ABSTRACT
Gastric cancer (GC) is lethal malignancy, which is associated with high mortality. Long noncoding RNA LINC01270 has been identified to act as a potential oncogene in several cancers. However, its role and related regulatory mechanism in GC are yet to be illustrated. The levels of lncRNA LINC01270, miR-326, and EphrinA3 (EFNA3) were assessed by quantitative reverse transcriptase polymerase chain reaction (qRT-PCR). Cell counting kit-8 (CCK-8) and colony formation assays were applied for analyzing cell proliferation. Transwell assay was used for measuring cellular migration and invasion. Western blot analysis was employed for evaluating the protein levels. Luciferase reporter and RNA pull-down assays were utilized to verify the binding ability between LINC01270 (or EFNA3) and miR-326. Our findings indicated that LINC01270 expression was significantly up-regulated in GC tissues and cell lines. Additionally, LINC01270 knockdown attenuated GC progression through inhibiting cell proliferation, migration, and invasion. Functional experiments identified that lncRNA LINC01270 could positively regulate EFNA3 expression by serving as a competing endogenous RNA (ceRNA) for miR-326. Through rescue assays, inhibition of GC progression caused by LINC01270 suppression was found to be reversed by the application of miR-326 inhibitor or EFNA3 overexpression. Overall, our work demonstrated that lncRNA LINC01270 can accelerate cell proliferation, migration, and invasion via modulating miR-326/EFNA3 axis. These findings might implicate the potential role of lncRNA LINC01270 in GC treatment.
Graphical abstract
Introduction
Gastric cancer (GC) is the fourth most commonly diagnosed malignancy and the second-leading cause for cancer death worldwide [Citation1,Citation2]. GC patients with early diagnosis have a good prognosis, but they are commonly diagnosed in the advanced stage at which surgery is not possible [Citation3]. At present, most advanced GC patients are treated with radiotherapy or chemotherapy, but the adverse reactions exhibited by these treatment modalities can significantly reduce the patients’ life quality, which is not beneficial for the prognosis of these patients [Citation4]. Although great advances have been achieved in the development of effective diagnosis and therapeutic methods for GC [Citation5,Citation6], the prognosis and survival rate of these patients, especially those who are diagnosed at an advanced stage, remain unsatisfactory. Therefore, strategies to develop novel biomarkers for improving the diagnosis and treatment of GC can be of great clinical value.
As one RNA molecular subset with transcriptional length over 200 nts, long non-coding RNAs (lncRNAs) have been reported to be involved in multiple aspects of tumorigenesis. For example, lncRNA HOTTIP can facilitate ovarian cancer cell proliferation and invasion [Citation7]. Moreover, lncRNA W42 can bind to DBN1 to accelerate tumor development in hepatocellular carcinoma [Citation8]. LncRNA SLNCR1 can effectively interact with secretory phospholipase A2 to modulate the migration, invasion, and stemness in non-small cell lung cancer [Citation9]. Additionally, lncRNA SNHG16 can significantly enhance sorafenib resistance in hepatocellular carcinoma [Citation10]. Importantly, lncRNAs have also been implicated in GC development. For instance, lncRNA MIR22HG can attenuate NOTCH2 signaling to affect GC progression [Citation11]. LncRNA NRON can modulate ALKBH5 (AlkB homolog-5) and Nanog to facilitate tumorigenesis in GC [Citation12]. In addition, LINC00355 can exacerbate the ubiquitination of P53 to affect GC cell proliferation and invasion [Citation13]. LncRNA LINC01270 has been identified to be involved in the development of several diseases. LncRNA LINC01270 was also associated with worse overall survival and expressed at higher levels in triple-negative breast cancer [Citation14]. Besides, LINC01270 knockdown can enhance the chemosensitivity to 5-fluorouracil by regulating glutathione S-transferase P1 (GSTP1) methylation in esophageal cancer progression [Citation15]. However, the function and detailed regulatory mechanism of lncRNA LINC01270 in GC have not been investigated in detail.
