ABSTRACT
The aim of this study was to investigate the expression of miRNA regulated by c-myc and its mechanism in three negative breast cancer (TNBC). We constructed MDA-MB-231 cell line with low expression of c-myc by lentivirus short hairpin RNA (shRNA), analyzed the miRNA expression profile of MDA-MB-231 cell line with different expression levels of c-myc by high-throughput sequencing technology, obtained differential miRNA by bioinformatics analysis and statistical analysis, and verified hsa-mir-4723-5p by Quantitative Real-time polymerase chain reaction(QRT-PCR). The target gene of hsa-mir-4723-5p was analyzed by miRDB and miRWalk database. The results showed that there were significant differences in 126 miRNAs in c-myc knockdown cell lines compared with the control group, of which 84 were significantly up-regulated and 42 were significantly down regulated. According to the results of miRNA sequencing, the miRNA closely related to the expression of c-myc was hsa-mir-4723-5p. QRT PCR showed that the expression of hsa-mir-4723-5p was down regulated in TNBC cell line MDA-MB-231 with low expression of c-myc, which was positively correlated with the expression. The target genes of hsa-mir-4723-5p were predicted according to mirdb and mirwalk database. A total of 112 target genes were obtained, and 107 target genes were related to hsa-mir-4723-5p. Through mirdb and mirwalk databases, it was found that the target gene TRAF4 of hsa-mir-4723-5p may be related to cancer pathway and affect tumor metastasis. In conclusion, the hsa-miR-4723-5p regulated by c-myc may be involved.
Introduction
Breast cancer accounts for the highest incidence of malignant tumors in women [Citation1]. Its morbidity and mortality rank first among all types of female malignant tumors, posing a serious threat to health [Citation2]. As a subtype of breast cancer, triple-negative breast cancer (TNBC) has the characteristics of high heterogeneity, poor histopathological grade, and strong tumor invasiveness, and it is prone to local recurrence and distant metastasis. Additionally, the distant metastasis of TNBC has an obvious organ tendency and is most commonly found in the lung, liver, and brain. Due to the lack of therapeutic targets, the prognosis of TNBC is extremely poor, and the recurrence rate can reach 40%–50% after 3 years [Citation3]. The deaths caused by TNBC account for about 25% of all breast cancer deaths [Citation4]. At present, the treatment of TNBC still presents many challenges.
The c-myc gene is an important target of a variety of cell signaling pathways and has an important impact on the occurrence and development of breast cancer [Citation4]. Studies have shown that, compared with other types of breast cancer, TNBC exhibits an abnormally high expression of c-myc [Citation5,Citation6]. The abnormal expression of c-myc causes the activation of oncogenes and promotes the occurrence and development of tumors [Citation7].
MicroRNAs (miRNAs) are endogenous 15–23-nt RNAs that can negatively regulate the expression of target genes [Citation8]. Studies have shown that miRNAs play an important role in the occurrence and development of cancer by targeting a variety of oncogenes and tumor suppressor genes [Citation9–12]. In addition, miRNAs may become biomarkers of various diseases, including cancer [Citation13–17]. Importantly, several studies have shown [Citation18–20] that c-myc can regulate the expression of miRNA through transcriptional activation or inhibition and further promote the occurrence and development of tumors through miRNA target genes.
We hypothesized that c-myc regulated hsa-mir-4723-5p may play an important role in the progression of TNBC. In this study, we measured the expression of hsa-mir-4723-5p in TNBC cell line MDA-MB-231. In addition, we also predicted the direct target gene TRAF4 of hsa-mir-4723-5p. Therefore, our results may provide new targets for the treatment of TNBC.
Methods
This study was approved by the Ethics Committee of the Affiliated Cancer Hospital of Xinjiang Medical University. (Approval number: HY-202191235)
Cells and cell culture
MDA-MB-231 cells were obtained from Procell Life Science & Technology Co., Ltd. (Cat.#CL-0290, Wuhan, China). They were kept in dulbecco’s modified eagle (DMEM) medium (C11965500BT, GIBCO) supplemented with 10% fetal bovine serum (FND500, Excell Bio) and 1% (10,000 U) penicillin–streptomycin (SC30010, GIBCO) at 37°C and 5% CO2.
