n = 10), and PCOS+sh-LINC00173 group (n = 10).
To induce PCOS, each SD rat received three-week dehydroepiandrosterone (DHEA; 60 mg/kg) injection. As for PCOS+sh-LINC00173 group, rats were injected with sh-LINC00173 through the caudal vein. Twenty-one days later, all SD rats were deprived of water and food for 12 hours and euthanized by cervical dislocation. Serum samples were collected for further experiments.
ELISA assay
Serum testosterone (T), luteinizing hormone (LH), estradiol (E2), and follicle‐stimulating hormone (FSH) levels were detected by commercial ELISA kits (Nanjing Jiancheng Bioengineering Institute, China) in accordance with the manufacturer’s instructions [Citation25] .
Statistical analysis
All data measured were shown as mean ± S.D. Statistical analyses were performed via GraphPad 6.0. For the data comparison, Student’s test or one-way ANOVA was applied. Pearson’s correlation analysis was employed to analyze the association between LINC00173, miR-124-3p, and JAG1. P < 0.05 was deemed significant in statistics.
Results
In this work, we mainly explore the role of LINC00173 in PCOS. Through molecular analysis as well as in vitro and in vivo experiments, the regulatory function of the LINC00173/miR-124-3p/JAG1 network in PCOS progression was further investigated.
LINC00173 inhibits viability and promotes apoptosis in KGN and GC cells
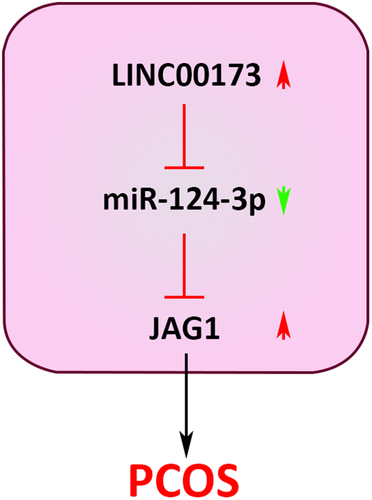
According to RT-qPCR assay, we observed that LINC00173 was highly expressed in PCOS patients (n = 32), relative to non-PCOS controls (n = 32; ), indicating that LINC00173 might be involved in PCOS pathogenesis. To further explore the role of LINC00173 in PCOS, GCs extracted from PCOS patients and KGN cells were used for functional assays. First of all, LINC00173 was overexpressed or knocked down in KGN and GC cells, with transfection efficiency evaluated by RT-qPCR (). The results of CCK-8 assay exhibited that KGN and GC cell viability were significantly decreased after LINC00173 upregulation but remarkably increased after LINC00173 knockdown (). Moreover, as indicated by flow cytometry assay, LINC00173 overexpression significantly upregulated the apoptotic rate of KGN and GC cells, while LINC00173 silencing manifestly suppressed apoptosis in KGN and GC cells (). Furthermore, we detected the protein levels of Bax and c-caspase-3 in KGN and GC cells from each group by Western blotting. The results exhibited that LINC00173 overexpression upregulated Bax and c-caspase-3 levels in KGN and GC cells, while LINC00173 knockdown exerted opposite effects on Bax and c-caspase-3 levels (). These results suggested that LINC00173 could impair biological functions of KGN and GC cells.
Figure 1. LINC00173 inhibits viability and promotes apoptosis in KGN and GC cells. (a) LINC00173 expression in PCOS patients (n = 32) and non-PCOS controls (n = 32) was detected by RT-qPCR. (b and c) LINC00173 expression in KGN and GC cells transfected with sh-NC, sh-LINC00173, Vector, or oe-LINC00173 was detected by RT-qPCR. (d and e) The viability of KGN and GC cells transfected with sh-NC, sh-LINC00173, Vector, or oe-LINC00173 was detected by CCK-8 assay. (f and g) The apoptotic rate of KGN and GC cells transfected with sh-NC, sh-LINC00173, Vector, or oe-LINC00173 was detected by flow cytometry. (h and i) Bax and c-caspase-3 levels in KGN and GC cells transfected with Vector, oe-LINC00173, sh-NC, or sh-LINC00173 were detected by Western blotting. *p < 0.05; **p < 0.01.
