ABSTRACT
Drug resistance has become the major obstacle for the treatment of non-small cell lung cancer (NSCLC). Circular RNAs (circRNAs) are tightly linked to the development of drug resistance of NSCLC. Herein, we tested the function of circ_0002360 in the Taxol resistance of NSCLC. Circ_0002360, microRNA (miR)-585-3p and G protein regulated inducer of neurite outgrowth 1 (GPRIN1) were quantified by quantitative real-time PCR (qRT-PCR). To identify the circular structure of circ_0002360, RNase R digestion was applied. To detect cell proliferation, colony formation and 5-ethynyl-2’-deoxyuridine (EdU) assays were used. For assessment of cell apoptosis, flow cytometry was adopted. For motility and invasion analyses, transwell assay was employed. Our data showed that circ_0002360 was mainly located in the cytoplasm and was highly expressed in the Taxol-resistant NSCLC. Silencing of circ_0002360 inhibited cell Taxol resistance, proliferation, motility, and invasiveness and induced apoptosis in vitro. MiR-585-3p was underexpressed in Taxol-resistant NSCLC and was targeted by circ_0002360. MiR-585-3p knockdown alleviated the influence of circ_0002360 silence on Taxol-resistant cells. GPRIN1 was directly targeted by miR-585-3p. The influence of miR-585-3p on cell Taxol resistance and functional behaviors was reversed by GPRIN1 overexpression. Moreover, circ_0002360 modulated GPRIN1 through miR-585-3p. Additionally, silencing of circ_0002360 weakened the growth of xenografts in vivo. Our study demonstrated that silencing of circ_0002360 enhanced the Taxol sensitivity and suppressed the malignant behaviors of Taxol-resistant NSCLC cells by miR-585-3p/GPRIN1 axis, providing novel targets for improving the anti-tumor efficacy of Taxol in NSCLC.
1 Introduction
Lung cancer ranks second in terms of morbidity, but first in terms of cancer mortality in 2020 all through the world [Citation1]. Non-small cell lung cancer (NSCLC) is the most prevalent type of lung cancer [Citation2]. Despite intense efforts, the prognosis of NSCLC remains poor because drug resistance frequently arises [Citation3]. Therefore, more and more researchers focus on the study of the resistance mechanism in NSCLC [Citation4–6]. The exploration of new biomarkers affecting the development and chemotherapy resistance of NSCLC is helpful to improve NSCLC management.
Circular RNAs (circRNAs) are a specific type of RNAs [Citation7] that can sequester microRNAs (miRNAs) to impact lung tumorigenesis and drug resistance [Citation8–10]. For example, Liu et al. demonstrated that depletion of circ_0001821 hampered the Taxol resistance, growth and metastasis of NSCLC cells depending on the control of the miR-526b-5p/GRK5 axis [Citation11]. Zhang et al. reported that circRNA SRY-box transcription factor 13 (circSOX13) functioned as a strong contributor to NSCLC cell cisplatin resistance and malignant behaviors via microtubule-associated protein RP/EB family member 1 (MAPRE1) upregulation by sponging miR-3194-3p [Citation12]. Human circ_0002360, generated by the back-spliced exons of RUNX family transcription factor 1 (RUNX1), is highly expressed in NSCLC and operates as a promoter in NSCLC [Citation13,Citation14]. Moreover, circ_0002360 can contribute to the cisplatin resistance and malignant phenotypes of lung cancer cells by upregulating zinc finger protein 300 (ZNF300) through miR-6751-3p competition [Citation15]. However, no studies proved the function of circ_0002360 in the Taxol resistance of NSCLC.
MiRNAs are closely associated with drug resistance of NSCLC cancer cells [Citation16]. For instance, elevated expression of miR-6734-3p sensitizes cisplatin-resistant NSCLC cells to cisplatin therapy by silencing zinc finger E-box binding homeobox 2 (ZEB2) [Citation17]. Abnormal expression of miR-888-5p actively participates in cisplatin resistance of NSCLC by targeting RB1 inducible coiled-coil 1 [Citation18]. Intriguingly, miR-585-3p has established an anti-tumor role in numerous cancers, such as ovarian cancer, colon cancer and gastric cancer [Citation19–21]. Moreover, Zhang and colleagues established that circ_0002360 directly sponged miR-585-3p to promote NSCLC cell growth and metastasis [Citation14]. Nonetheless, the relationship between miR-585-3p and circ_0002360 in Taxol resistance development of NSCLC has not been studied.
G protein regulated inducer of neurite outgrowth 1 (GPRIN1) is present at high levels in renal papillary cell carcinoma (KIRP) and lung adenocarcinoma (LUAD) [Citation22]. In LUAD, GPRIN1 may be an independent marker for this disease prognosis, and its upregulation promotes LUAD cell growth and motility [Citation23]. Furthermore, it remains undefined whether GPRIN1 represents a functionally effector of circ_0002360 in regulating NSCLC Taxol resistance.
In the preliminary survey, we found the potential relationship between miR-585-3p and circ_0002360 or GPRIN1 using bioinformatics analysis (circInteractome and TargetScan). We therefore hypothesized the implication of the circ_0002360/miR-585-3p/GPRIN1 axis in the Taxol resistance of NSCLC. In this work, we studied the expression, activity and mechanism of circ_0002360 activity in NSCLC resistance to Taxol.
2 Materials and methods
2.1 Tissue samples
The Ethics Committee of the First Affiliated Hospital of Xi’an Jiaotong University approved this study. In this study, 68 NSCLC patients (38 patients with recurrent tumor after treatment with Taxol-based chemotherapy and 30 patients with primary tumor) from the First Affiliated Hospital of Xi’an Jiaotong University were enrolled from March 2019 to December 2020, and the tumor specimens and adjacent lung samples were harvested. The clinicopathological features of these patients were provided in . We defined tumor tissues from recurrent patients as Taxol-resistant NSCLC (NSCLC-R) and primary patients as Taxol-resistant NSCLC (NSCLC-S). These tumor tissues were employed with the written informed consent of all patients.
Table 1. Correlation analysis for circ_0002360 expression and the clinicopathological parameters of patients with NSCLC (n = 68)
2.2 Cell lines
The Cell Bank of RIKEN BioResource Center (Tsukuba, Japan) provided two Taxol-resistant NSCLC cell lines (H1299/Taxol cells and A549/Taxol) and their parents (H1299 and A549) and one normal BEAS-2B cell line. All cells were cultured in a 37°C cell incubator with 5% CO2 (Panasonic, Osaka, Japan) with RPMI-1640 medium (BIOSUN, Shanghai, China) plus 1% penicillin-streptomycin and 10% fetal bovine serum (both from Bovogen, Melbourne, Australia).