Lentivirus transfection and construction of stable cell strain with low c-myc expression
MDA-MB-231 cells (5 × 104 cells/mL) were plated into 96-well plates. After 24 h, the cells were transfected with c-myc shRNA (5’-TGAGACAGATCAGCAACAA-3’) or shRNA negative control (5’-TTCTCCGAACGTGTCACGT-3’) lentiviral particles using polybrene. The shRNA lentiviral particles were obtained from Genechem Co., Ltd. (Shanghai, China). The optimal multiplicity of infection (MOI) was determined to be 10. After 72 h of lentivirus infection, the cells were cultured in the presence of 2-μg/mL puromycin to select the cell strain with a stable low expression of c-myc. The c-myc expression was verified using real-time PCRs.
miRNA sequencing
RNAs were extracted from cells using TRIzol (ET111, Transgene). Using the extracted RNA as a template, first-strand cDNA was synthesized with random hexamers. Then, second-strand cDNA was synthesized with dNTPs, DNA polymerase I, and RNase H. The double-stranded cDNA was purified using AMPure XP beads (Beckman, A63880). After ligating the adapters, the purified double-stranded cDNA was amplified by PCR. The cDNA library was obtained after purifying the PCR products with AMPure XP beads. Qubit 2.0 (Thermo Scientific) was used for the preliminary quantification of the cDNA library. The size of the insert in the library was detected with an Agilent 2100 Bioanalyzer (Thermo Scientific) system. Finally, the qualified cDNA library was sequenced on an Illumina high-throughput sequencing platform (Illumina, USA).
Screening of differential miRNAs
The study used Cutadapt software (National) [Citation21] to remove the Illumina 5’ and 3’ sequencing adapters, and BLASTN software was used to remove rRNA, tRNA, miscRNA, snRNA, and snoRNA. The processed sequences were searched in the miRBase 21.0 database (http://mirbase.org/) [Citation22–24] to identify new miRNAs. The miRNA expression levels were calculated using DEseq software [Citation25]. The screening criteria for differential miRNAs were P < 0.05 and fold change (FC) > 2.
miRNA target prediction and enrichment analysis
The miRDB (http://www.mirdb.org/), TargetScan (http://www.targetscan.org/) [Citation26] and miRWalk (http://mirwalk.umm.uni-heidelberg.de/) databases were used to predict the target genes of miRNAs. The Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses of the target genes were performed using the Blast2GO 5.1 database (https://www.biobam.com/blast2go-previous-versions/) [Citation27].
Real-time polymerase chain reaction
Total RNAs were extracted from cells using TRIzol as a reagent before being reverse transcribed into cDNA with PrimeScript™ RT Master Mix (Beyotime Biotechnology). The primer sequences were as follows: c-Myc forward primer 5’-GGCTCCTGGCAAAAGGTCA-3’ and reverse primer: 5’-CTGCGTAGTTGTGCTGATGT-3’; GAPDH forward primer 5’-CTGCGTAGTTGTGCTGATGT-3’ and reverse primer: TTCAGATCCCAGCGGTGC; hsa-miR-4723-5p primer TGGGGGAGCCATGAGATAAG; and U6 primer AGCACATATACTAAAATTGGAACGAT. GAPDH and U6 were internal references. A real-time PCR was performed on an ABI QuantStudioTM6 Flex (ABI, USA). The reaction program for c-Myc mRNA involved pre-denaturation at 95°C for 5 s followed by 40 cycles at 60°C for 30s and again at 72°C for 2 min. The reaction procedure for hsa-miR-4723-5p included pre-denaturation at 95°C for 15 min, 45 cycles of denaturation at 94°C for 20s, and annealing at 60°C for 34s. The relative mRNA/miRNA level was calculated using the 2−ΔΔCt method [Citation28].
Statistical analysis
All data were expressed as mean ± standard deviation and analyzed with Statistical Product and Service Solutions(SPSS)19.0 software. If the data conformed to a normal distribution, the single-factor analysis of variance was used for comparison; this was followed by the least significance difference (LSD) method (for data with homogeneity of variance) or the Tamhane method (for data with heterogeneity of variance). If the data did not conform to the normal distribution, it was converted to a logarithm to normalize the data before using the above-mentioned statistical methods for comparison. A value of P < 0.05 indicates a significant difference.
Results
We hypothesized that c-myc regulated hsa-mir-4723-5p may play an important role in the progression of TNBC. In this study, we measured the expression of hsa-mir-4723-5p in TNBC cell line MDA-MB-231. In addition, we also predicted the direct target gene TRAF4 of hsa-mir-4723-5p. Therefore, our results may provide new targets for the treatment of TNBC.