Thus, the present study was aimed to analyze the function and regulatory mechanism of lncRNA LINC01270 in GC. Our results revealed that LINC01270, miR-326, and EFNA3 together formed the lncRNA-miRNA-mRNA axis that affected the malignant phenotype GC. This finding may offer a novel potential target for GC treatment.
Methods
Samples
Forty GC tissues and normal adjacent tissues were collected from Chaohu Hospital of Anhui Medical University. The informed written consent was obtained from all patients. The collected tissues were promptly frozen at −80°C. The work was approved by the Ethics Committee of Chaohu Hospital of Anhui Medical University (No. 202105002).
Cell lines and cell culture
The normal gastric epithelial cell line (GES-1) and GC cell lines (HGC-27, AGS, and SGC-7901) were purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). The cells were cultured in Roswell Park Memorial Institute-1640 (RPMI-1640) medium (Sigma) with fetal bovine serum (FBS; 10%; Hyclone, USA), penicillin (100 units/ml), and streptomycin (100 mg/ml) at 37°C with 5% CO2.
Transfection
The short hairpin RNA targeting lncRNA LINC01270 (sh-LINC01270), miR-326 mimic/inhibitor, pcNDA3.1 targeting EFNA3 (pcDNA-EFNA3), and their respective controls (sh-NC, miR-NC, and pcDNA-NC) were obtained from GenePharma (Shanghai, China). These plasmids were thereafter transfected into GC cells through Lipofectamine 2000 (Invitrogen Life Technologies, USA).
Reverse transcription-quantitative PCR (RT-qPCR)
TRIzol reagent (Thermo Fisher Scientific) was used to extract the RNA from GC tissues and cells. Reverse PrimeScript RT Master Mix (TaKaRa, Tokyo, Japan) was then applied for cDNA reverse-transcription. RT-qPCR was done by using SYBR Green (Takara Bio, Japan). The lncRNA/mRNA (or miRNA) internal control was GAPDH (or U6). The expression of the different genes was calculated with 2−ΔΔCt method [Citation16].
RNA-fluorescence in-situ hybridization (FISH)
The tumor or normal tissues were hybridized with lncRNA LINC01270 oligodeoxynucleotide probe (GenePharma, Shanghai, China) in hybridization buffer overnight. The signal of the probe was assessed through the fluorescent FISH kit (GenePharma, China), then DAPI was employed to dye the nucleus. At last, the image was obtained through the fluorescence microscope (Olympus, Tokyo, Japan).
Cell counting kit-8 (CCK-8) assay
CCK-8 assay was performed for the assessment of cell viability as described previously [Citation17]. The cells were grown on a 96-well plate and cultured for 2 h. The addition of CCK-8 solution was carried out at 0, 24, 48, and 72 h. At the end, the absorbance at 450 nm was assessed through a microplate reader (Biotek Instruments, Winooski, VT, USA).
Colony formation assay
The cells were plated in a 6-well plate and then incubated in an appropriate incubator for 2 weeks. Thereafter, the cells were fixed using absolute methanol and stained by crystal violet dye (Beyotime). Finally, the colonies were photographed and counted through the microscope (Olympus, Japan) [Citation18].
Transwell assay
Transwell chambers (pore size, 8 μM; Corning, NY, USA) pre-coated with Matrigel (for the invasion assay) or without coating (for the migration assay) were obtained from BD Biosciences (Franklin Lakes, NJ, USA) and were utilized for analyzing cell invasion and migration. At first, 200 μl of serum-free medium and constructed cells were plated into the upper chamber, and 600 μl of medium with 20% FBS was placed into the lower chamber. After being cultured for 48 h, the cells were fixed by 4% paraformaldehyde and stained by 0.1% crystal violet. Ultimately, migrated or invaded cells were observed through a microscope (Leica, Wetzlar, Germany).
