ABSTRACT
Recently, kinesin family member 21B (KIF21B) has been reported to be an oncogene in non-small cell lung cancer and hepatocellular carcinoma. However, the functional role of KIF21B and related molecular mechanisms in gastric cancer (GC) remain largely uncovered. In this study, online bioinformatics analysis showed that KIF21B was overexpression in GC and predicted poor prognosis. Consistently, we found that the protein expression of KIF21B was upregulated in GC tissues compared with adjacent tissues by immunohistochemistry. Knockdown of KIF21B significantly suppressed cell proliferation, migration and invasion in GC cell lines (AGS and SNU-5) using Cell counting kit‑8 (CCK-8) assay, colony formation and transwell assay. KIF21B was confirmed as the target of miR-132-3p in GC cells by luciferase reporter assay. Moreover, miR-132-3p was down-regulated and KIF21B expression was upregulated in GC tissues. Overexpression of KIF21B reversed the miR-132-3p-mediated suppressive effects on GC cell proliferation, migration and invasion. Furthermore, miR-132-3p overexpression downregulated the protein levels of Wnt1, c-Myc, β-catenin, proliferating cell nuclear antigen (PCNA) and N-cadherin, and upregulated E-cadherin expression in GC cells, which were all alleviated after KIF21B overexpression. In conclusion, our findings indicate that down-regulation of KIF21B by miR-132-3p suppresses cellular functions in GC, which might be linked to reduced Wnt/β-catenin signaling.
Graphical abstract
Introduction
Gastric cancer (GC) is considered as the third leading cause of tumor-associated death worldwide, remaining a global health problem [Citation1,Citation2]. Despite advances in comprehensive therapies, including surgery, adjuvant chemotherapy and radiation interventions, the prognosis of GC patients remains poor primarily ascribed to uncontrolled metastasis [Citation3,Citation4]. Therefore, having a better understanding the molecular mechanisms associated with GC pathogenesis will be of great importance to improve the clinical outcomes of GC patients.
Kinesin superfamily proteins (KIFs), as microtubule-dependent molecular motors, play an important role in a series of cellular processes, including mitosis, motility and organelle transportation [Citation5–7]. KIFs have been recently found to be aberrantly expressed and participate in tumorigenesis by modulating chromosomal and spindle movement [Citation8–10]. As a member of the KIFs, kinesin family member 21B (KIF21B), located at 1q32.1, contains a motor domain, a stalk and a tail domain binding to microtubules, which is essential for neuronal morphology [Citation11]. Interestingly, accumulating evidence has demonstrated that KIF21B was involved in tumorigenesis. For instance, increased KIF21B expression is correlated with severe multiple sclerosis and Alzheimer’s disease pathology [Citation12,Citation13]. Zhao et al. [Citation14] showed that KIF21B expression level was associated with overall survival and disease-free survival in hepatocellular carcinoma. Sun et al. [Citation15] not only manifested that increased KIF21B expression was correlated with worse survival prognosis, but also demonstrated that KIF21B promoted cell proliferation and migration in non-small cell lung cancer (NSCLC). However, little is known about the functional role of KIF21B in GC.
MicroRNAs (miRNAs/miRs), a class of small non-coding RNAs, have been frequently identified as pivotal regulators in tumorigenesis though modulating various biological processes, including proliferation, invasion and differentiation via binding to the 3′-untranslated region (3′-UTR) of target genes [Citation16–19]. In recent years, miR-132-3p has been reported to be aberrantly expressed in tumor tissues and serves an important regulator in tumor cellular functions. For example, miR-132-3p suppressed breast carcinoma cell migration and invasion by targeting lysosomal-associated protein transmembrane 4 beta (LAPTM4B) [Citation20]. Ectopic miR-132-3p aggravated cell apoptosis and inhibited cell proliferation in mantle cell lymphoma [Citation21]. In addition, miR-132-3p functions as a tumor suppressor in colorectal cancer [Citation22,Citation23], osteosarcoma [Citation24,Citation25] and bladder carcinoma [Citation26]. Here, our previous investigation revealed that KIF21B was a potential target gene of miR-132-3p. Moreover, Zhang et al. [Citation27] and Wang et al. [Citation28] consistently demonstrated that miR-132-3p expression was significantly altered between tumor tissues and normal tissues derived from GC patients by microRNA profiling. Based on these evidences, we speculated that miR-132-3p/KIF21B axis plays a critical role in regulating cellular functions in GC cells.
To validate our hypothesis, we first determined the expression pattern of KIF21B in GC tissues by searching Oncomine database and collecting clinical specimens. Then, loss-of-function assays were performed to analyze the functional role of KIF21B in GC cells. Furthermore, we further explored whether KIF21B was the downstream regulator involved in miR-132-3p regulating GC cell functions
Materials and methods
Oncomine gene expression analysis
We first examined the expression of KIF21B gene in GC by searching microarray gene expression datasets derived from Oncomine database (www.oncomine.com). In brief, the Cancer Type was defined as Gastric Cancer, Data Type was mRNA, and Analysis Type was Cancer versus Normal Analysis. Total three datasets, including Wang Gastric [Citation29], Chen Gastric [Citation30] and DErrico Gastric [Citation31] were included in our analysis. The log-transformed, median-centered and normalized expression values of KIF21B were extracted, analyzed and read from the scatterplot accordingly.