Establishment of MDA-MB-231 stable cell line with low c-myc expression
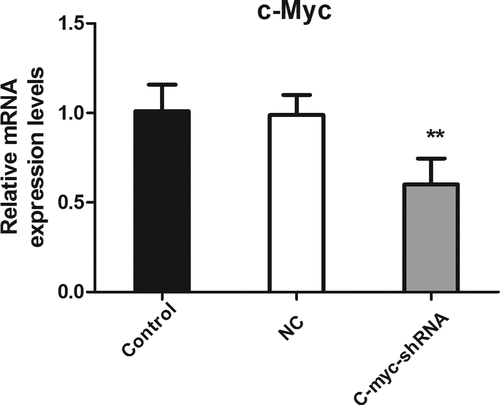
The establishment of an MDA-MB-231 stable cell line with low c-myc expression was verified using real-time PCR. As shown in , compared with the blank control group and the negative control group, the c-myc mRNA expression level of the c-myc shRNA lentiviral transfection group was significantly reduced (P < 0.01). This indicates the successful knockdown of c-myc.
Figure 1. The level of c-myc mRNA detected with real-time PCR after transfection of shRNA in MDA-MB-231 cells. **P < 0.01.
Differential miRNA screening and functional analysis
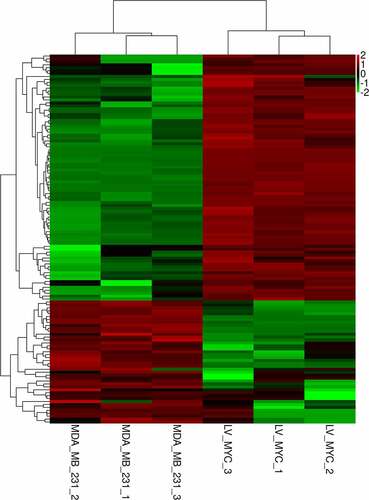
DEseq software was used to screen differentially expressed miRNAs with P < 0.05 and FC > 2. A total of 126 differentially expressed miRNAs were identified. Of these, 84 miRNAs were upregulated and 42 miRNAs were downregulated. The heat map of differentially expressed miRNAs is shown in . The top 10 differentially downregulated miRNAs are listed in . Of note, hsa-miR-4723-5p experienced the most substantial downregulation of the 42 downregulated miRNAs. Thus, hsa-miR-4723-5p was used for further analysis.
Table 1. The top 10 differentially up-regulated and down-regulated miRNAs
Figure 2. Heat map of upregulated and downregulated differentially expressed miRNAs after knocking down c-myc. Each column represents a sample, and each row represents a differential miRNA. Red indicates high expression, and green indicates low expression.
GO and KEGG pathway enrichment analyses of the differentially expressed miRNAs were performed. As shown in (), the GO enrichment analysis indicated that the differentially expressed miRNAs were primarily involved in processes such as endoplasmic reticulum localization and Ca2+ channel activity regulation. The KEGG pathway analysis showed the main enriched pathways include the cell adhesion signaling pathway, tumor necrosis factor (TNF) signaling pathway, transforming growth factor (TGF)-beta signaling pathway, and mitogen-activated protein kinase (MAPK) signaling pathway ().
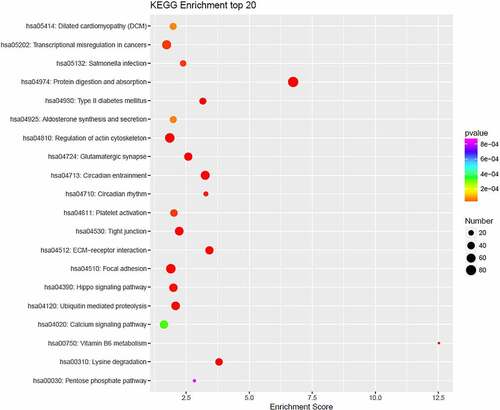
Figure 3. The top-10 enriched GO terms of target genes of differentially expressed miRNAs. The ordinate is the number of genes enriched by GO, and the abscissa is the name of the GO term.
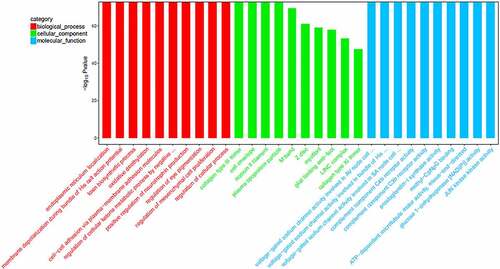
Figure 4. KEGG pathway enrichment bubble chart of top-20 enriched pathways of target genes of differentially expressed miRNAs. Each dot corresponds to a pathway. The color gradient, from red to purple, corresponds to the P value. The lower the P value, the more the color tends toward red. The larger the dot, the more genes in the pathway.