Luciferase reporter assay
Bioinformatics tools were utilized to predict the putative binding sites for miR-326 on lncRNA LINC01270 (or EFNA3 3’-UTR). Wild-type (WT) and mutant (Mut) of LINC01270 (LINC01270-WT/Mut) or EFNA3 (EFNA3-WT/Mut) were incorporated into pmirGLO dual-luciferase reporters (Promega, Madison, USA). LINC01270-WT/Mut or EFNA3-WT/Mut reporters were then co-transfected with miR-326 mimic (or miR-NC) in GC cells. Then, 48 h after the transfection, the luciferase activity was examined through the luciferase reporter assay system (Promega) [Citation19].
RNA pull-down assay
The biotinylated LINC01270 (or EFNA3) was incubated with GC cell extracts and M-280 streptavidin beads (S3762; Sigma-Aldrich Chemical Company, St. Louis, MO, USA) [Citation20]. Finally, RT-qPCR was applied to evaluate the miR-326 expression.
Western blot analysis
GC cells were first lysed in RIPA lysis buffer. The isolated proteins were then separated by polyacrylamide gel electrophoresis (PAGE) followed by transfer to polyvinylidene fluoride (PVDF) membranes (Amersham, USA). The membrane was blocked by nonfat milk for 1 h. Thereafter, various primary antibodies were added into the membranes including EFNA3 (ab64814, 1/1000, Abcam) and β-actin (ab8226, 1 µg/mL, Abcam) followed by incubation for 12 h at 4°C. Next, horseradish peroxidase-labeled secondary antibody (ab6721, 1:2000, Abcam) was mixed with the membranes at room temperature for 2 h. After washing, the visualization of protein bands was done through the ECL-detection system (PerkinElmer, Boston, MA, USA).
In vivo assay
6-week-old male BALB/c nude mice (n=6) were gained from Charles River (Beijing, China). GC cells transfected with sh-NC and sh-LINC01270 were inoculated into nude mice subcutaneously. After 30 days, mice were killed. The tumor volume was examined, and was calculated as 1/2 × (length) × (width)2. The tumor weight was also examined. Our work had adopted the approval of the Animal Care and Use Committee from our hospital.
Immunohistochemistry (IHC) assay
The mice tissues were sliced in to sections and followed by dewaxing by xylene. After being sealed with goat serum, the primary antibody of Ki-67 (0.1 µg/mL, ab15580, Abcam) and samples were mixed at 4°C for a night. Then the HRP binding protein secondary antibody (Abcam) was incubated with the sections for 1 h, followed by staining with diaminobenzidine and hematoxylin. Images were acquired through a light microscope (Olympus BX40F, Olympus, Tokyo, Japan).
Statistical analysis
The data has been represented as the mean ± standard deviation (SD). All experiments were repeated three times. SPSS version 20.0 software (SPSS, Chicago, USA) was employed to analyze the data. The comparisons between the two groups (or among multiple groups) were performed through the Student’s t-test (or one-way ANOVA). The value of p < 0.05 was considered as statistically significant.
Results
The main objective of this study was to investigate the function and regulatory mechanism of lncRNA LINC01270 in GC. Our findings indicated that LINC01270 expression was up-regulated in GC tissues and cell lines. Moreover, LINC01270 knockdown attenuated GC progression through inhibiting cell proliferation, migration, and invasion. Functional experiments identified that lncRNA LINC01270 could positively regulate EFNA3 expression by serving as a ceRNA for miR-326. Through rescue assays, the inhibition of GC progression caused by LINC01270 suppression could be reversed by use of a miR-326 inhibitor or EFNA3 overexpression. The above findings revealed that lncRNA LINC01270 accelerated cell proliferation, migration, and invasion through modulating miR-326/EFNA3 axis in GC.