Online data analysis
The analysis of relative KIF21B expression was performed using UALCAN (http://ualcan.path.uab.edu), a user-friendly, interactive web resource for analyzing cancer transcriptome data (TCGA and MET500 transcriptome sequencing) [Citation32]. The prognostic value of KIF21B expression in stomach adenocarcinoma (STAD) patients was evaluated using Kaplan–Meier Plotter database (http://kmplot.com/analysis/) by dividing all patients into high and low KIF21B expression group based on median KIF21B expression. Briefly, the overall survival information was extracted, which was applied to analyze the effect of KIF21B expression on the survival rate of GC patients by a Kaplan–Meier survival plot. The hazard ratio (HR) with 95% confidence intervals and log-rank P-value were computed.
Tissue specimens
Total 30 pairs of fresh tumor tissues and matched adjacent normal stomach mucosa tissues were collected from GC patients who received radical gastrectomy at the Second Hospital, Cheeloo College of Medicine, Shandong University (Shandong Province, China). All patients did not receive radiotherapy and chemotherapy before surgery. In addition, paraffin-embedded specimens of GC were also obtained to evaluate the KIF21B protein expression. Informed consent was obtained from all patients. This study was conducted in accordance with Helsinki Declaration and approved by the Medical Ethics Committee of the Second Hospital, Cheeloo College of Medicine, Shandong University (Approval number: CCM32-J2; 2018.6.3; Shandong, China).
Immunohistochemistry
Immunohistochemistry staining was performed to assess the protein expression of KIF21B in tissue samples using the EliVisionTMplus kit (Maixin, Fuzhou, China) according to the instructions provided. Briefly, paraffin-embedded tissues were sliced into 5-μm thick sections. The tissue sections were deparaffinized in xylene and rehydrated in gradient ethanol. After subjected to antigen retrieval by heated citrate buffer, sections were blocked in 3% (v/v) hydrogen peroxide for 30 min and incubated overnight at 4°C with anti-KIF21B antibody (ab121931; Abcam, Cambridge, UK). Afterward, the sections were washed with twice with Tris-buffered saline and then incubated with horseradish peroxidase-conjugated secondary antibody for 2 h at room temperature. The immunohistochemistry staining results were evaluated by two independent pathologists according to the multiplication of staining proportion score (0–4: 0, 0–5%; 1, 6–20%; 2, 21–60%; 3, 61–75%; or 4, 76–100%) and staining intensity score (0, no staining; 1, weak; 2, moderate; 3, strong staining) as the final score of KIF21B protein expression. Tissue sections with immunoreactivity score scaling 0–2, 2–4 and over 4 were considered to be weak staining (-+), moderate staining (+) and strong staining (++), respectively.
Cell culture and transfection
Two GC cell lines, including AGS and SNU-5 were purchased from the Cell Bank of China Academy of Sciences (Shanghai, China) and cultured RPMI-1640 medium (Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS, Gibco) at 37 C in the atmosphere containing 5% CO2. The sequences of small interfering RNA targeting KIF21B (si-KIF21B#1 and si-KIF21B#2), si-NC, miR-132-3p mimics and miR-NC were synthesized by GenePharma Co., Ltd. (Shanghai, China). The pcDNA3.1 containing the open reading frame of KIF21B or empty vector were purchased from GenScript (Nanjing, China). After incubating GC cells in 12-well plates at a density of 8 × 104 cells per well, the cells were transfected with the above oligonucleotides for 48 h using Lipofectamine 2000 (Thermo Fisher Scientific, Inc.).
Reverse transcription quantitative PCR analysis
Total RNA sample was extracted from tissue specimens or cell lines using TRIzol reagent (Invitrogen). Reverse transcription was conducted by One Step PrimeScript miRNA complementary DNA (cDNA) Synthesis Kit (Takara, Dalian, China) for miR-132-3p and a HiFiScript cDNA Synthesis Kit (CWBIO, Beijing, China) for KIF21B. Next, reverse transcription quantitative PCR was performed to determine the expression of miR-132-3p and KIF21B using SYBR Green Human miRNA Assay Kit and a SYBR Premix Ex Taq II kit (Takara, Japan), respectively. The primer sequences were used as follows: miR-132-3p (forward: 5ʹ-GCGCGCGTAACAGTCTACAGG-3ʹ and reverse: 5ʹ-GTCGTATCCAGTGCAGGGTCC-3ʹ); U6 (forward: 5ʹ-CTCGCTTCGGCAGCACATATACT-3ʹ and reverse: 5ʹ-CGCTTCACGAATTTGCGTGT-3ʹ); KIF21B (forward: 5′-CGA GGAGACGGATGAGAACG-3′ and reverse, 5′-CCACCAGGCTCTCTTCACTG-3′); β-actin (forward: 5′-CCCGAGCCGTGTTTCCT-3′ and reverse: 5′-GTCCCAGTTGGT GACGATGC-3′). Relative miR-132-3p and KIF21B mRNA expression were normalized against the endogenous control U6 and β-actin, respectively, using the 2− ΔΔCt method [Citation33].
Cell counting kit‑8 (CCK-8) assay
Transfected cells at a density of 4 × 103 cells per well were seeded into 96-well plates and cultured at 37°C. At 0, 24, 48 and 72 h, respectively, cells in each well were incubated with 10 μl of CCK-8 solution (Solarbio Science & Technology, Beijing, China) for another 2 h at 37°C. Afterward, the optical density (OD) value at 450 nm was measured with a microplate reader.
Colony formation assay
Transfected cells at the logarithmic growth phase were seeded onto six-well plates. After culture for 2 weeks, the cells were fixed with 4% paraformaldehyde and stained with crystal violet. Images were obtained and cells were counted.