Target gene prediction for hsa-miR-4723-5p
For hsa-miR-4723-5p, 124 target genes were predicted using the miRDB database, and 16,792 target genes were predicted using miWalk. In total, 107 common target genes of hsa-miR-4723-5p were predicted. Five representative target genes of hsa-miR-4723-5p are shown in .
Table 2. Some representative target genes of hsa-miR-4723-5p
Function analysis of the target genes of hsa-miR-4723-5p
The target genes of hsa-miR-4723-5p were significantly enriched in biological processes, such as the intra-S DNA damage checkpoint, positive regulation of Rho protein signal transduction, protein localization of the M band, migration guided by radial glial cells in the cerebral cortex, production of vascular endothelial growth factor, cytoskeleton anchoring on the nuclear membrane, actin filament movement, and mature behavior (P < 0.05); in cellular components, including the M band, Z disc, myofibril, glial cell restriction terminal foot, linker of nucleoskeleton and cytoskeleton complex (LINC) complex, type XI collagen trimer, chromosome, and histone deacetylase complex (P < 0.05); and in the molecular function of actin filament binding, ankyrin binding, actin binding, in the structural components of muscle, protein C-terminal binding, microfilament movement activity, calmodulin binding, and extracellular matrix binding (P < 0.05) ().
Table 3. The top 8 enriched GO terms and KEGG pathways of target genes of hsa-miR-4723-5p
The target genes of hsa-miR-4723-5p were related to protein digestion and absorption, diurnal entrainment, extracellular matrix (ECM)-receptor interaction, lysine degradation, glutamatergic synapses, vitamin B6 metabolism, type-II diabetes, and tight junctions (P < 0.05) ().
The verification of hsa-miR-4723-5p
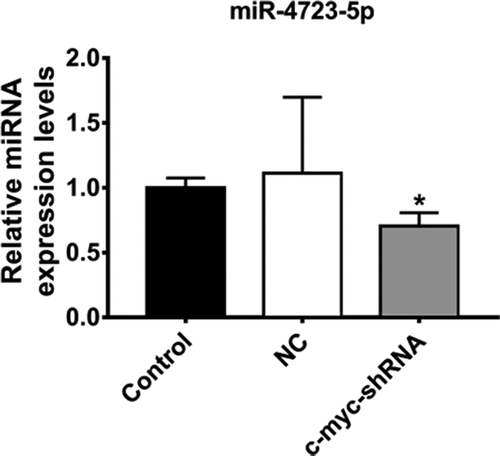
The expression of hsa-miR-4723-5p in the MDA-MB-231 cells was further verified by real-time PCR. The results indicated that the expression levels of has-miR-4723-5p was consistent with the results of the miRNA sequencing and bioinformatics analysis, showing the downregulated expression of hsa-miR-4723-5p (P < 0.05) ().
Discussion
TNBC is distinct from other types of breast cancer due to its characteristic high recurrence rate, easy metastasis, short survival time, and poor prognosis. Because it does not express estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER2), it cannot benefit from endocrine therapy or HER2-targeted therapy. The lack of therapeutic targets makes the treatment of TNBC difficult.
Previous studies have shown that c-myc regulates the expression of a variety of miRNAs, leading to extensive miRNA suppression [Citation29,Citation30]. Herein, we used miRNA sequencing to identify the expression profile of miRNAs related to c-myc in a TNBC cell line. Compared with the control, there were 126 differentially expressed miRNAs, including 84 upregulated miRNAs and 42 downregulated miRNAs. Among these, hsa-miR-4723-5p was the most differentially downregulated miRNA. Therefore, it was selected for further analysis, including target gene prediction. In total, 107 target genes were predicted for hsa-miR-4723-5p. The GO and KEGG analyses revealed that the target genes were related to proteoglycan expression and pathways in cancer. The target genes were also involved in protein digestion and absorption, diurnal entrainment, ECM-receptor interaction, lysine degradation, glutamatergic synapses, vitamin B6 metabolism, type-II diabetes, and tight junctions.