LINC01270 expression was up-regulated in GC
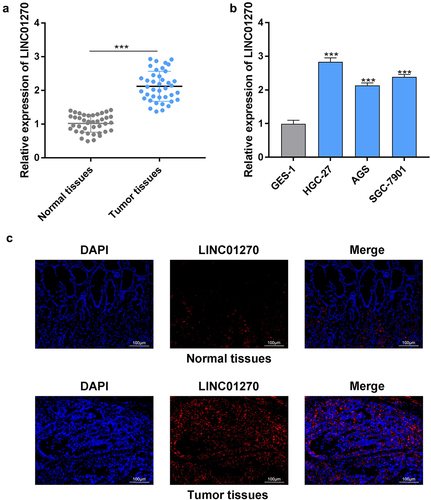
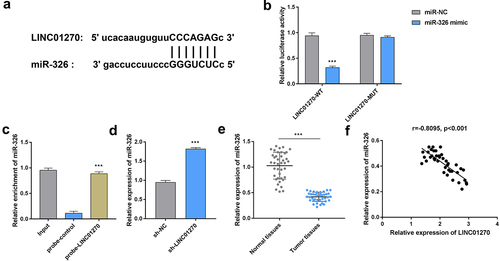
First, GC tissues were collected and the expression of LINC01270 was examined. The results from RT-qPCR demonstrated that LINC01270 exhibited higher expression in GC tissues compared with the normal adjacent tissues (). Additionally, LINC01270 expression was up-observed to be regulated in GC cell lines (HGC-27, AGS, and SGC-7901) in comparison with the gastric epithelial cell (GES-1) (). HGC-27 cell line was utilized for the rest of the experiments. Moreover, through RNA-FISH, it was demonstrated that LINC01270 was located in cytoplasm, and the expression of LINC01270 was enhanced in tumor tissues compared with the normal tissues (). Taken together, the increased expression of LINC01270 was found in GC.
Figure 1. LINC01270 expression was up-regulated in GC.
LINC01270 knockdown attenuated GC progression
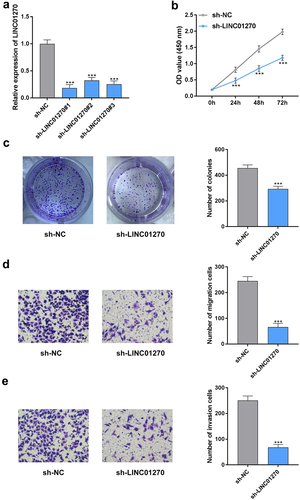
The knockdown efficiency of LINC01270 has been displayed in . The sh-LINC01270#1 with the highest knockdown efficiency was selected for the subsequent experiments. In addition, the cell proliferation ability was significantly reduced after silencing LINC01270 (). Moreover, both the rates of cellular migration and invasion were inhibited by suppressing LINC01270 (). Interestingly, similar changes in cell proliferation, migration, and invasion in AGS and SGC-7901 cells have been depicted in Figure S1A–C. These findings revealed that LINC01270 knockdown significantly abrogated GC progression.
Figure 2. LINC01270 knockdown retarded GC progression.
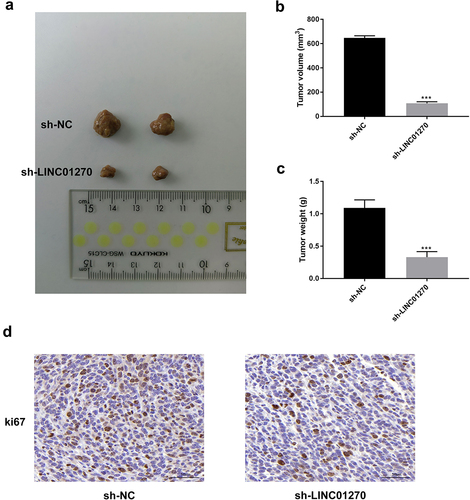
LINC01270 suppression reduced the tumor growth in vivo
As displayed in , the tumor size, volume, and weight were all markedly decreased after LINC01270 suppression. In addition, the Ki-67 expression was also reduced after silencing LINC01270 (). These findings indicated that LINC01270 suppression could effectively reduce the tumor growth in vivo.