Transwell migration and invasion assay
For migration assay, approximately 4 × 104 transfected cells in 200 µl of serum-free medium were plated into the upper chamber (24-well insert; 8-µm pore size; Corning Costar, Corning, NY, USA), while 600 µl complete medium (with 10% FBS) as chemoattractant was added into the lower chamber. After 24 h incubation, the cells that migrated to the lower chamber were fixed with 4% paraformaldehyde for 30 min and stained with 0.1% crystal violet for 10 min. The number of migrated cells was quantified under a microscope (magnification, ×200) in five random microscope fields. The procedure of invasion assay was similar to the migration assay except for the addition of 50 µl Matrigel on the membranes of transwell inserts and incubation time of 48 h.
Bioinformatic analysis and luciferase reporter assay
The potential miRNAs that target KIF21B were predicted by TargetScan (www.targetscan.org/vert_71/). From all these predictions, we selected miR-132-3p for further analysis. The mRNA 3′-UTR of KIF21B, carrying the predicted binding site or mutant binding site of miR-132-3p, was amplified by PCR and cloned into pmirGLO (Promega, Madison, Wisconsin), which were named as KIF21B wild-type (WT) and mutant (MUT) constructs, respectively. Next, cells were co-transfected with KIF21B WT or KIF21B MUT and miR-132-3p mimics or miR-NC using the transfection reagent Lipofectamine 2000. After 48 h incubation, cells were harvested and relative luciferase activity was measured via a Dual-luciferase reporter assay system (Promega).
Western blot analysis
Total protein sample was extracted with RIPA lysis buffer with protease inhibitor (Solarbio Science & Technology Co., Ltd., Beijing, China), and protein concentration was determined by a BCA kit (Beyotime, Shanghai, China). Then, equal amount of protein sample was separated by 10% SDS-PAGE and transferred onto PVDF membranes (Millipore, Billerica, MA, USA). After blocked with 5% nonfat milk, the membranes were incubated with primary antibodies against KIF21B, Wnt1, c-Myc, β-catenin, PCNA, E-cadherin, N-cadherin and GAPDH (All from Abcam, Cambridge, UK) at 4°C overnight. Subsequently, the membranes were incubated with horseradish peroxidase-coupled secondary antibody for 2 h at room temperature, followed by detection of protein signals with an enhanced chemiluminescence reagent (EMD Millipore, Billerica, MA, USA).
Statistical analysis
All quantitative data were presented as mean ± standard deviation (SD) from three independent experiments. All statistical analyses were performed using GraphPad Prism 6.0 (GraphPad Software, Inc., San Diego, California, USA). Differences between two groups were assessed by paired Student’s t-test. Differences among three groups were evaluated by one-way analysis of variance (ANOVA), followed by Dunnett’s test or Tukey’s test. The p-value less than 0.05 was considered to be statistically significant.
Results
Here, we hypothesized that miR-132-3p suppressed GC cell proliferation, migration and invasion by inhibiting KIF21B. First, the expression and survival prognosis of KIF21B were assessed through online bioinformatics analysis. Second, we investigated the effects of KIF21B on the proliferation, migration and invasion of GC cells. Then, we validated the target binding between miR-132-3p and KIF21B. Moreover, miR-132-3p was found to be abnormally expressed in GC tissues. In addition, we evaluated whether miR-132-3p regulated GC cellular functions, Wnt/β-catenin signaling and epithelial–mesenchymal transition (EMT) process by targeting KIF21B. The results suggest that targeting the miR-132-3p/KIF21B axis may be a new therapeutic strategy for GC.
KIF21B expression level was upregulated in GC tissues
We first searched the Oncomine microarray gene expression datasets to investigate the expression pattern of KIF21B in GC. As depicted in ), KIF21B expression level was significantly increased in gastric cancer tissues (p = 7.73E-4) compared with the corresponding normal tissues in the Wang Gastric dataset, in diffuse gastric adenocarcinoma (p = 0.002) and gastric intestinal-type adenocarcinoma (p = 0.005) compared with normal tissues in Chen Gastric dataset, as well as in gastric intestinal-type adenocarcinoma (p = 0.010) and gastric mixed adenocarcinoma (p = 0.016) compared with normal tissues in DErrico Gastric dataset. To confirm the upregulation of KIF21B in GC, reverse transcription quantitative PCR was performed to analyze the expression of KIF21B mRNA in 30 pairs of fresh GC tissue and matched adjacent normal tissues. We then examined the expression of KIF21B between STAD and normal gastric tissues in UALCAN database, discovering it was highly expressed in tumor tissues ()). Moreover, Kaplan–Meier plotter database was used to evaluate the prognostic value of KIF21B in GC. As shown in ), higher KIF21B expression was correlated with the poor survival prognosis in GC (HR = 1.31, 95% CI = 1.03 to 1.66, p = 0.026). Moreover, IHC analysis presented significantly elevated moderate and strong staining of KIF21B in GC tissues compared with adjacent normal tissues ().
Figure 1. Expression of KIF21B mRNA in GC tissues; (a) The mRNA expression of KIF21B in Oncomine datasets including Wang Gastric, Chen Gastric and DErrico Gastric. (b) KIF21B expression level comparison between normal and tumor tissues obtained from the UALCAN web tool (Wilcoxon test). TCGA: The Cancer Genome Atlas; STAD: stomach adenocarcinoma. (c) Kaplan-Meier survival curves comparing the high and low expression of KIF21B in GC in Kaplan-Meier plotter database.