2.3 Quantitative real-time PCR (qRT-PCR)
RNA was obtained with TRIzol reagent (Invitrogen, São Paulo, Brazil) and treated with DNase I (MedChemExpress, Shanghai, China) before cDNA transcription, which were performed using an iScript Kit (Bio-Rad, Gladesville, NSW, Australia) with 1 µg of RNA or BON-miR miRNA cDNA Synthesis Kit (Bonyakhteh, Tehran, Iran) with 100 ng of RNA. SYBR-based qRT-PCR was done using specific primer sets and SYBR Premix Ex Taq II as described by the manufacturers (TaKaRa, Dalian, China). We carried out PCR reaction using the ABI Prism 7700 System (Thermo Fisher Scientific, São Paulo, Brazil). Samples were amplified for 40 cycles using the following parameters: 95°C for 10 minutes, 95°C for 15 seconds, and 60°C for 1 minute. U6 snRNA (U6) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) served as the internal reference genes. We evaluated the fold changes using the 2−ΔΔCt method [Citation24]. Genscript Biotechnology Co., Ltd (Nanjing, China) synthesized the primers (Supplement ) used in our study.
2.4 Cell counting kit-8 (CCK-8) assay
The biochemical half maximal inhibitory concentration (IC50) value of Taxol was tested by the CCK-8 kit from Beyotime (Shanghai, China) as described [Citation25]. In brief, transfected cells (4,000 cells/well) in 96-well plates were stimulated with different doses of Taxol (Solarbio, Beijing, China) for 48 h and then incubated with 10 µL CCK-8 (Beyotime) for 4 h. Using the SpectraMax M5e microplate reader (Molecular Devices, Silicon Valley, California, USA), we gauged the absorbance of per well at 450 nm.
2.5 RNase R assay
RNase R [Citation26] and qRT-PCR were employed to identify the circular structure of circ_0002360. RNase R assay was done by treating RNA (1 µg) with or without RNase R (3 units) as per the protocols of manufacturers (Geneseed, Guangzhou, China).
2.6 CircRNA subcellular localization assay
We performed subcellular fractionation assays to examine the localization of circ_0002360 as reported [Citation26]. The expression of circ_0002360 in cytoplasmic and nuclear fractions was tested by qRT-PCR after RNA preparation using the Cytoplasmic and Nuclear RNA Purification Kit as described by the producers (Norgen Biotek, Ontario, Canada).
2.7 Cell transfection
Plasmids expressing shRNA-circ_0002360 (sh-circ_0002360), control shRNA (sh-NC), pcDNA3.1-based GPRIN1 expression plasmid (expressing the coding sequence without 3ʹUTR) GPRIN1-pcDNA, control vector, miR-585-3p mimic, mimic control miR-NC, anti-miR-585-3p and control anti-NC were gained from Kebai (Nanjing, China). For transfection, H1299/Taxol and A549/Taxol cells were plated in 12-well dishes at 1,000,000 cells per well. Next day, Lipofectamine 3000 liposome transfection reagent (Invitrogen) and the indicated plasmid (300 ng) or/and oligonucleotide (50 nM) were added into cells on the basis of the guidance of Lipofectamine 3000. 24 h later, we harvested the transfected cells for in vitro studies.
2.8 Colony formation assay
Transfected H1299/Taxol and A549/Taxol cells were cultured in 6-well dishes at a colony density of 150 cells per well for 10–14 days. The cells were then fixed with methanol (Tianjin Concord Technology Co., Ltd, Tianjin, China), dyed with 0.5% crystal violet, and observed using microscopy as described elsewhere [Citation26].
2.9 5-ethynyl-2’-deoxyuridine (EdU) cell proliferation assay
The EdU experiment was operated according to the instructions of the EdU Apollo488/567 in vitro kit (Solarbio) as reported [Citation27]. Briefly, transfected H1299/Taxol and A549/Taxol cells (4,000 cells/well) were seeded in 96-well dishes for 48 h, treated with 50 nM of EdU reagent for 2 h, and then dyed with 1× Apollo488/567 for 30 min, followed by the nucleus staining with 4’,6-diamidino-2-phenylindole (DAPI, Solarbio) for 30 min. The EdU positive cells were determined using fluorescence microscopy (Leica TCS SP5, Leica, Wetzlar, Germany) relative to total nuclei.
2.10 Flow cytometry
After transfection, ~1,000,000 H1299/Taxol and A549/Taxol cells were harvested and stained in accordance with the Annexin V-fluorescein isothiocyanate (FITC)/propidium iodide (PI) apoptosis detection kit (Solarbio) specifications as described elsewhere [Citation27]. A total of 100,000 events were analyzed by Cytoflex flow cytometry (BECKMAN COULTER, Pasadena, Texas, USA), and the apoptotic cells were defined as the cells that was positive with Annexin V.
2.11 Transwell assay
We performed transwell assays to assess cell motility and invasion under standard protocols [Citation26]. To test motility ability, the cells (50,000 cells/well) in FBS-free media were placed in the upper chamber of the chamber (Corning, Tewksbury, MA, USA) without matrigel (BD Biosciences, Franklin Lakes, New Jersey, USA), and the medium with 10% FBS (Bovogen) was appended in the lower cabinet. 24 h later, the number of cells on the lower surface of the supernatant chamber was counted by a microscope (BX51IW, Olympus, Tokyo, Japan) to evaluate the migration ability of cells after staining with crystal violet (0.5%). Similarly, the invasion experiment was performed in a chamber containing matrigel (BD Biosciences), and the cells (100,000 cells/well) were seeded into the upper chamber.
2.12 Western blot
Protein expression was evaluated by immunoblotting using standard methods [Citation28]. Total protein was prepared from tissues and cells using RIPA buffer enriched with protease inhibitor cocktails (Santa Cruz Biotechnology, Dallas, TX, USA) and then quantified by the bicinchoninic acid (BCA) assay using a BCA protein assay kit (Thermo Fisher Scientific). Proteins (30 µg) were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on 10–12% SDS-polyacrylamide gels and blotted to polyvinylidene fluoride (PVDF) membranes (Bio-Rad). After being blocked in 5% skim dry milk in TBS-T (Tris, NaCl, pH 7.6, 0.1% Tween-20), the membranes were probed with the primary and horseradish peroxidase (HRP)-conjugated IgG secondary antibodies (ab6789 or ab97051, 1:10,000) (all from Abcam, Cambridge, UK). Primary antibodies used were as follows: anti-proliferating cell nuclear antigen (anti-PCNA, ab29, 1:1,000), anti-Bcl2 associated X (anti-Bax, ab182733, 1:2,000), anti-multi-drug resistant associate protein 1 (anti-MRP1, ab260038, 1:1,000), anti-GRPIN1 (ab226201, 1:3,000), anti-P-glycoprotein (anti-P-gp, ab242104, 1:1,000), and anti-β-actin (ab8226, 1:1,000). Then, the bands were observed with HRP Substrate (Merck, Darmstadt, Hesse, Germany) and analyzed using an imaging analyzer (Alliance Ld2, UVITEC, Cambridge, UK) with GelDoc software (Bio-Rad).