LINC00173 absorbs miR-124-3p
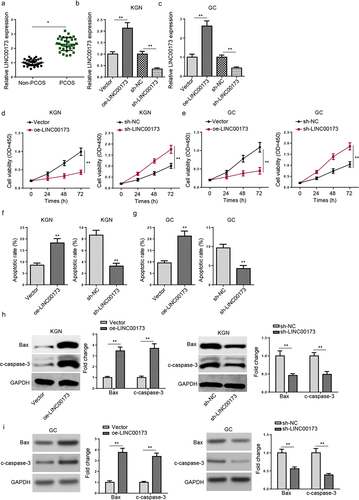
According to StarBase website, 12 candidate microRNAs (miRNAs) (miR-1286, miR-490-3p, miR-423-5p, miR-1252-5p, miR-506-3p, miR-124-3p, miR-493-3p, miR-130a-5p, miR-23c, miR-23a-3p, miR-23b-3p, and miR-182-5p) with latent binding sites for LINC00173 were predicted. According to a previous report by Ding et al., miR-124-3p expression was manifestly downregulated in PCOS patients [Citation26]. Subsequently, we tested miR-124-3p abundance in PCOS and normal samples. Consistent with the above study, it was revealed by RT-qPCR assay that miR-124-3p was remarkably lowly expressed in PCOS (). Therefore, we selected miR-124-3p for further experiments. Then, we found the binding sites between LINC00173 and miR-124-3p at StarBase website (). Afterward, miR-124-3p overexpression and inhibition efficiencies in KGN cells were tested (). It was shown that miR-124-3p upregulation led to a significant decrease in the luciferase activity of LINC00173-WT, while miR-124-3p inhibition increased the luciferase activity of LINC00173-WT; however, both miR-124-3p upregulation and miR-124-3p inhibition caused almost no influence on the luciferase activity of LINC00173-MUT, indicating the interaction between LINC00173 and miR-124-3p in KGN cells (). Besides, miR-124-3p enrichment in KGN cells was evidently decreased after LINC00173 overexpression and markedly increased after LINC00173 knockdown (). Furthermore, miR-124-3p expression was negatively related to LINC00173 enrichment in PCOS samples, as indicated by the result of Pearson’s correlation analysis (). To sum up, LINC00173 inversely modulated miR-124-3p level in PCOS.
Figure 2. LINC00173 absorbs miR-124-3p. (a) MiR-124-3p expression in PCOS patients (n = 32) and non-PCOS controls (n = 32) was detected by RT-qPCR. (b) The binding sites between LINC00173 and miR-124-3p were predicted by StarBase website. (c) MiR-124-3p expression in KGN cells transfected with NC mimics, miR-124-3p mimics, NC inhibitor, and miR-124-3p inhibitor. (d-e) The binding relationship between LINC00173 and miR-124-3p was validated by luciferase reporter assay. (f) MiR-124-3p expression in KGN cells transfected with sh-NC, sh-LINC00173, Vector, or oe-LINC00173 was detected by RT-qPCR. (g) Pearson’s correlation analysis to analyze the correlation between LINC00173 and miR-124-3p in PCOS patients. *p < 0.05; **p < 0.01.
LINC00173 regulates KGN cell viability and apoptosis via interaction with miR-124-3p
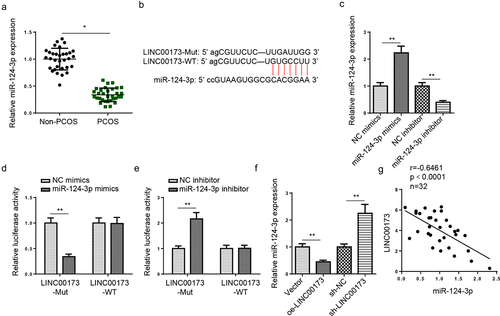
To confirm whether LINC00173 could modulate KGN cell viability and apoptosis via regulating miR-124-3p expression, a series of functional assays were performed. Firstly, KGN cells were transfected with Vector, oe-LINC00173, oe-LINC00173+ NC mimics, or oe-LINC00173+ miR-124-3p mimics. Also, sh-NC, sh-LINC00173, sh-LINC00173+ NC inhibitor, or sh-LINC00173+ miR-124-3p inhibitor were transfected into KGN cells. CCK-8 results showed that miR-124-3p overexpression partly offset the decreased cell viability of KGN cells overexpressing LINC00173 (). Conversely, miR-124-3p inhibition significantly abolished the increase in KGN cell viability caused by LINC00173 knockdown (). Also, LINC00173 amplification induced a significant increase in the apoptotic rate of KGN cells, while miR-124-3p upregulation partly offset such a promoting impact of LINC00173 overexpression on KGN cell apoptosis (). In contrast, miR-124-3p inhibition significantly abrogated the effect of LINC00173 knockdown on KGN cell apoptosis (). Taken together, LINC00173 inhibited proliferation and induced apoptosis in KGN cells via interaction with miR-124-3p.