LINC01270 sponged miR-326
Accumulating evidences have suggested that lncRNAs can act as decoys to sequester miRNAs, which can prevent them from binding to target mRNAs, and thereby regulating the functions of the various proteins through translation. Bioinformatics analysis was applied to predict the potential binding sequences between lncRNA LINC01270 and miR-326 (). The luciferase activity of LINC01270-WT reporter was found to be suppressed by miR-326 mimic (). However, the luciferase activity of LINC01270-WT reporter was not substantially affected by miR-326 mimic. As shown in , miR-326 in cell lysis was enriched by the LINC01270-specific probe. Additionally, the expression of miR-326 was found to be up-regulated after LINC01270 knockdown (). It was observed that miR-326 expression was significantly lower in GC tissues than that in the normal tissues (). Moreover, we discovered that LINC01270 expression was negatively correlated with miR-326 expression (). To sum up, LINC01270 could sponge and negatively regulate miR-326.
Figure 4. LINC01270 sponged miR-326.
Moreover, the cell proliferation was found to be increased in the pcDNA3.1/LINC01270-wt group but was not affected in the pcDNA3.1/LINC01270-mut. In addition, the miR-326 overexpression markedly reduced the increased cell proliferation caused by LINC01270-wt/mut overexpression (Figure S2A). Moreover, the cell migration and invasion was enhanced by overexpressing LINC01270-wt, but was not altered by overexpressing LINC01270-mut. Besides, the miR-326 overexpression was found to attenuate the enhanced cell migration and invasion mediated by LINC01270-wt/mut overexpression (Figure S2B-C). These results suggested that LINC01270 could directly bind to miR-326 to regulate GC progression, and miR-326 was not affected by LINC01270 to exhibit its anti-cancer functions.
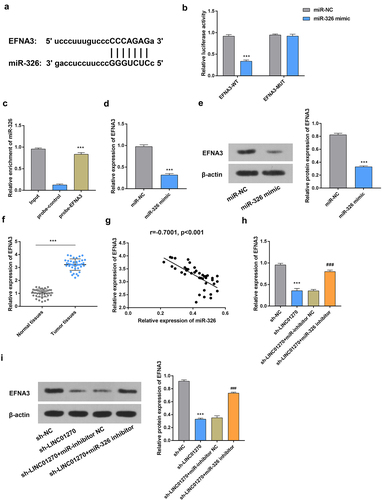
MiR-326 targeted and regulated EFNA3
Next, the binding sequences between miR-326 and EFNA3 have been shown in . In addition, the results of luciferase reporter and RNA pull-down assays indicated that miR-326 interacted with EFNA3 (). The mRNA and protein expressions of EFNA3 were decreased after overexpressing miR-326 (). The up-regulated expression of EFNA3 was found in GC tissues (). Furthermore, there was a negative correlation observed between miR-326 and EFNA3 expressions (). Both the mRNA and protein expressions of EFNA3 were significantly decreased after silencing LINC01270, but this effect was reversed by miR-326 inhibitor (). Taken together, miR-326 was found to target and regulate EFNA expression in GC.
Figure 5. MiR-326 targeted and regulated EFNA3.
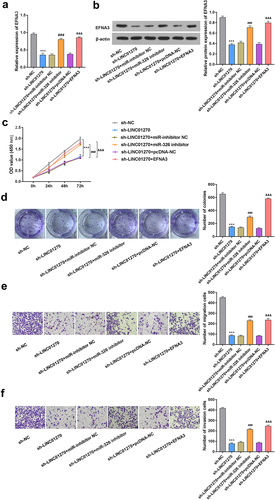
LINC01270 affected GC progression via miR-326/EFNA3 axis
The expression of EFNA3 at both mRNA and protein level was decreased after LINC01270 knockdown, but this effect was rescued by miR-326 inhibitor or EFNA3 overexpression (). Moreover, the decreased cell proliferation mediated by LINC01270 suppression was found to be reversed by miR-326 inhibitor or EFNA3 overexpression (). The reduced cell migration and invasion abilities caused by LINC01270 inhibition could be effectively counteracted by miR-326 inhibitor or EFNA3 overexpression (). These results indicated that LINC01270 can affect GC progression via modulating miR-326/EFNA3 axis.