2.13 Determination of caspase-3 activity
The activity of caspase-3 in transfected cells was gauged with the Caspase-3 Activity assay kit as per the manufacturing protocols (Abcam). We measured the fluorescence at Ex/Em = 400/505 nm using the Leica TCS SP5 fluorescence microscope.
2.14 Dual-luciferase reporter assay
For dual-luciferase reporter assay, we generated reporter constructs as described [Citation29]. The segments of circ_0002360 and GPRIN1 3ʹUTR containing the predicted miR-585-3p binding site were ligated in psiCHECK-2 vector (Promega, Barcelona, Spain) to create wild-type (WT) reporters (circ_0002360 WT and GPRIN1 3ʹUTR WT). Mutations (circ_0002360 MUT and GPRIN1 3ʹUTR MUT) were produced by the TaKaRa MutanBest kit (TaKaRa). For luciferase reporter assays, A549/Taxol and H1299/Taxol cells (100,000) were cotransfected with the reporter constructs (300 ng) and miR-585-3p mimics (50 nM) or mimic controls using Lipofectamine 3000. The luciferase activities were measured by the Dual Luciferase Kit (Vazyme, Nanjing, China) after 48 h transfection.
2.15 RNA immunoprecipitation (RIP) assay
Magna RIP kit (Geneseed) was engaged for RIP assay, which was conducted using methods as reported [Citation26]. A549/Taxol and H1299/ Taxol cells (100,000) were cleaved with 200 µL RIP lysis buffer and protease K (Vazyme). Then, lysates were subjected to the incubation with Protein A + G magnetic beads-coupled with antibody against Ago2 or IgG (Millipore) overnight at 4°C. Ago2-associated RNA was harvested, and circ_0002360 and miR-585-3p were quantified by qRT-PCR.
2.16 Tumor xenograft mice model construction
Animal experimental procedures were done according to the methods reported by Chu et al. [Citation27]. 1 × 106 A549/Taxol cells infected with a lentiviral sh-NC or sh-circ_0002360 construct were injected into the axilla of BALB/c nude mice (female, 5–7 weeks old, Beijing Vital River Laboratory Animal Technology Co., Ltd., Beijing, China). Each group included six mice. One week later, Taxol was intraperitoneally injected every 3 days (15 mg/kg) into nude mice. The volume of tumor was tested every 7 days and calculated by the 0.5 × (length × width2) formula. The mice were euthanized after 28 days, and the tumors were weighed. Animal procedures were authorized by Ethics Committee at the First Affiliated Hospital of Xi’an Jiaotong University.
2.17 Immunohistochemistry (IHC) assay
Tumor tissues were fixed with 4% paraformaldehyde (Solarbio) and encapsulated in paraffin. IHC analysis was used to analyze PCNA expression as reported [Citation30]. The sections (4 µm) of the tumors were hatched with diluted anti-PCNA antibody (ab29, 1:1000, Abcam) and biotinylated IgG secondary antibody (ab64256, 1:500 dilution, Abcam), stained with 3,3’-diaminobenzidine substrate (Invitrogen), and observed under microscopy (Leica TCS SP5) and photographed.
2.18 Statistical analysis
Data analysis was conducted with GraphPad Prism 8.0 software (GraphPad Inc., LaJolla, California, USA). Differences were gauged by ANOVA with Tukey’s post hoc test or Student’s t-test. P < 0.05 indicated significant difference. Pearson’s correlation coefficient was applied to test the expression correlation of variables in NSCLC tissues.
3 Results
3.1 Circ_0002360 was highly expressed in Taxol-resistant NSCLC
To elucidate the involvement of circ_0002360 in Taxol resistance of NSCLC, we first evaluated its expression. The up-regulated expression of circ_0002360 in NSCLC was shown by the GEO database (GSE158695 and GSE112214) ()). Circ_0002360 expression in a cohort of NSCLC tissues was measured by qRT-PCR, and results confirmed that circ_0002360 was highly expressed in human NSCLC tissues ()). We then analyzed the expression pattern of circ_0002360 in NSCLC tissues from 38 patients with recurrent tumor after treatment with Taxol-based chemotherapy (defined as Taxol-resistant NSCLC, NSCLC-R) compared with tumor tissues from 30 patients with primary tumor (defined as Taxol-sensitive NSCLC, NSCLC-S). Through qRT-PCR, circ_0002360 expression was augmented in Taxol-resistant tissues of NSCLC (NSCLC-R) compared to the sensitive tissues (NSCLC-S) ()). Additionally, CCK-8 assays revealed that the IC50 value of Taxol in H1299/Taxol and A549/Taxol cells was higher than that in their parent cells ()), indicating the successful establishment of Taxol-resistant NSCLC cells. In agreement with tumor tissues, circ_0002360 was up-regulated in NSCLC cells compared with normal BEAS-2B cells and significantly overexpressed in H1299/Taxol and A549/Taxol cells compared with their parents ()). Additionally, circ_0002360 expression was correlated with tumor size, differentiation, and lymphatic metastasis of the tumors ().
Figure 1. The expression of circ_0002360 was increased in Taxol-resistant NSCLC. (a) GSE158695 and GSE112214 datasets were used to analyze the expression of circ_0002360 in NSCLC tissues. (b) The expression of circ_0002360 in NSCLC tissues and normal tissues was detected by qRT-PCR. (c) The expression of circ_0002360 in Taxol-sensitive and resistant tissues of NSCLC was detected by qRT-PCR. (d) The IC50 values of Taxol in H1299/Taxol and A549/Taxol cells were tested via CCK-8 assay. (e) The expression of circ_0002360 in normal cells, Taxol-sensitive and resistant cells of NSCLC was assessed. (f and g) The abundance of circ_0002360 and RUNX1 mRNA levels after treatment with RNase R. (h) The existence of circ_0002360 was detected in cell lines by qRT-PCR using cDNA or gDNA as templates with divergent or convergent primers and verified by Gel electrophoresis. ◄►: divergent primers, ►◄: convergent primers. (i) Schematic diagram showed the formation of circ_0002360. ***P < 0.001, ****P < 0.0001.
