ABSTRACT
Lung adenocarcinoma (LUAD) severely threatens the health of people owing to its lethality. Nonetheless, the underlying mechanisms on LUAD development remain unclear to a great extent. This work aimed to probe the functions of miR-218-5p in LUAD. MiR-218-5p and endoplasmic reticulum oxidoreductase 1 alpha (ERO1A) were screened as differently downregulated and upregulated RNAs in LUAD, respectively, by bioinformatics analyses. The results of cell functional assays stated that enforced expression of miR-218-5p notably restrained cell viability, invasion, and migration in LUAD. MiR-218-5p may interact with 3’-untranslated region of ERO1A mRNA as analyzed by bioinformatics. Afterward, western blot and dual-luciferase reporter gene analyses were introduced to identify their interaction. ERO1A overexpression reversed the suppressive impacts of miR-218-5p on LUAD cell progression, indicating the implication of miR-218-5p/ERO1A axis in suppressing cancer development. We also observed that this regulatory axis suppressed angiogenesis in LUAD. Taken together, miR-218-5p/ERO1A axis exerted an imperative role in LUAD cell progression, which provides a valuable clue for the development of LUAD therapeutic regimen.
Graphic Abstract
Introduction
Lung cancer is a prevalent and lethal malignancy worldwide [Citation1], the death from which composes 18.4% of all deaths triggered by cancers according to the statistics [Citation2]. Lung adenocarcinoma (LUAD) constitutes the major pathological subtype of lung cancer [Citation3]. Surgery, chemotherapy, radiotherapy, immune therapy, and targeted therapy are the existing options for LUAD, with chemotherapy relatively potent for the advanced-stage LUAD [Citation4,Citation5]. But internal or acquired chemotherapy has restrained its use. Despite many achievements in treatment methods of LUAD, the survival outcomes of this disease remain unsatisfactory [Citation6]. Hence, much more in-depth understanding on the molecular mechanism and biomarkers of LUAD is urgently needed to optimize therapeutic strategies for this disease.
MicroRNAs (miRNAs) are small endogenous non-coding RNAs that play a role in the post-transcriptional modulation of target gene expression [Citation7,Citation8]. Investigations suggested that various miRNAs are abnormally expressed in cancers and are pivotal for cancer progression [Citation9,Citation10]. For instance, miR-218-5p represses cancer cell migration and proliferation via modulating EGFR [Citation11]. Similarly, downregulated miR-218-5p in LUAD [Citation12] inhibits SHMT1 and restrains natural killing cells to exert the killing effect [Citation13]. Nevertheless, whether miR-218-5p can affect the pathogenesis of LUAD remains to be further explored.
Endoplasmic reticulum oxidoreductase 1 alpha (ERO1A) is an oxidortase in the endoplasmic reticulum and the primary regulator of protein disulfide isomerase (PDI) oxidation [Citation14,Citation15]. The overexpression of ERO1A in cancers, like breast cancer [Citation16], gastric cancer [Citation17], pancreatic cancer [Citation18], and LUAD [Citation19], is linked with poor prognoses of patients. Besides, ERO1A upregulation induces vascular endothelial growth factor (VEGF) expression and angiogenesis [Citation20–22]. However, the way ERO1A modulates LUAD development is uncertain.
In this study, to comprehensively reveal the underlying mechanisms of miR-218-5p in LUAD, we assayed its expression and effects on cell functions in LUAD. Additionally, combining bioinformatics methods, we screened the target of miR-218-5p and identified miR-218-5p/ERO1A regulatory axis in LUAD. The findings here might open novel horizons and offer novel targets for LUAD treatment.
1. Materials and methods
1.1 Bioinformatics methods
Bioinformatics prediction was introduced as previously denoted [Citation23]. Data of mature miRNA (normal: 46, cancer: 521) and mRNA (normal: 59, cancer: 535) of The Cancer Genome Atlas (TCGA)-LUAD were accessed from TCGA database (https://portal.gdc.cancer.gov/). Differential expression analysis (|logFC|>1.5, padj<0.01) on miRNAs and mRNAs between normal and cancer samples was introduced by using ‘EdgeR’ package. High- and low-expression groups from LUAD samples were constructed based on median ERO1A expression value, and a survival analysis was conducted on the groups using ‘survival’ package. The mirDIP (http://ophid.utoronto.ca/mirDIP/index.jsp#r), TargetScan (http://www.target scan.org/vert_72/), and starBase (http://starbase.sysu.edu. cn/index.php) databases were utilized to predict the targets of miR-218-5p. Pearson correlation analysis was carried out to determine the target of miR-218-5p.