Figure 6. LINC01270 affected GC progression via miR-326/EFNA3 axis.
Discussion
GC is a dreaded malignancy, and it is important to identify useful biomarkers for improving GC treatment. LncRNA LINC01270 has been implicated in the progression of both triple-negative breast and esophageal cancers [Citation14,Citation15], but the detailed role and regulatory mechanism of it in GC were not fully elucidated. In this study, we report for the first time that LINC01270 exhibited higher expression in GC tissues and cell lines. Additionally, LINC01270 knockdown attenuated GC progression through inhibiting cell proliferation, migration, and invasion.
MicroRNAs (miRNAs) are a kind of small-molecule RNAs with 18–24 nts length that can effectively bind with the mRNAs’ 3’-Untranslated Region (UTR) to modulate their expression [Citation21]. MiR-326 has been reported to target ZEB1 and mediate malignant phenotypes in lung adenocarcinoma [Citation22]. MiR-326 can affect Bcl-2 expression to modulate endometrial cancer cell proliferation and apoptosis [Citation23]. Additionally, miR-326 can target MDK to mediate the cardiac hypertrophy progression via inhibiting JAK/STAT and MAPK pathways [Citation24]. As such, miR-326 plays a pivotal regulatory role in the development of GC. For instance, circ_0000467/miR-326-3p can drive the progression of GC [Citation25]. Moreover, lncRNA DDX11-AS1/miR-326/IRS1 axis can contribute to cancer progression and oxaliplatin resistance in GC [Citation26]. Furthermore, miR-326 can effectively target FSCN1 to suppress the tumor growth and metastasis in GC [Citation27]. In addition, miR-326 can negative regulate NOB1 to retard GC cell growth [Citation28].
EphrinA3 (EFNA3) can play a pivotal role in tumorigenesis. For instance, EFNA3 can affect lung adenocarcinoma clinical prognosis and immune checkpoint therapy efficacy [Citation29]. MiR-210 has been found to target EFNA3 to accelerate the peripheral nerve sheath tumor cell proliferation and invasion [Citation30]. Additionally, OSCC exosomes can modulate miR-210-3p/EFNA3 axis to aggravate angiogenesis in oral cancer [Citation31].
A number of previous studies have proposed the hypothesis of competitive endogenous RNA (ceRNA), which revealed that endogenous lncRNAs could regulate mRNAs levels via sponging miRNAs [Citation32,Citation33]. Moreover, multiple evidences have illustrated that this ceRNA network can participate in the process of GC development. For example, lncRNA DLGAP1-AS1/miR-628-5p/AEG-1 ceRNA axis can facilitate the aggressive behavior in GC [Citation34]. In addition, lncRNA TINCR can act as a ceRNA to modulate PDK1 expression by absorbing miR-375 in GC [Citation35]. LncRNA UCA1 can sponge miR-495 to affect PRL-3 expression and thereby aggravate GC progression [Citation36]. Nevertheless, whether lncRNA LINC01270/miR-326/EFNA3 can regulate GC progression is largely unknown. Thus, we have first identified that lncRNA LINC01270 can positively regulate EFNA3 expression by serving as a ceRNA for miR-326. Finally, rescue assays revealed that inhibition of GC progression caused by LINC01270 suppression could be effectively neutralized by miR-326 inhibitor or EFNA3 overexpression.
Conclusion
We have demonstrated the potential role of ceRNA axis (LINC01270/miR-326/EFNA3) in GC progression. In summary, this study establishes that lncRNA LINC01270/miR-326/EFNA3 axis can aggravate GC progression, which might provide novel insights on the possible role of lncRNA LINC01270 in GC treatment.