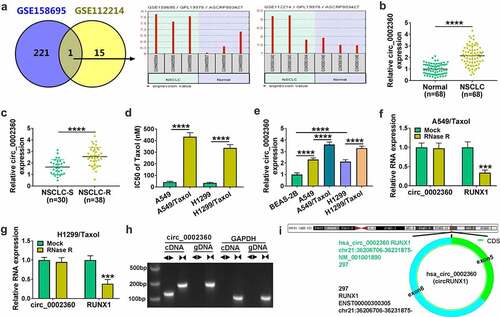
To confirm the stability of circ_0002360, we adopted RNase R treatment. RNase R assays indicated that the level of the linear RUNX1 mRNA was reduced by RNase R digestion, while circ_0002360 expression was not affect by RNase R (), indicating that circ_0002360 was RNase R resistance. When we performed qRT-PCR analysis using cDNA or genomic DNA (gDNA) from A549/Taxol or H1299/Taxol cells as templates and divergent (◄►) or Convergent (►◄) primers, we found that circ_0002360 was only detectable in cDNA rather than gDNA from NSCLC cells when divergent primers were used ()), validating the circular structure of circ_0002360. As shown in ), circ_0002360 was formed by back-splicing of exons 5 and 6 of RUNX1 located at chr21:36,206,706–36,231,875. In short, circ_0002360 was up-regulated in human NSCLC and might be associated with Taxol tolerance of NSCLC.
3.2 Circ_0002360 depletion hindered cell Taxol-resistance, proliferation, motility, and invasiveness and accelerated cell apoptosis
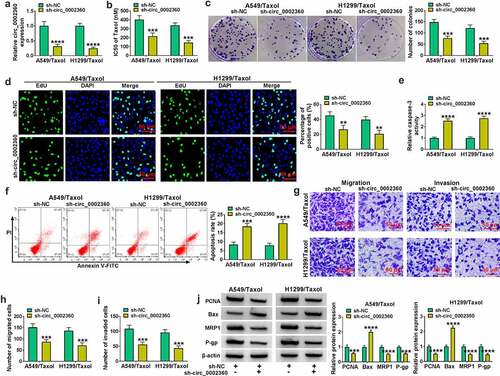
To test the function of circ_0002360 in Taxol-resistant NSCLC cells, we performed phenocopy silencing by shRNAs targeting circ_0002360 (sh-circ_0002360#1/#2/#3). We selected sh-circ_0002360#2 (also called sh-circ_0002360) for further experiments in our study because the transfection of sh-circ_0002360#2 resulted in the most significant (****P < 0.0001) and durable (>14 days) downregulation in circ_0002360 expression (Supplement and data not shown). In H1299/Taxol and A549/Taxol cells, qRT-PCR results showed that circ_0002360 expression was reduced by sh-circ_0002360 introduction, indicating the high tranfection efficiency of sh-circ_0002360 in reducing circ_0002360 expression ()). CCK-8 results revealed that the IC50 value of Taxol was diminished in the sh-circ_0002360 group compared with the sh-NC group ()). The results of colony formation and EdU assays displayed that the proliferation of H1299/Taxol and A549/Taxol cells was hampered by circ_0002360 depletion (). Silencing circ_0002360 also elevated the caspase-3 activity of H1299/Taxol and A549/Taxol cells and promoted cell apoptosis (). We next performed transwell assays to evaluate the effect on cell motility and invasion. Results showed that downregulation of circ_0002360 retarded cell motility and invasiveness compared with the control group (). Additionally, western blot results revealed that the expression levels of proliferating marker PCNA, resistant-related MRP1 and P-gp proteins were reduced by downregulation of circ_0002360 in H1299/Taxol and A549/Taxol cells, while the expression of anti-apoptotic protein Bax was enhanced by sh-circ_0002360 introduction ()). In summary, silencing of circ_0002360 enhanced the Taxol sensitivity and apoptosis as well as hindered cell proliferation, motility and invasiveness in Taxol-resistant NSCLC cells.
Figure 2. Circ_0002360 knockdown regulated the behaviors of Taxol-resistant cells. (a) The transfection efficiency of sh-circ_0002360 in H1299/Taxol and A549/Taxol cells. (b) IC50 value of Taxol in cells transfected as indicated by CCK-8. (c) Colony formation assay performed with cells transfected as indicated. (d) EdU assay for proliferation of cells transfected as indicated. (e) The activity of caspase-3 in cells transfected as indicated using an assay kit. (f) Flow cytometry performed with cells transfected as indicated to evaluate apoptosis. (G to I) The ability of cells transfected as indicated to migrate and invade was detected by the Transwell assay. (j) PCNA, Bax, MRP1 and P-gp levels in cells transfected as indicated. **P < 0.01, ***P < 0.001, ****P < 0.0001.
3.3 Circ_0002360 targeted miR-585-3p in Taxol-resistant NSCLC cells
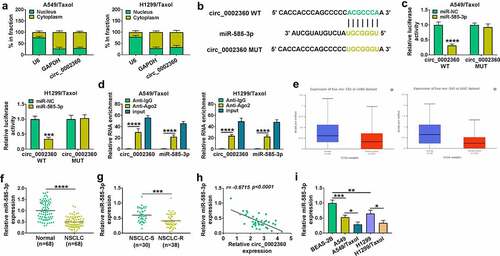
We then asked the mechanism of circ_0002360 regulation in the Taxol sensitivity and functional behaviors of Taxol-resistant NSCLC cells. The results of subcellular localization assays exhibited that circ_0002360 was mainly present in the cytoplasm of H1299/Taxol and A549/Taxol cells ()). To search the targeted miRNAs of circ_0002360, we used the online tool circInteractome and observed a putative complementary sequence for miR-585-3p in circ_0002360 ()). To ascertain this, we constructed circ_0002360 wild-type (WT) or mutant-type (MUT) luciferase reporters and tested them by luciferase assays. Reporter experiments indicated that the luciferase activities of H1299/Taxol and A549/Taxol cells were reduced in the circ_0002360 WT and miR-585-3p co-transfected group ()). However, no reduction in luciferase was found in the circ_0002360 MUT and miR-585-3p co-transfected group compared with the control group ()). MiRNAs exert gene regulatory activity in the RNA-induced silencing complex (RISC), which also contains Ago2, a core component of the RISC [Citation31]. We thus performed RIP experiments using the anti-Ago2 antibody. RIP results showed that miR-585-3p and circ_0002360 enrichment levels were elevated in Ago2-associating complexes compared with the IgG controls, implying the association between circ_0002360 and RISC ()). Moreover, the UALCAN database showed that miR-585-3p was down-regulated in the tissues of LUAD and lung squamous cell carcinoma (LUSC) ()). qRT-PCR also validated that miR-585-3p was down-regulated in NSCLC tissues compared with normal tissues ()) and underexpressed in Taxol-resistant NSCLC samples (NSCLC-S) compared with the sensitive samples (NSCLC-R) ()). In Taxol-resistant NSCLC samples, an inverse expression correlation of miR-585-3p and circ_0002360 was observed ()). Furthermore, miR-585-3p was decreased in NSCLC cells compared with normal BEAS-2B cells and suppressed in H1299/Taxol and A549/Taxol cells compared with their sensitive parents ()). The data together suggested that circ_0002360 directly targeted miR-585-3p.