Through mirdb and mirwalk databases, it was found that the target gene TRAF4 of hsa-mir-4723-5p may be related to cancer pathway and affect tumor metastasis. It has been reported that lncrna hcg18 promotes the proliferation, migration and EMT of epithelial ovarian cancer by up regulating TRAF4/TRAF5 [Citation31]. It is also reported that TRAF4 may be associated with esophageal squamous cell carcinoma [Citation32]. Other studies have shown that TRAF4 mediated ubiquitination of IRS-1 is physiologically related to IGF signal transduction and is very important [Citation33]. Through site directed mutagenesis, TRAF4 was also found to promote the ubiquitination of atypical k29 junction at the C-terminal of IRS-1 [Citation34]. Yao w et al found that TRAF4 was highly expressed in osteosarcoma [Citation35].
The expression of hsa-miR-4723-5p was verified by real-time PCR. The results showed that in the MDA-MB-231 cells with low c-myc expression, the expression of hsa-miR-4723-5p was significantly decreased. It has been shown that hsa-miR-4723-5p is abnormally expressed in a variety of tumors, including those related to multiple myeloma [Citation18] and prostate cancer [Citation36]. However, the mechanism of c-myc/hsa-miR-4723-5p/TRAF4 regulation in TNBC is largely unknown and deserves further study.
Conclusions
In summary, we constructed an MDA-MB-231 cell strain with a low expression of c-myc and analyzed the miRNA expression profile using high-throughput sequencing. In vitro cell analysis showed that knocking down c-myc could significantly reduce the expression of hsa-miR-4723-5p. Therefore, we speculate that hsa-miR-4723-5p regulated by c-myc may be involved in the occurrence and metastasis of TNBC. Our data may help to better elucidate the specific molecular mechanism of TNBC and to identify potential targets related to the diagnosis, treatment, and prognosis of TNBC.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Beiki O, Hall P, Ekbom A, et al. Breast cancer incidence and case fatality among 4.7 million women in relation to social and ethnic background: a population-based cohort study. Bcr. 2012;14:R5.
- Shaw JE, Sicree RA, Zimmet PZ. Global estimates of the prevalence of diabetes for 2010 and 2030. Diabetes Res Clin Pract. 2010;87:4–14.
- Rhodes LV, Tate CR, Hoang VT, et al. Regulation of triple-negative breast cancer cell metastasis by the tumor-suppressor liver kinase B1. Oncogenesis. 2015;4:e168.
- Shah AN, Metzger O, Bartlett CH, et al. Hormone receptor-positive/human epidermal growth receptor 2-negative metastatic breast cancer in young women: emerging data in the era of molecularly targeted agents. Oncologist. 2020;25:e900–e908.
- Anders CK, Abramson V, Tan T, et al. The evolution of triple-negative breast cancer: from biology to novel therapeutics. American Society of Clinical Oncology educational book American Society of Clinical Oncology Annual Meeting, San Francisco, USA 2016; 35: 34–42
- Karagoz K, Sinha R, Arga KY. Triple negative breast cancer: a multi-omics network discovery strategy for candidate targets and driving pathways. OMICS. 2015;19:115–130.
- Mathsyaraja H, Eisenman RN. Parsing Myc Paralogs in Oncogenesis. Cancer Cell. 2016;29:1–2.
- Shah SP, Roth A, Goya R, et al. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature. 2012;486:395–399.
- Diehl F, Schmidt K, Choti MA, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med. 2008;14:985–990.
- Gwak JM, Kim HJ, Kim EJ, et al. MicroRNA-9 is associated with epithelial-mesenchymal transition, breast cancer stem cell phenotype, and tumor progression in breast cancer. Breast Cancer Res Treat. 2014;147:39–49.
- Gravgaard KH, Lyng MB, Laenkholm AV, et al. The miRNA-200 family and miRNA-9 exhibit differential expression in primary versus corresponding metastatic tissue in breast cancer. Breast Cancer Res Treat. 2012;134:207–217.
- Kolacinska A, Morawiec J, Pawlowska Z, et al. Association of microRNA-93, 190, 200b and receptor status in core biopsies from stage III breast cancer patients. DNA Cell Biol. 2014;33:624–629.
- Sai S, Ichikawa D, Tomita H, et al. Quantification of plasma cell-free DNA in patients with gastric cancer. Anticancer Res. 2007;27:2747–2751.
- Vinayanuwattikun C, Winayanuwattikun P, Chantranuwat P, et al. The impact of non-tumor-derived circulating nucleic acids implicates the prognosis of non-small cell lung cancer. J Cancer Res Clin Oncol. 2013;139:67–76.