Supplemental Material
Download Zip (2.5 MB)Data availability statement
The authors confirm that the data supporting the findings of this study are available within the article.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary material
Supplemental data for this article can be accessed here
Additional information
Funding
References
- Correa P. Gastric cancer: overview. Gastroenterol Clin North Am. 2013;42(2):211–217.
- Guggenheim DE, Shah MA. Gastric cancer epidemiology and risk factors. J Surg Oncol. 2013;107(3):230–236.
- Karimi P, Islami F, Anandasabapathy S, et al. Gastric cancer: descriptive epidemiology, risk factors, screening, and prevention. Cancer Epidemiol Biomarkers Prev. 2014;23(5):700–713.
- Song Z, Wu Y, Yang J, et al. Progress in the treatment of advanced gastric cancer. Tumour Biol. 2017;39(7):1010428317714626.
- Digklia A, Wagner AD. Advanced gastric cancer: current treatment landscape and future perspectives. World J Gastroenterol. 2016;22(8):2403–2414.
- Sexton RE, Al Hallak MN, Diab M, et al. Gastric cancer: a comprehensive review of current and future treatment strategies. Cancer Metastasis Rev. 2020;39(4):1179–1203.
- Zou T, Wang PL, Gao Y, et al. Long noncoding RNA HOTTIP is a significant indicator of ovarian cancer prognosis and enhances cell proliferation and invasion. Cancer Biomark. 2019;25(2):133–139.
- Lei GL, Niu Y, Cheng S-J, et al. Upregulation of long noncoding RNA W42 promotes tumor development by binding with DBN1 in hepatocellular carcinoma. World J Gastroenterol. 2021;27(20):2586–2602.
- Xu W, Xu Q, Kuang D, et al. Long non‑coding RNA SLNCR1 regulates non‑small cell lung cancer migration, invasion and stemness through interactions with secretory phospholipase A2. Mol Med Rep. 2019;20(3):2591–2596.
- Guo Z, Zhang J, Fan L, et al. Long noncoding RNA (lncRNA) small nucleolar RNA host gene 16 (SNHG16) predicts poor prognosis and sorafenib resistance in hepatocellular carcinoma. Med Sci Monit. 2019;25:2079–2086.
- Li H, Wang Y. Long noncoding RNA (lncRNA) MIR22HG suppresses gastric cancer progression through attenuating NOTCH2 signaling. Med Sci Monit. 2019;25:656–665.
- Wang S, Wang Y, Zhang Z, et al. Long non-coding RNA NRON promotes tumor proliferation by regulating ALKBH5 and nanog in gastric cancer. J Cancer. 2021;12(22):6861–6872.
- Zhao W, Jin Y, Wu P, et al. LINC00355 induces gastric cancer proliferation and invasion through promoting ubiquitination of P53. Cell Death Discov. 2020;6(1):99.
- Ping J, Huang S, Wu J, et al. Association between lincRNA expression and overall survival for patients with triple-negative breast cancer. Breast Cancer Res Treat. 2021;186(3):769–777.
- Li N, Zhao Z, Miao F, et al. Silencing of long non-coding RNA LINC01270 inhibits esophageal cancer progression and enhances chemosensitivity to 5-fluorouracil by mediating GSTP1methylation. Cancer Gene Ther. 2021;28(5):471–485.
- Liu S, Xie X, Lei H, et al. Identification of key circRNAs/lncRNAs/miRNAs/mRNAs and pathways in preeclampsia using bioinformatics analysis. Med Sci Monit. 2019;25:1679–1693.
- Zhang Q, Zheng J, Liu L. The long noncoding RNA PCGEM1 promotes cell proliferation, migration and invasion via targeting the miR-182/FBXW11 axis in cervical cancer. Cancer Cell Int. 2019;19(1):304.
- Wang Y, Lei X, Gao C, et al. MiR-506-3p suppresses the proliferation of ovarian cancer cells by negatively regulating the expression of MTMR6. J Biosci. 2019;44(6). DOI:10.1007/s12038-019-9952-9.