1.2 Culture and transfection of cells
Human LUAD cell lines PC-9 (BNCC330767), A549 (BNCC337696), SPC-A-1 (BNCC100120), Calu-3 (BNCC359757), and human umbilical vein endothelial cells (HUVEC, BNCC337616) were supplied by BeNa Culture Collection (Beijing, China). Human lung cell line HLF-a was obtained from Procell (CL-0359, Wuhan, China). Dulbecco’s Modified Eagle Medium (DMEM, Gibco, USA) containing 10% fetal bovine serum (FBS) (Thermo Scientific HyClone, USA) was utilized for cell culture at 37°C with 5% CO2.
The miR-218-5p mimics and negative control (NC micmic) were designed by GenePharma Biotech (Shanghai, China). In the present study, all transfections were transient. JetPRIME reagent (Polyplus-transfection) was added to cells. After transfected with RNA oligonucleotides for 48 h, cells were gathered for subsequent detection.
1.3 Quantitative reverse-transcription polymerase chain reaction (qRT-PCR)
As elaborated previously [Citation24], total RNA was separated from cells by using miRNeasy Mini kit (Qiagen, Hilden, Germany). Complementary DNA of miRNA was obtained by using QuantiMir RT kit (SBI). To confirm the results of bioinformatics analysis, ABI 7500 Fast System Thermocycler (Applied Biosystems Life Technologies, Foster City, CA, USA) was applied to perform PCR, and SYBR-Green qPCR Master Mix (Takara, Japan) was used to quantify mRNA and miRNA samples. Transfection efficiency of oligonucleotides was also testified by qRT-PCR. Relative expression was calculated by comparing cycle threshold (Ct) method. GAPDH and U6 snRNA were utilized as the internal controls for detection of ERO1A and miR-218-5p, respectively. Primers utilized were synthesized by Shanghai GenePharma Company. lists the primer sequences.
Table 1. Primer sequence designed in this study
1.4 Western blot
Total proteins were isolated from cultured cells using Radio Immunoprecipitation Assay lysis buffer (Thermo Fisher, USA) and the protein concentration was assayed with bicinchoninic acid kit (Thermo, USA), as described previously [Citation25]. After the isolated proteins were separated on sodium dodecyl sulfate-polyacrylamide gel electrophoresis, protein samples were transferred onto a polyvinylidene fluoride (PVDF) membrane and blocked with 5% skim milk for 1 h. Subsequently, the PVDF membrane was incubated with primary antibodies overnight: rabbit anti-ERO1A (1:1000, ab177156, Abcam, UK), rabbit anti-VEGFA (1:10,000, ab52917, Abcam, UK), and rabbit anti-GAPDH (1:10,000, ab181602, Abcam, UK), followed by culturing with secondary antibody IgG (ab6721, Abcam, UK) at room temperature for 30 min. Protein bands were measured by enhanced chemiluminescence (Amersham, Piscataway, NJ, USA) and protein expression was quantified by ImageJ software. Protein level of each sample was standardized to GAPDH. Each sample in the experiments was set with three parallel wells.
1.5 Cell Counting Kit-8 (CCK-8) assay
To evaluate cell survival rate [Citation26], transfected cells (5 × 103 cells/well) were planted into 96-well plates and cultured at 37°C with 5% CO2. At 0, 24, 48, and 72 h, 10 μL CCK-8 reagent (Dojindo, Japan) was respectively added to each well. The cells were then cultured for another 2 h, and the absorbance was detected at 450 nm using a Microplate Reader (Bio-Rad, USA).
1.6 Cell migration and invasion evaluation
For cell migration examination [Citation27], A549 or PC-9 cells (5 × 103 cells/well) were seeded into 24-well plates. Cell single layer was scraped with a sterile microtubule tip, and cells were rinsed with serum-free medium 3 times. Images of each wound were taken at 0 and 24 h at the same location. Wound healing area was then measured. Wound images were analyzed by ImageJ software 3.2 (National Institutes of Health, Bethesda, MD).
Transwell invasion assay [Citation25] was conducted in a 24-well Transwell chamber (8 μm, Corning, NY, USA) precoated with 100 μg Matrigel (Becton-Dickinson, Bedford, USA). Simply, cells were seeded into the upper chamber and cultivated in 500 μl FBS-free DMEM. The lower chamber was placed with 500 μl medium + 10% FBS. Next, cells were incubated in a moist incubator with 5% CO2 at 37°C for 24 h. Cells in the upper chamber were removed and then the chamber was washed with phosphate buffer saline (PBS). Cells in the lower chamber were treated with 4% formaldehyde and dyed with 1% crystal violet for 5 min, which were defined as invading cells. Lastly, invading cells were counted in five random fields (×100).