Figure 3. MiR-585-3p was targeted by circ_0002360. (a) The localization of circ_0002360 in cells was detected by subcellular localization experiment. (b) Predicted duplex formation between circ_0002360 and miR-585-3p by CircInteractome. (c and d) Dual-luciferase report assay in H1299/Taxol and A549/Taxol cells. (d) RIP assay in H1299/Taxol and A549/Taxol cells. (e) MiR-585-3p expression in LUAD samples and healthy samples in and LUSC. (f) MiR-585-3p level in NSCLC samples and normal samples. (g) MiR-585-3p level in Taxol-resistant and sensitive tissues in NSCLC. (h) Correlation analysis between circ_0002360 and miR-585-3p expression in NSCLC tissues. (i) MiR-585-3p level in normal cells, NSCLC cells and Taxol-resistant cells in NSCLC. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
3.4 Inhibition of miR-585-3p partially reversed the effect of sh-circ_0002360 on Taxol-resistant NSCLC cells
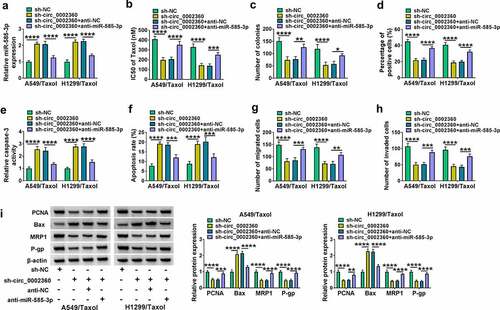
To determine whether the effects of circ_0002360 depletion were due to the alteration of miR-585-3p expression, we performed recovery experiments in vitro. Through qRT-PCR, miR-585-3p expression was enhanced by circ_0002360 knockdown in H1299/Taxol and A549/Taxol cells, and the effect was abolished by anti-miR-585-3p introduction ()). CCK-8 assays showed that circ_0002360 depletion-imposed reduction on the IC50 of Taxol and cell proliferation in H1299/Taxol and A549/Taxol cells were reversed by miR-585-3p knockdown ()). Moreover, silencing of circ_0002360 boosted caspase-3 activity and cell apoptosis, while reduced expression of miR-585-3p reversed these effects (). Reduced expression of miR-585-3p also rescued circ_0002360 depletion-driven cell motility and invasiveness defects (). Additionally, downregulation of miR-585-3p reversed the regulatory effects of sh-circ_0002360 on PCNA, Bax, MRP1 and P-gp expression in H1299/Taxol and A549/Taxol cells ()). Together, these findings demonstrated that circ_0002360 targeted miR-585-3p to regulate the Taxol resistance and biological behaviors of Taxol-resistant NSCLC cells.
Figure 4. Anti-miR-585-3p reversed the impact of sh-circ_0002360 in H1299/Taxol and A549/Taxol cells. (a) MiR-585-3p level in H1299/Taxol and A549/Taxol cells after transfection by sh-NC, sh-circ_0002360, sh-circ_0002360+ anti-NC or sh-circ_0002360+ anti-miR-585-3p. (b) IC50 value of Taxol in cells transfected as indicated by CCK-8. (c) Colony formation assay performed with cells transfected as indicated. (d) EdU assay for proliferation of cells transfected as indicated. (e) The activity of caspase-3 in cells transfected as indicated using an assay kit. (f) Flow cytometry performed with cells transfected as indicated to evaluate apoptosis. (g and h) The ability of cells transfected as indicated to migrate and invade was detected by the Transwell assay. (i) PCNA, Bax, MRP1 and P-gp levels in cells transfected as indicated. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
3.5 GPRIN1 was directly targeted by miR-585-3p
To establish the downstream effectors of miR-585-3p, the online software TargetScan was employed to search the downstream targets of miR-585-3p. The predicted data showed a putative pairing site between miR-585-3p and GPRIN1 3ʹUTR ()). Compared with the miR-NC control, overexpression of miR-585-3p downregulated the luciferase activity of GPRIN1 3ʹUTR WT reporter, but not GPRIN1 3ʹUTR MUT construct ()), validating the targeting of GPRIN1 by miR-585-3p. GEPIA database showed that GPRIN1 was augmented in cancer tissues of LUAD and LUSC ()). qRT-PCR confirmed that GPRIN1 mRNA expression was elevated in NSCLC samples compared with normal tissues ()). When comparing with the Taxol-sensitive NSCLC tissues (NSCLC-S), GPRIN1 was overexpressed in the Taxol-resistant NSCLC samples (NSCLC-R) ()). Expression correlation analysis revealed that in Taxol-resistant NSCLC samples, GPRIN1 negatively associated with miR-585-3p and positively associated with circ_0002360 (). Western blot exhibited that GPRIN1 protein was highly expressed in Taxol-sensitive NSCLC samples and cells compared with the corresponding normal controls and was increased in Taxol-resistant NSCLC samples and cells compared with their Taxol-sensitive counterparts (). To examine whether miR-585-3p could regulate GPRIN1 expression, we manipulated miR-585-3p expression in H1299/Taxol and A549/Taxol cells by introducing miR-585-3p mimics or anti-miR-585-3p. By contrast, miR-585-3p expression was augmented by miR-585-3p mimics and inhibited by anti-miR-585-3p ()). As expected, the expression levels of GPRIN1 protein in H1299/Taxol and A549/Taxol cells were diminished by overexpression of miR-585-3p (). Conversely, GPRIN1 protein expression was enhanced by miR-585-3p inhibition (). Moreover, GPRIN1 protein was downregulated by sh-circ_0002360 in H1299/Taxol and A549/Taxol cells, while knockdown of miR-585-3p attenuated the effect (). In summary, circ_0002360 regulated GPRIN1 expression by miR-585-3p.