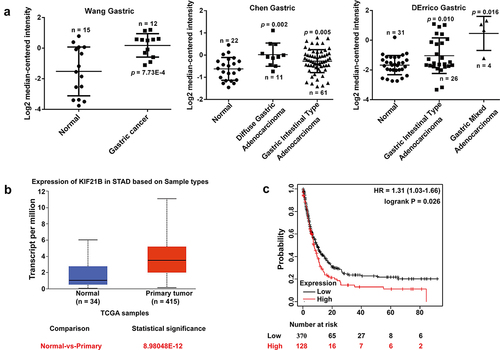
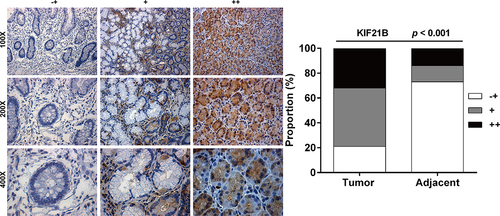
Figure 2. KIF21B protein level was highly expressed in GC tissues. Representative images of different degrees of KIF21B immunohistochemistry staining (-+, weak staining, + moderate staining, ++ strong staining) (left panel) and the percentage of KIF21B high in GC tissues was significantly higher than that in adjacent normal tissues (right panel).
Knockdown of KIF21B inhibited CC cell proliferation, migration and invasion
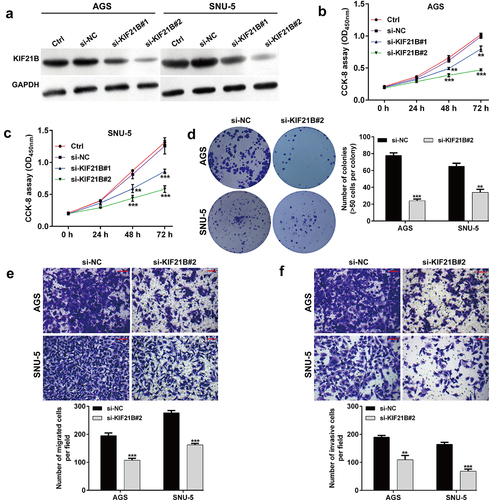
Next, we explored the functional role of KIF21B in GC cells by performing loss-of-function assays in AGS and SNU-5 cells. At first, the protein expression of KIF21B was suppressed by transfecting with two different siRNAs in AGS and SNU-5 cells, as demonstrated by western blot analysis ()). CCK-8 assay showed that either si-KIF21B#1 or si-KIF21B#2 transfection significantly suppressed cell proliferation ability in AGS and SNU-5 cells (). Notably, si-KIF21B#2 generated more powerful suppressive effects on KIF21B expression and cell proliferation ability, in comparison with si-KIF21B#1, which was thus selected for the subsequent in vitro experiments. Consistent with CCK-8 assay, colony formation assay further confirmed the suppressive effects of si-KIF21B#2 on cell proliferation in AGS and SNU-5 cells ()). Subsequently, transwell assay was applied to analyze the effects of KIF21B knockdown on GC cell migration and invasion. Our data showed that the number of migratory ()) and invasive ()) cells was remarkedly decreased in si-KIF21B#2 group compared with si-NC group in both AGS and SNU-5 cells.
Figure 3. Effects of KIF21B knockdown on cell proliferation, migration and invasion in GC cells. (a) Western blot analysis was performed to detect KIF21B protein expression in AGS and SNU-5 cells after transfection with si-NC, si-KIF21B#1 or si-KIF21B#2 for 48 h. (b-c) CCK-8 assay was used to evaluate cell proliferation ability in AGS and SNU-5 cells. (d) Colony formation was assessed in AGS and SNU-5 cells. (e) Cell migration and (f) invasion in the analyzed groups. Magnification, ×200; scale bar, 100 μm. All data are expressed as the means ± SD. **p < 0.01, ***p < 0.001, compared with si-NC group.
KIF21B as a target gene of miR-132-3p
TargetScan 7.1 database was used to predict miRNAs that target KIF21B expression, which led to the identification of miR-132-3p as a direct target gene. As depicted in ), miR-132-3p exhibited complementary binding to the 3′-UTR of KIF21B. Then, the results from luciferase reporter assay showed that miR-132-3p mimics significantly suppressed the luciferase activity in AGS ()) and SNU-5 ()) cells transfected with KIF21B WT plasmid. However, the luciferase activity remained unchanged when the cells were transfected with KIF21B MUT without the miR-132-3p binding sites. What’s more, miR-132-3p overexpression significantly downregulated the expression levels of KIF21B mRNA ()) and protein ()) in both AGS and SNU-5 cells. These findings suggested that miR-132-3p could negatively regulate KIF21B expression through binding its 3′-UTR. In addition, we analyzed the expression of miR-132-3p in GC tissues. As shown in ), miR-132-3p expression level was significantly down-regulated in GC tissues compared to that in adjacent tissues. On the contrast, the expression of KIF21B mRNA was significantly upregulated in GC tissues compared to that in adjacent tissues ()).
Figure 4. KIF21B was a target gene of miR-132-3p. (a) The potential binding sites of miR-132-3p and KIF21B mRNA, as well as the sequences in potential binding sites of mutant-type plasmid. Luciferase reporter assays were performed in (b) AGS and (c) SNU-5 cells with vectors including the putative miR-132-3p target sites in the 3’-UTR of KIF21B mRNA (wild type) and mutant-type. Data were normalized by Renilla/firefly luciferase activity. (d) Reverse transcription quantitative PCR and (e) western blot analysis was used to determine the expression levels of KIF21B in AGS and SNU-5 cells transfected with miR-132-3p mimics or miR-NC. (f) Lower expression of miR-132-3p was observed in 30 pairs of GC tissues compared with matched adjacent tissues by reverse transcription quantitative PCR. (g) Expression of KIF21B mRNA was determined using reverse transcription quantitative PCR in 30 pairs of GC tissues and adjacent tissues. All data are expressed as the means ± SD. **p < 0.01, compared with miR-NC group.