Figure 3. LINC00173 regulates KGN cell viability and apoptosis via interaction with miR-124-3p. (a) The viability of KGN cells transfected with Vector, oe-LINC00173, oe-LINC00173+ NC mimics, or oe-LINC00173+ miR-124-3p mimics was detected by CCK-8 assay. (b) The viability of KGN cells transfected with sh-NC, sh-LINC00173, sh-LINC00173+ NC inhibitor, or sh-LINC00173+ miR-124-3p inhibitor was detected by CCK-8 assay. (c) The apoptotic rate of KGN cells transfected with Vector, oe-LINC00173, oe-LINC00173+ NC mimics, or oe-LINC00173+ miR-124-3p mimics was detected by flow cytometry. (d) The apoptotic rate of KGN cells transfected with sh-NC, sh-LINC00173, sh-LINC00173+ NC inhibitor, or sh-LINC00173+ miR-124-3p inhibitor was detected by flow cytometry. *p < 0.05; **p < 0.01.
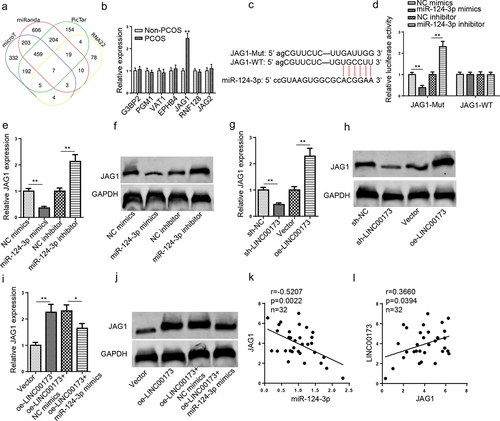
JAG1 is directly targeted by miR-124-3p
Four databases (microT, miRanda, RNA22, and PicTar) were applied to predict target messenger RNAs (mRNAs) for miR-124-3p. Finally, seven mRNAs (G3BP2, PGM1, VAT1, EPHB4, JAG1, RNF128, and JAG2) were screened by Venn diagram (). It was demonstrated that JAG1 was prominently elevated in PCOS, while the other six genes did not present an obvious upregulation (). Therefore, JAG1 was chosen as the direct target for miR-124-3p. The binding site between miR-124-3p and JAG1 predicted by StarBase is as shown in . The dual luciferase reporter experiment results indicated that the luciferase activity in JAG1-WT group was evidently decreased or increased in KGN cells after miR-124-3p upregulation or downregulation, whereas the luciferase activity in JAG1-MUT group showed no obvious changes (). Next, JAG1 mRNA and protein expressions were detected under the condition of aberrant LINC00173 or miR-124-3p expression. As indicated in , it was observed that JAG1 expression was substantially increased after miR-124-3p inhibition or LINC00173 upregulation, while miR-124-3p overexpression or LINC00173 depletion manifestly reduced JAG1 expression. Moreover, JAG1 expression was significantly elevated after LINC00173 overexpression; however, miR-124-3p upregulation partially reversed such an effect (). Similarly, JAG1 expression was negatively correlated to miR-124-3p expression in PCOS samples (). Also, JAG1 expression was positively correlated to LINC00173 expression in PCOS samples (). To sum up, JAG1 expression was oppositely correlated with miR-124-3p abundance and positively related to LINC00173 expression, indicating a competing endogenous RNA (ceRNA) regulatory network among LINC00173, miR-124-3p, and JAG1.