- Luo J, Wang K, Yeh S, et al. LncRNA-p21 alters the antiandrogen enzalutamide-induced prostate cancer neuroendocrine differentiation via modulating the EZH2/STAT3 signaling. Nat Commun. 2019;10(1):2571.
- Xue K, Li J, Nan S, et al. Downregulation of LINC00460 decreases STC2 and promotes autophagy of head and neck squamous cell carcinoma by up-regulating microRNA-206. Life Sci. 2019;231:116459.
- Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79(1):351–379.
- Liu M, Wu H, Liu Y, et al. MiR-326 mediates malignant biological behaviors of lung adenocarcinoma by targeting ZEB1. Sci Prog. 2021;104(2):368504211009379.
- Cai L, Chen -J-J, Deng F-M, et al. MiR-326 regulates the proliferation and apoptosis of endometrial cancer by targeting Bcl-2. J Obstet Gynaecol Res. 2021;47(2):621–630.
- Zhang\ J, Wei X, Zhang W, et al. MiR-326 targets MDK to regulate the progression of cardiac hypertrophy through blocking JAK/STAT and MAPK signaling pathways. Eur J Pharmacol. 2020;872:172941.
- Mo WL, Jiang JT, Zhang L, et al. Circular RNA HSA_circ_0000467 promotes the development of gastric cancer by competitively binding to MicroRNA miR-326-3p. Biomed Res Int. 2020;2020:4030826.
- Song W, Qian Y, Zhang M-H, et al. The long non-coding RNA DDX11-AS1 facilitates cell progression and oxaliplatin resistance via regulating miR-326/IRS1 axis in gastric cancer. Eur Rev Med Pharmacol Sci. 2020;24(6):3049–3061.
- Li Y, Gao Y, Xu Y, et al. Down-regulation of miR-326 is associated with poor prognosis and promotes growth and metastasis by targeting FSCN1 in gastric cancer. Growth Factors. 2015;33(4):267–274.
- Ji S, Zhang B, Kong Y, et al. miR-326 inhibits gastric cancer cell growth through downregulating NOB1. Oncol Res. 2017;25(6):853–861.
- Deng M, Tong R, Zhang Z, et al. EFNA3 as a predictor of clinical prognosis and immune checkpoint therapy efficacy in patients with lung adenocarcinoma. Cancer Cell Int. 2021;21(1):535.
- Wang Z, Yin B, Wang B, et al. MicroRNA-210 promotes proliferation and invasion of peripheral nerve sheath tumor cells targeting EFNA3. Oncol Res. 2013;21(3):145–154.
- Wang H, Wang L, Zhou X, et al. OSCC exosomes regulate miR-210-3p targeting EFNA3 to promote oral cancer angiogenesis through the PI3K/AKT pathway. Biomed Res Int. 2020;2020:2125656.
- Qi X, Zhang D-H, Wu N, et al. ceRNA in cancer: possible functions and clinical implications. J Med Genet. 2015;52(10):710–718.
- Karreth FA, Pandolfi PP. ceRNA cross-talk in cancer: when ce-bling rivalries go awry. Cancer Discov. 2013;3(10):1113–1121.
- Deng J, Zhang Q, Lu L, et al. Long noncoding RNA DLGAP1-AS1 promotes the aggressive behavior of gastric cancer by acting as a ceRNA for microRNA-628-5p and raising astrocyte elevated gene 1 expression. Cancer Manag Res. 2020;12:2947–2960.
- Chen Z, Liu H, Yang H, et al. The long noncoding RNA, TINCR, functions as a competing endogenous RNA to regulate PDK1 expression by sponging miR-375 in gastric cancer. Onco Targets Ther. 2017;10:3353–3362.
- Cao Y, Xiong J-B, Zhang G-Y, et al. Long noncoding RNA UCA1 regulates PRL-3 expression by sponging MicroRNA-495 to promote the progression of gastric cancer. Mol Ther Nucleic Acids. 2020;19:853–864.