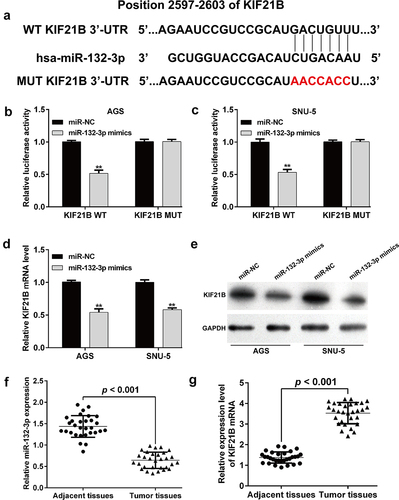
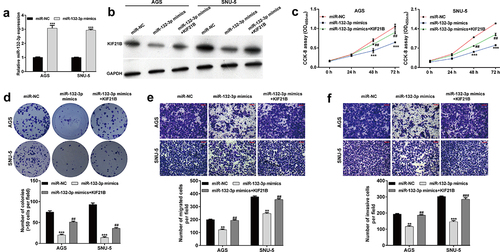
KIF21B overexpression reversed the suppressive effects of miR-132-3p on GC cells
Since miR-132-3p was identified to target KIF21B and negatively correlated with KIF21B expression in GC tissues, we thus explored whether miR-132-3p regulated GC cellular functions by targeting KIF21B. At first, the expression of miR-132-3p was overexpressed in AGS and SNU-5 cells after transfection with miR-132-3p mimics, as demonstrated by reverse transcription quantitative PCR ()). Then, AGS and SNU-5 cells were co-transfected with miR-132-3p mimics and KIF21B. As shown in ), downregulated KIF21B protein level induced by miR-132-3p overexpression was attenuated after KIF21B overexpression plasmid transfection in both AGS and SNU-5 cells. Furthermore, ectopic KIF21B expression effectively reversed miR-132-3p overexpression-mediated suppressive effects on cell proliferation ()), colony formation ()), migration ()) and invasion ()) abilities in AGS and SNU-5 cells.
Figure 5. KIF21B alleviated the effects of miR-132-3p on cell proliferation, migration and invasion in GC cells. AGS and SNU-5 cells were transfected with miR-NC, miR-132-3p mimics or miR-132-3p mimics + KIF21B, respectively for 48 h. (a) Reverse transcription quantitative PCR was used to determine miR-132-3p expression with U6 as internal control. (b) Western blot was used to determine the expression levels of KIF21B with GAPDH as a loading control. (c) CCK-8 assay and (d) colony formation assay was utilized to determine the cell proliferation in different groups. (e) Cell migration and (f) invasion ability was assessed by transwell assay in the above transfected cells. Magnification, ×200; scale bar, 100 μm; All data are expressed as the means ± SD. **p < 0.01, ***p < 0.001, compared with miR-NC group; ##p < 0.01, ###p < 0.001, compared with miR-132-3p mimics group.
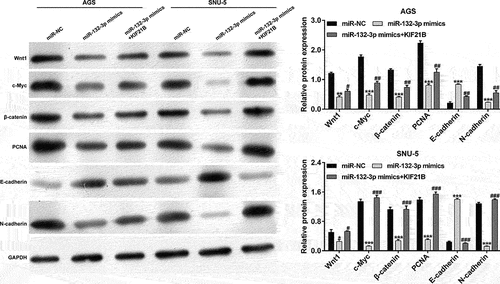
The effects of miR-132-3p/KIF21B axis on Wnt/β-catenin signaling and EMT process in GC cells
It has been reported that Wnt/β-catenin signaling pathway participates in regulating tumor cell proliferation and migration. Then, we investigated whether Wnt/β-catenin signaling pathway was involved in miR-132-5p-mediated suppressive effects on GC cells. As shown in , miR-132-3p overexpression obviously suppressed the expression of Wnt1, c-Myc and β-catenin, which was reversed after KIF21B overexpression in AGS and SNU-5 cells. In addition, we found that miR-132-3p overexpression downregulated the protein expression of PCNA and N-cadherin, and upregulated E-cadherin expression in AGS and SNU-5 cells, which was reversed after co-transfection with miR-132-3p mimics and KIF21B.
Figure 6. The effects of miR-132-3p/KIF21B axis on Wnt/β-catenin signaling and EMT process in GC cells. AGS and SNU-5 cells were transfected with miR-NC, miR-132-3p mimics or miR-132-3p mimics plus KIF21B, respectively. Western blot assay was used to compare the protein levels of Wnt1, c-Myc, β-catenin, PCNA, E-cadherin and N-cadherin in transfected AGS and SNU-5 cells. GAPDH was used as an internal control. All data are expressed as the means ± SD. **p < 0.01, ***p < 0.001, compared with miR-NC group; #p < 0.05, ##p < 0.01, ###p < 0.001, compared with miR-132-3p mimics group.
Discussion
In the current study, we first observed that KIF21B expression was upregulated in GC tissues compared with adjacent normal tissues by analysis of the Oncomine microarray gene expression datasets and clinical specimens. Next, we demonstrated that silencing of KIF21B suppressed the proliferation, migration and invasion of AGS and SNU-5 cells. Actually, KIFs play crucial roles in mitotic cell division, which make them involved in tumorigenesis [Citation34,Citation35]. Similarly, upregulation of kinesin family member 14 (KIF14) could promote cancer metastasis in gastric cancer [Citation36] and promote cell proliferation in colorectal cancer [Citation37]. Kinesin family member 11 (KIF11) knockdown significantly inhibited cell proliferation, migration and invasion in breast cancer [Citation38]. Knockdown of kinesin family member 23 (KIF23) resulted in a marked inhibition of cell proliferation in GC, with significant downregulation of Ki67 and PCNA expression [Citation39]. Consistent with our data, KIF21B was upregulated in hepatocellular carcinoma and NSCLC tissues, which promoted the corresponding tumor cell proliferation, migration and invasion [Citation14,Citation15]. Low KIF21B expression indirectly increased the apoptosis and inhibited the proliferation of osteosarcoma cells [Citation40]. A recent study by Yang et al. [Citation41] also indicated that the overexpression of KIF21B was associated with the immunological and oncogenic pathway activation in pancreatic cancer. Herein, our data highlighted the oncogenic role of KIF21B in GC.