Figure 4. JAG1 is directly targeted by miR-124-3p. (a) The downstream targets of miR-124-3p were predicted by microT, miRanda, RNA22, and PicTar databases. (b) G3BP2, PGM1, VAT1, EPHB4, JAG1, RNF128, and JAG2 expressions in PCOS patients (n = 32) and non-PCOS controls (n = 32) were detected by RT-qPCR. (c) The binding sites between miR-124-3p and JAG1 were predicted by StarBase website. (d) The binding relationship between JAG1 and miR-124-3p was validated by luciferase reporter assay. (e and f) JAG1 mRNA and protein expressions in KGN cells transfected with NC mimics, miR-124-3p mimics, NC inhibitor, and miR-124-3p inhibitor were detected by RT-qPCR and Western blotting. (g and h) JAG1 mRNA and protein expressions in KGN cells transfected with sh-NC, sh-LINC00173, Vector, or oe-LINC00173 were detected by RT-qPCR and Western blotting. (i and j) JAG1 mRNA and protein expressions in KGN cells transfected with Vector, oe-LINC00173, oe-LINC00173+ NC mimics, and oe-LINC00173+ miR-124-3p mimics were detected by RT-qPCR and Western blotting. (k) Pearson’s correlation analysis to analyze the correlation between miR-124-3p and JAG1 in PCOS patients. (l) Pearson’s correlation analysis to analyze the correlation between LINC00173 and JAG1 in PCOS patients. *p < 0.05; **p < 0.01.
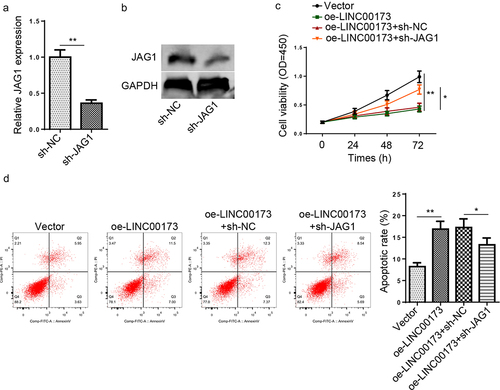
JAG1 depletion rescues the changes in KGN cell growth and apoptosis caused by LINC00173 overexpression
To further verify the regulatory functions of LINC00173/miR-124-3p/JAG1 ceRNA network in KGN cells, the cell viability and apoptotic rate of KGN cells transfected with Vector, oe-LINC00173, oe-LINC00173+ sh-NC, or oe-LINC00173+ sh-JAG1 were detected. Firstly, JAG1 was knocked down in KGN cells, and the transfection efficiency was confirmed (). As shown in , KGN cell viability was restrained by LINC00173 overexpression, while JAG1 deletion blocked the inhibitory effect of LINC00173 on KGN cell viability. Besides, flow cytometry assay demonstrated that JAG1 downregulation significantly restrained KGN cell apoptosis triggered by LINC00173 (). In sum, the above results revealed that LINC00173 suppressed KGN cell growth and accelerated KGN cell apoptosis by regulating JAG1.
Figure 5. JAG1 depletion rescues the changes in KGN cell growth and apoptosis caused by LINC00173 overexpression. (a and b) JAG1 mRNA and protein expressions in KGN cells transfected with sh-NC or sh-JAG1 were detected by RT-qPCR and Western blotting. (c) KGN cells were transfected with Vector, oe-LINC00173, oe-LINC00173+ sh-NC, or oe-LINC00173+ sh-JAG1. The viability of KGN cells in each group was detected by CCK-8 assay. (d) The apoptotic rate of KGN cells in each group was detected by flow cytometry. *p < 0.05; **p < 0.01.