Figure 5. GPRIN1 was targeted by miR-585-3p. (a) Predicted duplex formation between miR-585-3p in GPRIN1. (b) Dual-luciferase reporting assay in H1299/Taxol and A549/Taxol cells. (c) GPRIN1 mRNA level in LUAD and LUSC. (d) GPRIN1 mRNA level in NSCLC samples compared with normal human samples. (e) GPRIN1 level in Taxol-sensitive and resistant samples. (f) Correlation analysis between GPRIN1 and miR-585-3p expression. (g) Correlation analysis between circ_0002360 expression and GPRIN1 expression. (h) GPRIN1 protein level in Taxol-resistant and sensitive samples. (i) GPRIN1 protein level in normal cells, NSCLC cells and Taxol-resistant cells in NSCLC. (J to L) GPRIN1 protein level in H1299/Taxol and A549/Taxol cells after miR-585-3p or anti-miR-585-3p transfection. (m and n) GPRIN1 protein level in cells transfected as indicated. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
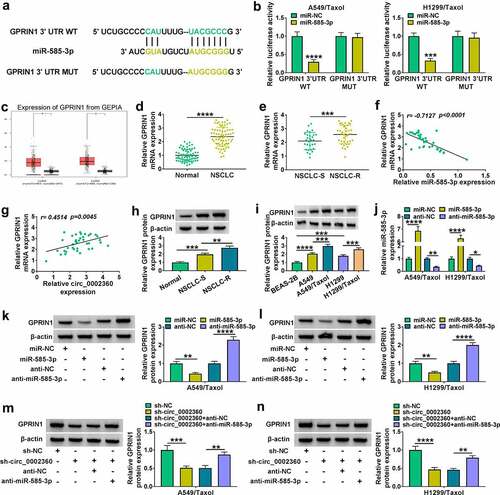
3.6 Overexpression of GPRIN1 overturned the influence of miR-585-3p on cell Taxol-resistance, proliferation, motility, invasiveness and apoptosis
We next elucidated whether GPRIN1 represented a functional target of miR-585-3p in regulating cell Taxol-resistance and biological behaviors. The protein levels of GPRIN1 in H1299/Taxol and A549/Taxol cells were enhanced by GPRIN1 overexpression plasmid transfection ()). Overexpression of miR-585-3p reduced the IC50 value of Taxol and suppressed colony formation and proliferation of H1299/Taxol and A549/Taxol cells, while GPRIN1 upregulation reversed these effects (). The promoting effects of miR-585-3p on caspase-3 activity and cell apoptosis were reversed by GPRIN1 upregulation (). Moreover, increase of GPRIN1 relieved the inhibitory effects of miR-585-3p on cell motility and invasiveness (). The impact of miR-585-3p on PCNA, Bax, MRP1 and P-gp expression in H1299/Taxol and A549/Taxol cells were also overturned by GPRIN1 upregulation ()). These data indicated that the influence of miR-585-3p on Taxol-resistant cells was abolished by GPRIN1 augmentation.
Figure 6. GPRIN1 overexpression reversed the influence of miR-585-3p in H1299/Taxol and A549/Taxol cells. (a) GPRIN1 level in H1299/Taxol and A549/Taxol cells after transfection by vector and GPRIN1. (b-i) H1299/Taxol and A549/Taxol cells were introduced with miR-NC mimics, miR-585-3p mimics, miR-585-3p mimics+vector or miR-585-3p mimics+GPRIN1. (b) IC50 value of Taxol in cells transfected as indicated by CCK-8. (c) Colony formation assay performed with cells transfected as indicated. (d) EdU assay for proliferation of cells transfected as indicated. (e) The activity of caspase-3 in cells transfected as indicated using an assay kit. (f) Flow cytometry performed with cells transfected as indicated to evaluate apoptosis. (g and h) The ability of cells transfected as indicated to migrate and invade was detected by the Transwell assay. (i) PCNA, Bax, MRP1 and P-gp levels in cells transfected as indicated. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
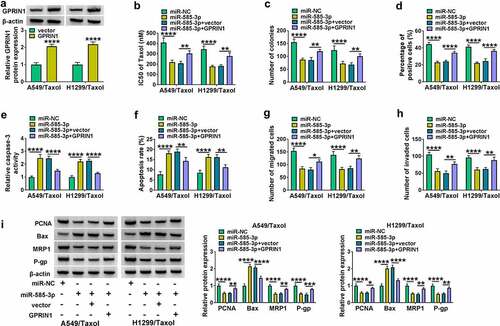
3.7 Circ_0002360 depletion suppressed the growth of xenografts in vivo
Tumor xenograft mice models were constructed to explore the role of circ_0002360 in vivo. By contrast, tumor growth was markedly decreased by sh-circ_0002360 transduction with Taxol treatment, as presented by the decreased tumor volume, size, and weight (). qRT-PCR results revealed that the levels of circ_0002360 and GPRIN1 mRNA were decreased in the Taxol+sh-circ_0002360 group, while miR-585-3p expression in the Taxol+sh-circ_0002360 group was obviously increased ()). Western blot data showed that the levels of GPRIN1, MRP1 and P-gp were reduced in the Taxol+sh-circ_0002360 tumor tissues compared to the Taxol+sh-NC group ()). The data of IHC assay elucidated that the number of the cells stained with PCNA was lower in the Taxol+sh-circ_0002360 tumors than that in the Taxol+sh-NC controls ()), supporting the suppression of circ_0002360 depletion on tumor growth. Hence, these data suggested that circ_0002360 depletion might enhance cell Taxol sensitivity in vivo.
Figure 7. Circ_0002360 inhibited the growth of NSCLC cells and enhanced their sensitivity to Taxol in vivo. (a) Tumor volume was periodically monitored. (b and c) Images and average weight of the xenograft tumors in each group at the endpoint. (d) The expression of circ_0002360, miR-585-3p and GPRIN1 in each group. (e) Western blot for GPRIN1, MRP1 and P-gp levels in xenograft tumors. (f) PCNA staining cells in xenograft tumors detected by IHC. **P < 0.01, ***P < 0.001, ****P < 0.0001.
4 Discussion
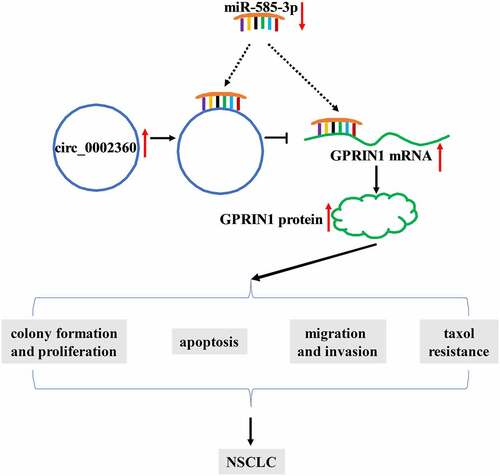
NSCLC is a prevalent malignant tumor around the world [Citation32]. Dysregulation of circRNAs has been shown to be associated with the Taxol resistance of various cancers, including NSCLC, ovarian cancer and esophageal squamous cell carcinoma [Citation10,Citation33,Citation34]. In this work, we defined the expression and activity of circ_0002360 in Taxol-resistant NSCLC. Furthermore, we provided a novel molecular explanation, the circ_0002360/miR-585-3p/GPRIN1 axis, for the promoting activity of circ_0002360 in NSCLC Taxol resistance ().