- Shin VY, Siu JM, Cheuk I, et al. Circulating cell-free miRNAs as biomarker for triple-negative breast cancer. Br J Cancer. 2015;112:1751–1759.
- Mathe A, Scott RJ, Avery-Kiejda KA. MiRNAs and other epigenetic changes as biomarkers in triple negative breast cancer. Int J Mol Sci. 2015;16:28347–28376.
- Hu Z, Chen X, Zhao Y, et al. Serum microRNA signatures identified in a genome-wide serum microRNA expression profiling predict survival of non-small-cell lung cancer. J Clin Oncol. 2010;28:1721–1726.
- Li J, Zhang M, Wang C. Circulating miRNAs as diagnostic biomarkers for multiple myeloma and monoclonal gammopathy of undetermined significance. J Clin Lab Anal. 2020;34:e23233.
- Yue S, Ye X, Zhou T, et al. PGRN(-/-) TAMs-derived exosomes inhibit breast cancer cell invasion and migration and its mechanism exploration. Life Sci. 2021;264:118687.
- Wei Z, Lyu B, Hou D, et al. Mir-5100 mediates proliferation, migration and invasion of oral squamous cell carcinoma cells via targeting SCAI. J Invest Surg. 2021;34:834–841.
- Avery-Kiejda KA, Mathe A, Scott RJ. Genome-wide miRNA, gene and methylation analysis of triple negative breast cancer to identify changes associated with lymph node metastases. Genom Data. 2017;14:1–4.
- Schultz DJ, Muluhngwi P, Alizadeh-Rad N, et al. Genome-wide miRNA response to anacardic acid in breast cancer cells. PloS One. 2017;12:e0184471.
- Mitra S. MicroRNA therapeutics in triple negative breast cancer. Archives of Pathology and Clinical Research. 2017; Arch Pathol Clin Res. 1:009–017.
- Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17:10–12.
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140.
- Wong N, Wang X, Wang X. miRDB: an online resource for microRNA target prediction and functional annotations. Nucleic Acids Res. 2015;43:D146–152.
- Conesa A, Gotz S, Garcia-Gomez JM, et al. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–3676.
- Pratama MY, Cavalletto L, Tiribelli C, et al. Selection and validation of miR-1280 as a suitable endogenous normalizer for qRT-PCR Analysis of serum microRNA expression in Hepatocellular Carcinoma. Sci Rep. 2020 Feb 21; 10(1):3128.
- Horiuchi D, Kusdra L, Huskey NE, et al. MYC pathway activation in triple-negative breast cancer is synthetic lethal with CDK inhibition. J Exp Med. 2012;209:679–696.
- Gravina GL, Festuccia C, Popov VM, et al. c-Myc sustains transformed phenotype and promotes radioresistance of embryonal rhabdomyosarcoma cell lines. Radiat Res. 2016;185:411–422.
- Zhang F, Luo BH, Wu QH, et al. LncRNA HCG18 upregulates TRAF4/TRAF5 to facilitate proliferation, migration and EMT of epithelial ovarian cancer by targeting miR-29a/b. Mol Med. 2022 Jan 4;28(1):2. PMID: 34983361; PMCID: PMC8725507.
- Qiu H, Song H, Luo M, et al. Dysfunction of apoptosis and autophagy correlates with local recurrence in esophageal squamous cell carcinoma after definitive chemoradiation. Cancer Cell Int. 2021 Sep 6;21(1):466. PMID: 34488754; PMCID: PMC8419897.
- Yu W, Singh R, Wang Z, et al. The E3 ligase TRAF4 promotes IGF signaling by mediating atypical ubiquitination of IRS-1. J Biol Chem. 2021Jan-Jun; 296: 100739 Epub 2021 May 13. PMID: 33991522; PMCID: PMC8191236
- Singh R, Karri D, Shen H, et al. TRAF4-mediated ubiquitination of NGF receptor TrkA regulates prostate cancer metastasis. J Clin Invest. 2018 Jul 2;128(7):3129–3143. Epub 2018 Jun 18. PMID: 29715200; PMCID: PMC6026011.
- Yao W, Wang X, Cai Q, et al. TRAF4 enhances osteosarcoma cell proliferation and invasion by Akt signaling pathway. Oncol Res. 2014;22(1):21–28. PMID: 25700355; PMCID: PMC7592778.
- Arora S, Saini S, Fukuhara S, et al. MicroRNA-4723 inhibits prostate cancer growth through inactivation of the Abelson family of nonreceptor protein tyrosine kinases. PloS One. 2013;8:e78023.