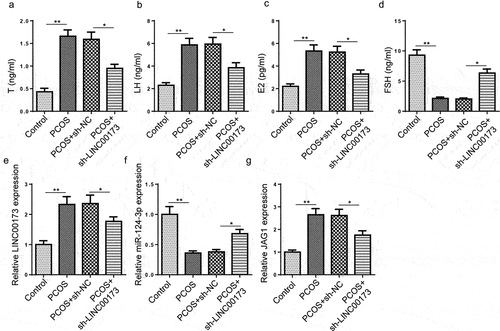
LINC00173 deletion regulates pathological features of PCOS in rats
To further affirm the function of LINC00173 in PCOS in vivo, SD rats were injected with DHEA to induce PCOS. These rats were divided to four groups: Control group (n = 10), PCOS group (n = 10), PCOS+sh-NC (n = 10), and PCOS+sh-LINC00173 group (n = 10). As revealed by ELISA assay, serum T, LH, and E2 levels were markedly increased in PCOS group, while FSH level was dramatically decreased in PCOS rats; however, LINC00173 inhibition led to a distinct suppression of T, LH, and E2 production and a significant increase in FSH level (), indicating LINC00173 knockdown might regulate PCOS-induced abnormal hormone secretion in rats. Furthermore, LINC00173 and JAG1 expressions were obviously upregulated, while miR-124-3p level was downregulated in PCOS rats, which was partly abated by LINC00173 inhibition (). Therefore, it was demonstrated that LINC00173 knockdown might regulate PCOS progression in vivo by modulating miR-124-3p and JAG1 expressions.
Figure 6. LINC00173 deletion regulates pathological features of PCOS in rats. (a-d) 40 female SD rats were randomly divided to 4 groups: Control group (n = 10), PCOS group (n = 10), PCOS+sh-NC (n = 10), and PCOS+sh-LINC00173 group (n = 10). Determination of serum T, LH, E2 and FSH levels by ELISA. (e-g) LINC00173, miR-124-3p, and JAG1 expressions in ovarian samples by RT-qPCR. *p < 0.05; **p < 0.01.
Discussion
PCOS, a heterogeneous endocrine disease featured with insulin resistance and hyperinsulinaemia, is a major cause for anovulatory infertility [Citation27,Citation28]. Recent studies have revealed that several lncRNAs may be complicated in PCOS development by regulating GC proliferation and apoptosis [Citation12,Citation29]. It has been proved that LINC00173 exerts opposite roles in regulating cell proliferation and apoptosis in different diseases [Citation30–32]. As reported by Zhang et al. in a former study, LINC00173 impaired inhibits the proliferation of cervical cancer cells through regulating FBXW7 expression via interaction with miR-182-5p [Citation33]. Yang et al. revealed that LINC00173 repressed cell proliferation and triggered cell apoptosis in non-small-cell lung cancer [Citation34]. Also, Li et al. elaborated that LINC00173 accelerated cell apoptosis and cell viability in pancreatic cancer [Citation35]. In this study, LINC00173 expression level was remarkably elevated in PCOS samples and PCOS rats. In addition, LINC00173 overexpression significantly suppressed cell proliferation and induced apoptosis in GCs and KGN cells, while LINC00173 depletion evidently increased GC and KGN cell viability and reduced GC and KGN cell apoptosis. In vivo, LINC00173 deletion could restore serum hormone levels in PCOS rats. These results indicated that LINC00173 might regulate PCOS development.
Mechanically, a lncRNA could competitively serve as a ceRNA to target specific miRNAs, thereby exerting effects on specific functions of target genes [Citation36]. In PCOS, lncRNAs may also play regulatory roles via interacting with miRNAs as specific ceRNAs. For example, Qin et al. demonstrated that lncRNA H19 high expression was closely correlative to PCOS in Chinese women in a former study [Citation37]. Then, Sun et al. further clarified that H19 regulated CTGF level via targeting miR-19b in GCs, in which the ceRNA regulating mechanism was applied to study PCOS pathogenesis for the first time [Citation38]. In our work, the involvement of LINC00173 in PCOS progression via the miR-124-3p/JAG1 pathway was explored. It has been widely reported that miR-124-3p plays a vital role in physiological processes in numerous diseases [Citation39–41]. For example, Li et al. disclosed that miR-124-3p promoted cell proliferation and accelerated cell apoptosis in nephrolithiasis [Citation42]. Liang et al. demonstrated that miR-124-3p considerably inhibited LPS-induced macrophage cell apoptosis [Citation43]. Also, Tao et al. revealed that miR-124-3p increased cell viability and repressed apoptosis in HTR-8/SVneo cells [Citation44]. Herein, we discovered that miR-124-3p overexpression was lowly expressed in PCOS samples and PCOS rats. MiR-124-3p expression was adversely correlated to LINC00173 expression in PCOS samples. Also, miR-124-3p abundance was decreased after LINC00173 overexpression and increased after LINC00173 downregulation. In addition, miR-124-3p overexpression or downregulation reversed the impacts of LINC00173 upregulation or knockdown on KGN cells viability and apoptosis.