Figure 8. Schematic model of the circ_0002360/miR-585-3p/GPRIN1 axis in NSCLC Taxol resistance. In Taxol-resistant NSCLC cells, circ_0002360 was upregulated and miR-585-3p was underexpressed and thus GPRIN1 expression was enhanced, thereby promoting Taxol resistance, cell proliferation, metastasis and diminishing cell apoptosis.
Chen et al. highlighted that circ_0002360 was associated with the pathogenesis of tongue squamous cell carcinoma (TSCC) [Citation35]. Circ_0002360 in LUAD has been confirmed to be obviously overexpressed [Citation13]. Circ_0002360 can function as a potent oncogenic promoter in NSCLC by elevating the expression of matrix metalloproteinase 16 (MMP16) by sponging multiple miRNAs [Citation14]. Moreover, circ_0002360 accelerates the cisplatin resistance and malignant phenotypes of lung cancer cells through the miR-6751-3p/ZNF300 axis [Citation15]. In this work, our data found that circ_0002360 was evidently overexpressed in Taxol-resistant NSCLC. Using in vitro and in vivo function experiments, we first demonstrated that silencing of circ_0002360 promoted cell sensitivity to Taxol and suppressed cell growth, motility and invasiveness in Taxol-resistant NSCLC cells. The data suggested that circ_0002360 might be a potential molecular regulator in affecting NSCLC Taxol resistance.
Further, we first identified that miR-585-3p worked as a targeted miRNA of circ_0002360 function. Numerous studies have documented the anti-tumor activity of miR-585-3p. For instance, miR-585-3p can hinder ovarian cancer cell growth and motility via the modulation of calpain 9 [Citation19]. In colon cancer cells, reduced expression of miR-585-3p induces cell proliferation by upregulating proteasome activator subunit 3 [Citation20]. Additionally, long noncoding RNA LINC01436 can enhance gastric tumorigenesis by inhibiting miR-585-3p expression [Citation21]. Ma et al. discovered the down-regulated plasma expression of miR-585-3p in patients with NSCLC using microRNA microarray detection [Citation36]. Our data also proved that the effects of circ_0002360 silencing were due to miR-585-3p upregulation, suggesting that miR-585-3p can work as a molecular mediator of circ_0002360 function in affecting NSCLC Taxol resistance. Consistently, circ_0002360 functions as a promoter in NSCLC progression by acting as a sponge of miR-585-3p [Citation14].
GPRIN1 is overexpressed in LUAD and kidney renal papillary cell carcinoma [Citation22]. In LUAD, GPRIN1 expression is associated with overall survival and poor prognosis [Citation22,Citation23]. Moreover, GPRIN1 may be a LUAD-related immune biomarker candidate to activate the immune response [Citation37]. Conversely, Zhou et al. reported that miR-654-5p contributed to gastric tumorigenesis by targeting GPRIN1 [Citation38]. In this work, GPRIN1 was first proved as a miR-585-3p target. Our data indicated that GPRIN1 was pronouncedly enhanced in Taxol-resistant NSCLC. We also ascertained that miR-585-3p-mediated inhibition of GPRIN1 promoted cell Taxol sensitivity and impeded the malignant behaviors of Taxol-resistant NSCLC cells. More importantly, we first uncovered that GPRIN1 modulated GPRIN1 expression through miR-585-3p, suggesting the regulatory effect of the circ_0002360/miR-585-3p/GPRIN1 axis on the Taxol resistance of NSCLC. Zhang et al. highlighted the implication of the circ_0002360/miR-585-3p/MMP16 axis in NSCLC pathogenesis [Citation14]. In NSCLC, the circ_0002360/miR-585-3p/GPRIN1 and circ_0002360/miR-585-3p/MMP16 axes may be two interactional networks. Whether the circ_0002360/miR-585-3p/MMP16 axis is involved in the Taxol resistance of NSCLC will be performed in further work. One circRNA can target many miRNAs, and one miRNA can regulate many target genes [Citation39]. There may be other miRNA/mRNA axes that remain to be defined in the regulation of circ_0002360 in NSCLC.
5 Conclusion
In this work, we demonstrated that upregulated circ_0002360 was associated with the Taxol resistance of NSCLC. Inhibition of circ_0002360 sensitized cells to Taxol and hindered cell malignant behaviors. The data of function rescue experiments disclosed that circ_0002360 regulated the development and Taxol resistance of NSCLC by miR-585-3p/GPRIN1 axis. This provided evidence for the notion that circ_0002360 inhibition might be used to sensitize NSCLC to Taxol therapy.
Highlights:
Circ_0002360 is overexpressed in Taxol-resistance NSCLC.
GPRIN1 is a miR-585-3p target.
GPRIN1 is regulated by circ_0002360 via miR-585-3p.
Circ_0002360 impacts the behaviors of Taxol-resistant NSCLC cells.
Supplemental Material
Download MS Word (16.3 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary material
Supplemental data for this article can be accessed here
Additional information
Funding
References
- Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249.
- Planchard D, Popat S, Kerr K, et al. Metastatic non-small cell lung cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2018;29(Suppl4):iv192–iv237.
- Duma N, Santana-Davila R, Molina JR. Non-Small Cell Lung Cancer: epidemiology, Screening, Diagnosis, and Treatment. Mayo Clin Proc. 2019;94(8):1623–1640.
- Terlizzi M, Colarusso C, Pinto A, et al. Drug resistance in non-small cell lung Cancer (NSCLC): impact of genetic and non-genetic alterations on therapeutic regimen and responsiveness. Pharmacol Ther. 2019;202:(140–148.
- Leonetti A, Assaraf YG, Veltsista PD, et al. MicroRNAs as a drug resistance mechanism to targeted therapies in EGFR-mutated NSCLC: current implications and future directions. Drug Resist Updat. 2019;42:(1–11.
- Meador CB, Hata AN. Acquired resistance to targeted therapies in NSCLC: updates and evolving insights. Pharmacol Ther. 2020;210:107522.
- Wang Y, Lu T, Wang Q, et al. Circular RNAs: crucial regulators in the human body (Review). Oncol Rep. 2018;40(6):3119–3135.
- Song L, Cui Z, Guo X. Comprehensive analysis of circular RNA expression profiles in cisplatin-resistant non-small cell lung cancer cell lines. Acta Biochim Biophys Sin (Shanghai). 2020;52(9):944–953.
- Liang ZZ, Guo C, Zou MM, et al. circRNA-miRNA-mRNA regulatory network in human lung cancer: an update. Cancer Cell Int. 2020;20(1):(173.