We further demonstrated that KIF21B was a potential target of miR-132-3p in AGS and SNU-5 cells. What’s more, decreased miR-132-3p expression was inversely correlated with KIF21B mRNA levels in 30 cases of GC tissues. The rescue experiments manifested that KIF21B overexpression significantly alleviated the suppressive effects of miR-132-3p on GC cell proliferation, migration and invasion. These data suggested that miR-132-3p inhibited the cellular functions by directly targeting KIF21B in GC cells. In fact, miR-132-3p expression was frequently downregulated in human cancers, including colorectal cancer [Citation42], pancreatic carcinoma [Citation43] and glioma [,Citation44]. Functionally, the suppressive effects of miR-132-3p on cell proliferation, migration and invasion have been reported in breast cancer [Citation20], mantle cell lymphoma [Citation21], colorectal cancer [Citation22,Citation23], osteosarcoma [Citation24,Citation25] and bladder carcinoma [Citation26]. Notably, He et al. showed that inhibition of miR‑132‑3p enhanced GC cell proliferation and migration by targeting mucin 13 (MUC13) [Citation45]. Despite the suppressive role of miR-132-3p has been already reported in GC, there are some major differences compared with our study as follows: 1) We performed gain-of-function assay to investigate the functional role of miR-132-3p, rather than loss-of-function assay in the above study from He et al. 2) Different target genes of miR-132-3p were identified. 3) Different molecular mechanisms underlying miR-132-3p regulating GC cell functions. 4) In addition to migration and invasion, we analyzed cell proliferation here. Until now, several KIFs have been reported to be directly regulated by miRNAs involved in the development of tumors. For instance, miR-338-3p could directly bind to the 3’-UTR of kinesin family member C1 (KIFC1) in renal cell carcinoma cells [Citation46]. MiR-30a could specifically suppress KIF11 by targeting its 3’-UTR in breast cancer [Citation47]. Additionally, miR-206 inhibited the cell proliferation, migration, and invasion of ovarian cancer cells by directly targeting kinesin family protein 2A (KIF2A) [Citation48]. To our best knowledge, KIF21B has not been demonstrated to be a target of certain miRNA, which made the identification of KIF21B as the target of miR-132-3p provided a novel therapeutic target against the progression of GC.
Wnt/β-catenin signaling pathway has been widely reported to be over-activated in multiple types of tumors, which plays a fundamental role in regulation of cellular proliferation and invasion [Citation49]. What’s more, Wnt/β-catenin pathway plays a critical role in metastasis and is closely associated with EMT process [Citation50]. Here, we found that miR-132-3p overexpression inhibited the activity of Wnt1, c-Myc and β-catenin, as key effector of Wnt/β-catenin signaling pathway and inactivated EMT markers (E-cadherin and N-cadherin), which were all reversed after KIF21B overexpression in GC cells. Similar with our data, miR-132-3p suppressed the migration and invasion of tumor cells by regulating the EMT-correlated molecules in lung cancer [Citation51,Citation52] and cervical cancer [Citation53]. It has been confirmed that kinesin superfamily member 26b (KIF26B) regulates cell invasion in breast cancer through driving EMT [Citation54]. KIF11 knockdown significantly upregulated E-cadherin expression and downregulated the expression of N-cadherin and Vimentin in breast cancer cells [Citation38]. KIF23 promoted GC cell proliferation by directly activating the Wnt/β-catenin signaling pathway [Citation55]. Based on these evidences, we thus speculated that miR-132-3p-mediated suppressive role in GC cell proliferation, migration and invasion through targeting KIF21B, which might be correlated with reduced Wnt/β-catenin signaling. It has to be mentioned that there are several limitations in this study, including Lacking of relevant results in vivo; Lacking of stronger evidence that presented the correlation between reduced Wnt/β-catenin signaling through miR-132-3p overexpression and the observed cellular phenotypes; Lacking the direct evidence that KIF21B regulating the expression of Wnt/β-catenin signaling and EMT process.
Conclusions
In summary, we first demonstrated that KIF21B knockdown suppressed the proliferation, migration and invasion in GC cells. Moreover, we showed that KIF21B was the downstream regulator underlying miR-132-3p exerted suppressive effects on GC cells, which might be correlated with modulation of Wnt/β-catenin pathway. Thus, we findings help us better understand targeting miR-132-3p/KIF21B axis as a promising therapeutic target for GC treatment.
Abbreviations
GC: gastric cancer; KIF21B, kinesin family member 21B; GC, gastric cancer; 3′-UTR, 3′ untranslated region; LAPTM4B, lysosomal-associated protein transmembrane 4 beta; FBS, fetal bovine serum; CCK-8, Cell counting kit‑8; OD, optical density; WT, wild-type; EMT, epithelial–mesenchymal transition
Ethics approval and consent to participate
The present study was conducted in accordance with Helsinki Declaration and approved by the Ethics Committee of the Second Hospital, Cheeloo College of Medicine, Shandong University (Approval number: CCM32-J2; 2018.6.3; Shandong, China).
Availability of data and materials
All results and data generated or analyzed during the present study are included in this published article or are available from the corresponding author on reasonable request.