Accumulated evidence has demonstrated that jagged canonical Notch ligand 1 (JAG1) is extensively implicated in the molecular regulation of cell growth and apoptosis in several diseases [Citation45–47]. As reported by Shi et al. in a former study, JAG1 could induce apoptosis in basal cell carcinoma cells [Citation48]. More importantly, Zeng et al. demonstrated that JAG1 was highly expressed in PCOS [Citation49]. Consistent with the above findings, we discovered that JAG1 expression was distinctly upregulated in PCOS samples and PCOS rats. Besides, JAG1 level was decreased in KGN cells after miR-124-3p upregulation or LINC00173 knockdown and increased after miR-124-3p inhibition or LINC00173 overexpression. Similarly, JAG1 expression was negatively correlated to miR-124-3p expression and positively related to LINC00173 expression in PCOS samples. Moreover, JAG1 depletion evidently alleviated the adverse effects of LINC00173 overexpression on KGN cells by promoting proliferation and suppressing apoptosis.
Conclusion
To sum up, our results revealed that LINC00173 might contribute to PCOS development by modulating the biological functions of GCs via the miR-124-3p/JAG1 axis via in vitro and in vivo experiments. However, our study still has several limitations. In future investigations, more phenotypic experiments will be performed to further validate our findings.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Azziz R. PCOS in 2015: new insights into the genetics of polycystic ovary syndrome. Nat Rev Endocrinol. 2016;12(3):183.
- Escobar-Morreale HF. Polycystic ovary syndrome: definition, aetiology, diagnosis and treatment. Nat Rev Endocrinol. 2018;14(5):270–284.
- Rotterdam EA-SPCWG. Revised 2003 consensus on diagnostic criteria and long-term health risks related to polycystic ovary syndrome. Fertil Steril. 2004;81(1):19–25.
- Norman RJ, Dewailly D, Legro RS, et al. Polycystic ovary syndrome. Lancet. 2007;370(9588):685–697.
- Hollinrake E, Abreu A, Maifeld M, et al. Increased risk of depressive disorders in women with polycystic ovary syndrome. Fertil Steril. 2007;87(6):1369–1376.
- De Diego MV, Gómez-Pardo O, Groar JK, et al. Metabolic impact of current therapeutic strategies in polycystic ovary syndrome: a preliminary study. Arch Gynecol Obstet. 2020;302(5):1169–1179.
- Baerwald AR, Adams GP, Pierson RA. Ovarian antral folliculogenesis during the human menstrual cycle: a review. Hum Reprod Update. 2012;18(1):73–91.
- Munakata Y, Kawahara-miki R, Shiratsuki S, et al. Gene expression patterns in granulosa cells and oocytes at various stages of follicle development as well as in in vitro grown oocyte-and-granulosa cell complexes. J Reprod Dev. 2016;62(4):359–366.
- Jiang JY, Cheung CK, Wang Y, et al. Regulation of cell death and cell survival gene expression during ovarian follicular development and atresia. Front Biosci. 2003;8(4):d222–37.
- Das M, Djahanbakhch O, Hacihanefioglu B, et al. Granulosa cell survival and proliferation are altered in polycystic ovary syndrome. J Clin Endocrinol Metab. 2008;93(3):881–887.
- Guo H, Li T, Sun X. LncRNA HOTAIRM1, miR-433-5p and PIK3CD function as a ceRNA network to exacerbate the development of PCOS. J Ovarian Res. 2021;14(1):19.
- Zheng Q, Li Y, Zhang D, et al. ANP promotes proliferation and inhibits apoptosis of ovarian granulosa cells by NPRA/PGRMC1/EGFR complex and improves ovary functions of PCOS rats. Cell Death Dis. 2017;8(10):e3145.
- Nishi Y, Yanase T, Mu Y-M, et al. Establishment and characterization of a steroidogenic human granulosa-like tumor cell line, KGN, that expresses functional follicle-stimulating hormone receptor. Endocrinology. 2001;142(1):437–445.