- Xu N, Chen S, Liu Y, et al. Profiles and Bioinformatics Analysis of Differentially Expressed Circrnas in Taxol-Resistant Non-Small Cell Lung Cancer Cells. Cell Physiol Biochem. 2018;48(5):2046–2060.
- Liu Y, Li C, Liu H, et al. Circ_0001821 knockdown suppresses growth, metastasis, and TAX resistance of non-small-cell lung cancer cells by regulating the miR-526b-5p/GRK5 axis. Pharmacol Res Perspect. 2021;9(4):e00812.
- Zhang L, Peng H, Xu Z, et al. Circular RNA SOX13 promotes malignant behavior and cisplatin resistance in non-small cell lung cancer through targeting microRNA-3194-3p/microtubule-associated protein RP/EB family member 1. Bioengineered. 2022;13(1):1814–1827.
- Yan Y, Zhang R, Zhang X, et al. RNA-Seq profiling of circular RNAs and potential function of hsa_circ_0002360 in human lung adenocarcinom. Am J Transl Res. 2019;11(1):160–175.
- Zhang Y, Zeng S, Wang T. Circular RNA hsa_circ_0002360 promotes non-small cell lung cancer progression through upregulating matrix metalloproteinase 16 and sponging multiple micorRNAs. Bioengineered. 2021;12(2):12767–12777.
- Ding L, Li L, Tang Z. Cisplatin resistance and malignant behaviors of lung cancer cells are promoted by circ_0002360 via targeting miR-6751-3p to regulate the expression of ZNF300. Thorac Cancer. 2022. DOI:10.1111/1759-7714.14342
- Tutar Y. miRNA and cancer; computational and experimental approaches. Curr Pharm Biotechnol. 2014;15(5):429.
- Wei L, Jiang J. Targeting the miR-6734-3p/ZEB2 axis hampers development of non-small cell lung cancer (NSCLC) and increases susceptibility of cancer cells to cisplatin treatment. Bioengineered. 2021;12(1):2499–2510.
- Chen H, Li F, Xue Q. Circ-CUL2/microRNA-888-5p/RB1CC1 axis participates in cisplatin resistance in NSCLC via repressing cell advancement. Bioengineered. 2022;13(2):2828–2840.
- Lu X, Li G, Liu S, et al. MiR-585-3p suppresses tumor proliferation and migration by directly targeting CAPN9 in high grade serous ovarian cancer. J Ovarian Res. 2021;14(1):90.
- Liu C, Yang J, Wu H, et al. Downregulated miR-585-3p promotes cell growth and proliferation in colon cancer by upregulating PSME3. Onco Targets Ther. 2019;12:6525–6534.
- Xu Y, Dong M, Wang J, et al. LINC01436 Inhibited miR-585-3p Expression and Upregulated MAPK1 Expression to Promote Gastric Cancer Progression. Dig Dis Sci. 2021;66(6):1885–1894.
- Zhou Q, Li D, Zheng H, et al. A novel lncRNA-miRNA-mRNA competing endogenous RNA regulatory network in lung adenocarcinoma and kidney renal papillary cell carcinoma. Thorac Cancer. 2021;12(19):2526–2536.
- Zhuang MQ, Li J, Han X, et al. G protein regulated inducer of neurite outgrowth 1 is a potential marker for lung cancer prognosis. J Biol Regul Homeost Agents. 2020;34(3):853–864.
- Ng EK, Tsang WP, Ng SS, et al. MicroRNA-143 targets DNA methyltransferases 3A in colorectal cancer. Br J Cancer. 2009;101(4):699–706.
- Zhou C, Liu C, Liu W, et al. SLFN11 inhibits hepatocellular carcinoma tumorigenesis and metastasis by targeting RPS4X via mTOR pathway. Theranostics. 2020;10(10):4627–4643.
- Chen L, Li H, Yao D, et al. The novel circ_0084904/miR-802/MAL2 axis promotes the development of cervical cancer. Reprod Biol. 2022;22(1):100600.
- Chu D, Li P, Li Y, et al. Identification of circ_0058357 as a regulator in non-small cell lung cancer cells resistant to cisplatin by miR-361-3p/ABCC1 axis. Thorac Cancer. 2021;12(21):2894–2906.
- D’Souza A, Pearman CM, Wang Y, et al. Targeting miR-423-5p Reverses Exercise Training–Induced HCN4 Channel Remodeling and Sinus Bradycardia. Circ Res. 2017;121(9):1058–1068.
- Zhu S, Pan W, Song X, et al. The microRNA miR-23b suppresses IL-17-associated autoimmune inflammation by targeting TAB2, TAB3 and IKK-α. Nat Med. 2012;18(7):1077–1086.
- Korpal M, Ell BJ, Buffa FM, et al. Direct targeting of Sec23a by miR-200s influences cancer cell secretome and promotes metastatic colonization. Nat Med. 2011;17(9):1101–1108.
- Michlewski G, Cáceres JF. Post-transcriptional control of miRNA biogenesis. Rna. 2019;25(1):1–16.
- Chen Z, Fillmore CM, Hammerman PS, et al. Non-small-cell lung cancers: a heterogeneous set of diseases. Nat Rev Cancer. 2014;14(8):535–546.
- Wei S, Qi L, Wang L. Overexpression of circ_CELSR1 facilitates paclitaxel resistance of ovarian cancer by regulating miR-149-5p/SIK2 axis. Anticancer Drugs. 2021;32(5):496–507.
- Qu F, Wang L, Wang C, et al. Circular RNA circ_0006168 enhances Taxol resistance in esophageal squamous cell carcinoma by regulating miR-194-5p/JMJD1C axis. Cancer Cell Int. 2021;21(1):273.
- Chen Z, Jie Y, Qiu Z, et al. Expression profile and bioinformatics analysis of circular RNAs in tongue squamous cell carcinoma. Oral Dis. 2021. DOI:10.1111/odi.13933.
- Ma Y, Pan X, Xu P, et al. Plasma microRNA alterations between EGFR-activating mutational NSCLC patients with and without primary resistance to TKI. Oncotarget. 2017;8(51):88529–88536.
- Wang Y, Tan H, Yu T, et al. Potential Immune Biomarker Candidates and Immune Subtypes of Lung Adenocarcinoma for Developing mRNA Vaccines. Front Immunol. 2021;12:(755401.
- Zhou W, Li P, Jin P. miR-654-5p promotes gastric cancer progression via the GPRIN1/NF-κB pathway. Open Med (Wars). 2021;16(1):1683–1695.
- Anastasiadou E, Jacob LS, Slack FJ. Non-coding RNA networks in cancer. Nat Rev Cancer. 2018;18(1):5–18.