Consent for publication
We have obtained consents to publish this paper from all the participants of this study.
Authors’ contributions
Supervision: Zhipeng Li; Interpretation of analysis of data: Bingtian Liu and Ling Qiang; Preparation of the manuscript: Bingxin Guan; Revision for important intellectual content: Ling Qiang. All authors read and approved the final manuscript.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Bozkurt O, Firat ST, Dogan E, et al. The prognostic value of the change in neutrophil-to-lymphocyte ratio during first-line palliative chemotherapy in patients with metastatic gastric cancer: a retrospective study. J Buon. 2019;24:1992–1999.
- Sitarz R, Skierucha M, Mielko J, et al. Gastric cancer: epidemiology, prevention, classification, and treatment. Cancer Manag Res. 2018;10:239–248.
- Baba H, Kuwabara K, Ishiguro T, et al. Prognostic factors for stage IV gastric cancer. Int Surg. 2013;98:181–187.
- Zhou Q, Lan X, Li N, et al. Analysis of prognostic factors and design of prognosis model for patients with stage IV gastric cancer following first-line palliative chemotherapy. Cancer Manag Res. 2020;12:10461–10468.
- Hirokawa N, Tanaka Y. Kinesin superfamily proteins (KIFs): various functions and their relevance for important phenomena in life and diseases. Exp Cell Res. 2015;334:16–25.
- Hirokawa N, Noda Y. Intracellular transport and kinesin superfamily proteins, KIFs: structure, function, and dynamics. Physiol Rev. 2008;88:1089–1118.
- Hirokawa N, Noda Y, Tanaka Y, et al. Kinesin superfamily motor proteins and intracellular transport. Nat Rev Mol Cell Biol. 2009;10:682–696.
- Myers SM, Collins I. Recent findings and future directions for interpolar mitotic kinesin inhibitors in cancer therapy. Future Med Chem. 2016;8:463–489.
- Liu X, Gong H, Huang K. Oncogenic role of kinesin proteins and targeting kinesin therapy. Cancer Sci. 2013;104:651–656.
- Liu Y, Zhan P, Zhou Z, et al. The overexpression of KIFC1 was associated with the proliferation and prognosis of non-small cell lung cancer. J Thorac Dis. 2016;8:2911–2923.
- Muhia M, Thies E, Labonte D, et al. The Kinesin KIF21B regulates microtubule dynamics and is essential for neuronal morphology, synapse function, and learning and memory. Cell Rep. 2016;15:968–977.
- Kreft KL, van Meurs M, Wierenga-Wolf AF, et al. Abundant kif21b is associated with accelerated progression in neurodegenerative diseases. Acta Neuropathol Commun. 2014;2:144.
- Hares K, Miners JS, Cook AJ, et al. Overexpression of Kinesin superfamily motor proteins in Alzheimer’s Disease. J Alzheimers dis. 2017;60:1511–1524.
- Zhao HQ, Dong BL, Zhang M, et al. Increased KIF21B expression is a potential prognostic biomarker in hepatocellular carcinoma. World J Gastrointest Oncol. 2020;12:276–288.
- Sun ZG, Pan F, Shao JB, et al. Kinesin superfamily protein 21B acts as an oncogene in non-small cell lung cancer. Cancer Cell Int. 2020;20:233.
- Kunej T, Godnic I, Ferdin J, et al. Epigenetic regulation of microRNAs in cancer: an integrated review of literature. Mutat Res. 2011;717:77–84.
- Cortez MA, Anfossi S, Ramapriyan R, et al. Role of miRNAs in immune responses and immunotherapy in cancer. Genes Chromosomes Cancer. 2019;58:244–253.
- Jia XN, Yin SD, Wei Y, et al. MiR-182-5p inhibited proliferation and migration of ovarian cancer cells by targeting BNIP3. Eur Rev Med Pharmacol Sci. 2020;24:7912.
- Wang L, Wang Y, Du X, et al. MiR-216b suppresses cell proliferation, migration, invasion, and epithelial-mesenchymal transition by regulating FOXM1 expression in human non-small cell lung cancer. Onco Targets Ther. 2019;12:2999–3009.
- Li S, Xu JJ, Zhang QY. MicroRNA-132-3p inhibits tumor malignant progression by regulating lysosomal-associated protein transmembrane 4 beta in breast cancer. Cancer Sci. 2019;110:3098–3109.
- Wu B, Li J, Wang H, et al. MiR-132-3p serves as a tumor suppressor in mantle cell lymphoma via directly targeting SOX11. Naunyn-Schmiedeberg’s Arch Pharmacol. 2020;393:2197–2208.
- He X, Ma J, Zhang M, et al. Long non-coding RNA SNHG16 activates USP22 expression to promote colorectal cancer progression by sponging miR-132-3p. Onco Targets Ther. 2020;13:4283–4294.
- Zhang M, Li Y, Wang H, et al. LncRNA SNHG5 affects cell proliferation, metastasis and migration of colorectal cancer through regulating miR-132-3p/CREB5. Cancer Biol Ther. 2019;20:524–536.
- Guan H, Shang G, Cui Y, et al. Long noncoding RNA APTR contributes to osteosarcoma progression through repression of miR-132-3p and upregulation of yes-associated protein 1. J Cell Physiol. 2019;234:8998–9007.
- Li G, Liu K, Du X. Long non-coding RNA TUG1 promotes proliferation and inhibits apoptosis of osteosarcoma cells by sponging miR-132-3p and upregulating SOX4 expression. Yonsei Med J. 2018;59:226–235.
- Liu P, Li X, Guo X, et al. Circular RNA DOCK1 promotes bladder carcinoma progression via modulating circDOCK1/hsa-miR-132-3p/Sox5 signalling pathway. Cell Prolif. 2019;52:e12614.