- Jandura A, Krause HM. The new RNA world: growing evidence for long noncoding RNA functionality. Trends Genet. 2017;33(10):665–676.
- Ding L, Ren J, Zhang D, et al. A novel stromal lncRNA signature reprograms fibroblasts to promote the growth of oral squamous cell carcinoma via LncRNA-CAF/interleukin-33. Carcinogenesis. 2018;39(3):397–406.
- Huang X, Hao C, Bao H, et al. Aberrant expression of long noncoding RNAs in cumulus cells isolated from PCOS patients. J Assist Reprod Genet. 2016;33(1):111–121.
- Zhu H-L, Chen Y-Q, Zhang Z-F. Downregulation of lncRNA ZFAS1 and upregulation of microRNA-129 repress endocrine disturbance, increase proliferation and inhibit apoptosis of ovarian granulosa cells in polycystic ovarian syndrome by downregulating HMGB1. Genomics. 2020;112(5):3597–3608.
- Liu G, Liu S, Xing G, et al. lncRNA PVT1/MicroRNA-17-5p/PTEN axis regulates secretion of E2 and P4, proliferation, and apoptosis of ovarian granulosa cells in PCOS. Mol Ther Nucleic Acids. 2020;20:205–216.
- Xu J, Wang H, Hu Y, et al. Inhibition of CaMKIIα activity enhances antitumor effect of fullerene C60 nanocrystals by suppression of autophagic degradation. Adv Sci (Weinheim, Baden-Wurttemberg, Germany). 2019;6(8):1801233.
- Wang L, Fan H, Zou Y, et al. Aberrant expression of long non-coding RNAs in exosomes in follicle fluid from PCOS patients. Front Genet. 2020;11:608178.
- Brower M, Brennan K, Pall M, et al. The severity of menstrual dysfunction as a predictor of insulin resistance in PCOS. J Clin Endocrinol Metab. 2013;98(12):E1967–71.
- Kaur S, Archer KJ, Devi MG, et al. Differential gene expression in granulosa cells from polycystic ovary syndrome patients with and without insulin resistance: identification of susceptibility gene sets through network analysis. J Clin Endocrinol Metab. 2012;97(10):E2016–21.
- Yu L, Gong X, Sun L, et al. The circular RNA Cdr1as act as an oncogene in hepatocellular carcinoma through targeting miR-7 expression. PloS One. 2016;11(7):e0158347.
- Guohua H Hongyang L, Zhiming J, et al. Study of small-cell lung cancer cell-based sensor and its applications in chemotherapy effects rapid evaluation for anticancer drugs. Biosens Bioelectron. 2017;97:184–195.
- Jiang B, Xue M, Xu D, et al. Down-regulated lncRNA HOTAIR alleviates polycystic ovaries syndrome in rats by reducing expression of insulin-like growth factor 1 via microRNA-130a. J Cell Mol Med. 2020;24(1):451–464.
- Ding CF, Chen W-Q, Zhu Y-T, et al. Circulating microRNAs in patients with polycystic ovary syndrome. Hum Fertil (Camb). 2015;18(1):22–29.
- McCartney CR, Marshall JC. CLINICAL PRACTICE. Polycystic ovary syndrome. N Engl J Med. 2016;375(1):54–64.
- Ben-Shlomo I, Younis JS. Basic research in PCOS: are we reaching new frontiers? Reprod Biomed Online. 2014;28(6):669–683.
- Tu J, Chen Y, Li Z, et al. Long non-coding RNAs in ovarian granulosa cells. J Ovarian Res. 2020;13(1):63.
- Li Q, Chen X, Chen L, et al. LINC00173 promotes the apoptosis of hypertrophic scar fibroblasts through increasing beta-catenin expression. Mol Cell Biochem. 2021;476(2):1005–1014.
- Zeng F, Wang Q, Wang S, et al. Linc00173 promotes chemoresistance and progression of small cell lung cancer by sponging miR-218 to regulate Etk expression. Oncogene. 2020;39(2):293–307.