- Zhang T, Liu C, Huang S, et al. A downmodulated MicroRNA profiling in patients with gastric cancer. Gastroenterol Res Pract. 2017;2017:1526981.
- Wang J, Zhang H, Zhou X, et al. Five serum-based miRNAs were identified as potential diagnostic biomarkers in gastric cardia adenocarcinoma. Cancer Biomarkers. 2018;23:193–203.
- Wang Q, Wen YG, Li DP, et al. Upregulated INHBA expression is associated with poor survival in gastric cancer. Med Oncol. 2012;29:77–83.
- Chen X, Leung SY, Yuen ST, et al. Variation in gene expression patterns in human gastric cancers. Mol Biol Cell. 2003;14:3208–3215.
- D’Errico M, de Rinaldis E, Blasi MF, et al. Genome-wide expression profile of sporadic gastric cancers with microsatellite instability. Eur J Cancer. 2009;45:461–469.
- Chandrashekar DS, Bashel B, Balasubramanya SAH, et al. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19:649–658.
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR. Methods. 2002;25:402–408.
- Huszar D, Theoclitou ME, Skolnik J, et al. Kinesin motor proteins as targets for cancer therapy. Cancer Metastasis Rev. 2009;28:197–208.
- Lucanus AJ, Yip GW. Kinesin superfamily: roles in breast cancer, patient prognosis and therapeutics. Oncogene. 2018;37:833–838.
- Yang Z, Li C, Yan C, et al. KIF14 promotes tumor progression and metastasis and is an independent predictor of poor prognosis in human gastric cancer. Biochimica et biophysica acta Mol basis dis. 2019;1865:181–192.
- Wang ZZ, Yang J, Jiang BH, et al. KIF14 promotes cell proliferation via activation of Akt and is directly targeted by miR-200c in colorectal cancer. Int J Oncol. 2018;53:1939–1952.
- Zhou J, Chen WR, Yang LC, et al. KIF11 functions as an oncogene and is associated with poor outcomes from breast cancer. Cancer Res Treat. 2019;51:1207–1221.
- Li XL, Ji YM, Song R, et al. KIF23 promotes gastric cancer by stimulating cell proliferation. Dis Markers. 2019;2019:9751923.
- Ni S, Li J, Qiu S, et al. KIF21B expression in osteosarcoma and its regulatory effect on osteosarcoma cell proliferation and apoptosis through the PI3K/AKT pathway. Front Oncol. 2020;10:606765.
- Yang Y, Gao L, Weng NN, et al. Identification of novel molecular therapeutic targets and their potential prognostic biomarkers among kinesin superfamily of proteins in pancreatic ductal adenocarcinoma. Front Oncol. 2021;11:708900.
- Kara M, Yumrutas O, Ozcan O, et al. Differential expressions of cancer-associated genes and their regulatory miRNAs in colorectal carcinoma. Gene. 2015;567:81–86.
- Zhang H, Liu A, Feng X, et al. MiR-132 promotes the proliferation, invasion and migration of human pancreatic carcinoma by inhibition of the tumor suppressor gene PTEN. Prog Biophys Mol Biol. 2019;148:65–72.
- Zhou K, Zhang C, Yao H, et al. Knockdown of long non-coding RNA NEAT1 inhibits glioma cell migration and invasion via modulation of SOX2 targeted by miR-132. Mol Cancer. 2018;17:105.
- He L, Qu L, Wei L, et al. Reduction of miR‑132‑3p contributes to gastric cancer proliferation by targeting MUC13. Mol Med Rep. 2017;15:3055–3061.
- Li G, Chong T, Yang J, et al. Kinesin motor protein KIFC1 is a target protein of miR-338-3p and is associated with poor prognosis and progression of renal cell carcinoma. Oncol Res. 2018;27:125–137.
- Wang B, Yu J, Sun Z, et al. Kinesin family member 11 is a potential therapeutic target and is suppressed by microRNA-30a in breast cancer. Mol Carcinog. 2020;59:908–922.
- Sheng N, Xu YZ, Xi QH, et al. Overexpression of KIF2A is suppressed by miR-206 and associated with poor prognosis in ovarian cancer. Cell Physiol Biochem. 2018;50:810–822.
- Logan CY, Nusse R. The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol. 2004;20:781–810.
- Talbot LJ, Bhattacharya SD, Kuo PC. Epithelial-mesenchymal transition, the tumor microenvironment, and metastatic behavior of epithelial malignancies. Int J Biochem Mol Biol. 2012;3:117–136.
- Guo H, Zhang X, Chen Q, et al. miR-132 suppresses the migration and invasion of lung cancer cells by blocking USP9X-induced epithelial-mesenchymal transition. Am J Transl Res. 2018;10:224–234.
- You J, Li Y, Fang N, et al. MiR-132 suppresses the migration and invasion of lung cancer cells via targeting the EMT regulator ZEB2. PloS one. 2014;9:e91827.
- Zhao JL, Zhang L, Guo X, et al. miR-212/132 downregulates SMAD2 expression to suppress the G1/S phase transition of the cell cycle and the epithelial to mesenchymal transition in cervical cancer cells. IUBMB Life. 2015;67:380–394.
- Gu S, Liang H, Qi D, et al. Knockdown of KIF26B inhibits breast cancer cell proliferation, migration, and invasion. Onco Targets Ther. 2018;11:3195–3203.
- Liu Y, Chen H, Dong P, et al. KIF23 activated Wnt/β-catenin signaling pathway through direct interaction with Amer1 in gastric cancer. Aging (Albany NY). 2020;12:8372–8396.