- Hu CH, Yang X-J, Yu L, et al. Long non-coding RNA LINC00173 serves as sponge for miR-338-3p to promote prostate cancer progression via regulating Rab25. Eur Rev Med Pharmacol Sci. 2020;24(18):9290–9302.
- Zhang J, Zhou M, Zhao X, et al. Long noncoding RNA LINC00173 is downregulated in cervical cancer and inhibits cell proliferation and invasion by modulating the miR-182-5p/FBXW7 axis. Pathol Res Pract. 2020;216(8):152994.
- Yang Q, Tang Y, Tang C, et al. Diminished LINC00173 expression induced miR-182-5p accumulation promotes cell proliferation, migration and apoptosis inhibition via AGER/NF-kappaB pathway in non-small-cell lung cancer. Am J Transl Res. 2019;11(7):4248–4262.
- Li Q, Li X, Yang X, et al. Long intergenic nonprotein coding RNA 173 inhibits tumor growth and promotes apoptosis by repressing sphingosine kinase 1 protein expression in pancreatic cancer. DNA Cell Biol. 2021;40(6):757–775.
- Zhao Y, Wang H, Wu C, et al. Construction and investigation of lncRNA-associated ceRNA regulatory network in papillary thyroid cancer. Oncol Rep. 2018;39(3):1197–1206.
- Qin L, Huang -C-C, Yan X-M, et al. Long non-coding RNA H19 is associated with polycystic ovary syndrome in Chinese women: a preliminary study. Endocr J. 2019;66(7):587–595.
- Sun X, Yan X, Liu K, et al. lncRNA H19 acts as a ceRNA to regulate the expression of CTGF by targeting miR-19b in polycystic ovary syndrome. Braz J Med Biol Res. 2020;53(11):e9266.
- Geng L, Liu W, Chen Y. miR-124-3p attenuates MPP(+)-induced neuronal injury by targeting STAT3 in SH-SY5Y cells. Exp Biol Med (Maywood). 2017;242(18):1757–1764.
- Zhaohui W, Yingli N, Hongli L, et al. Amentoflavone induces apoptosis and suppresses glycolysis in glioma cells by targeting miR-124-3p. Neurosci Lett. 2018;686:1–9.
- Jin J, Jia Z-H, Luo X-H, et al. Long non-coding RNA HOXA11-AS accelerates the progression of keloid formation via miR-124-3p/TGFbetaR1 axis. Cell Cycle. 2020;19(2):218–232.
- Wu J, Niu S, Bremner DH, et al. A tumor microenvironment-responsive biodegradable mesoporous nanosystem for anti-inflammation and cancer theranostics. Adv Healthc Mater. 2020;9(2):e1901307.
- Liang Y, Xie J, Che D, et al. MiR-124-3p helps to protect against acute respiratory distress syndrome by targeting p65. Biosci Rep. 2020;40(5). DOI:10.1042/BSR20192132.
- Tao J, Xia L-Z, Liang L, et al. MiR-124-3p promotes trophoblast cell HTR-8/SVneo pyroptosis by targeting placental growth factor. Placenta. 2020;101:176–184.
- Jin Y, Li C, Xu D, et al. Jagged1-mediated myeloid Notch1 signaling activates HSF1/Snail and controls NLRP3 inflammasome activation in liver inflammatory injury. Cell Mol Immunol. 2020;17(12):1245–1256.
- Sun W, Zhao J, Li C. Dexmedetomidine provides protection against hippocampal neuron apoptosis and cognitive impairment in mice with Alzheimer’s disease by mediating the miR-129/YAP1/JAG1 Axis. Mol Neurobiol. 2020;57(12):5044–5055.
- Radi E, Formichi P, Di Maio G, et al. Oxidative stress-induced apoptosis in two patients with Alagille syndrome. J Neurol Sci. 2011;308(1–2):49–56.
- Shi FT, Yu M, Zloty D, et al. Notch signaling is significantly suppressed in basal cell carcinomas and activation induces basal cell carcinoma cell apoptosis. Mol Med Rep. 2017;15(4):1441–1454.
- Zeng Z, Lin X, Xia T, et al. Identification of crucial lncRNAs, miRNAs, mRNAs, and potential therapeutic compounds for polycystic ovary syndrome by bioinformatics analysis. Biomed Res Int. 2020;2020:1817094.