1.7 Dual-luciferase assay
As described previously [Citation25], ERO1A mRNA 3’-untranslated region (3’-UTR) containing potential target region of miR-218-5p was PCR amplified. PCR was also applied to amplify mutant (MUT) ERO1A mRNA 3’-UTR. Thereafter, the region was cloned into pmirGLO dual-luciferase miRNA target expression vectors (Promega Corp., Madison, WI, USA), identified as dual-luciferase reporter vectors. The inserted fragments were identified by commercial sequencing in Sango Biotech (Shanghai, China). Dual-luciferase reporter plasmids of ERO1A-wild type (WT) and ERO1A-MUT were transfected with miR-218-5p mimics (50 nM) or NC mimics (50 nM) into cancer cells by using JetPRIME kit (Polyplus-transfection). 48 h later, cells were collected. Luciferase intensity was evaluated by Dual-Luciferase® Reporter Assay System (Promega Corp). Three replicates were performed for all transfection experiments.
1.8 HUVEC tube formation assay
To research whether miR-218-5p regulates angiogenesis through targeting ERO1A, HUVEC tube formation experiment was performed [Citation21]. The pre-iced Matrigel (Merck Millipore, USA) was coated onto 96-well plates, followed by polymerizing at room temperature. HUVECs (1 × 104 cells) were plated onto the plates and grown in differently transfected tumor cell-conditioned medium. After 7 h, angiogenesis statuses were observed with an inverted microscope (Olympus, Japan). Each treatment required three separate experiments.
1.9 Statistical analysis
Data were analyzed using GraphPad Prism 5.0 software, and displayed as mean ± standard deviation. The difference between two groups was determined by Student’s two-tailed test, while one-way analysis of variance followed by Tukeys post hoc test was used for comparisons among more than three groups. Survival was measured by Kaplan–Meier. A statistical significance was reached when P < 0.05, ** meaned p <0.01, and *** meaned p <0.001.
2. Results
To comprehensively reveal the mechanism of miR-218-5p in LUAD, we assayed its expression and effects on cell functions in LUAD. Additionally, combining bioinformatics methods, we screened the target of miR-218-5p and identified miR-218-5p/ERO1A regulatory axis in LUAD. The findings here might open novel horizons and generate novel targets for LUAD management.
2.1 Remarkable miR-218-5p downregulation in LUAD
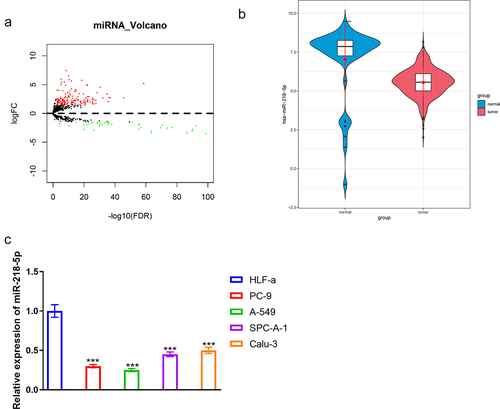
To screen the differentially expressed genes, a differential expression analysis was introduced on the downloaded miRNA profile, obtaining 184 DEmiRNAs (upregulated: 145, downregulated: 39), in which miR-218-5p was notably downregulated (). Several studies mentioned that miR-218-5p overexpression restrained cell proliferation in vitro and tumor growth in vivo in LUAD [Citation13,Citation28]. Therefore, combining the results of the bioinformatics analyses and literature review, miR-218-5p was determined as the research object. Next, expression detection presented markedly lower miR-218-5p level in LUAD cell lines (A549, PC-9, Calu-3, and SPC-A-1) than that in normal cell line (HLF-a) (P < 0.05, ). Thus, PC-9 and A549 were utilized for the following assays.
Figure 1. MiR-218-5p expression status in LUAD. (a) Volcano plot of DEmiRNAs in TCGA-LUAD. Red: upregulated miRNAs, green: downregulated ones. (b) Violin of miR-218-5p level in normal and tumor groups. (v) Relative miR-218-5p expression in HLF-a, PC-9, A549, SPC-A-1 and Calu-3 cell lines. *** P < 0.001.
2.2 MiR-218-5p suppresses proliferation, migration, and invasion of LUAD cells
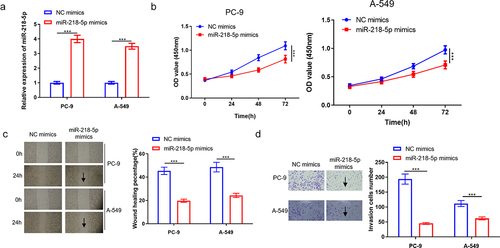
After cell transfection, in vitro functional assays were performed for determination of the function of miR-218-5p in LUAD. As exhibited in , miR-218-5p mimics transfection notably elevated miR-218-5p expression level in PC-9 and A549 cell lines (P < 0.05). Importantly, miR-218-5p mimics resulted in the significant suppression of survival of PC-9 and A549 cells in a time-dependent manner (P < 0.05, ). Moreover, it was observed that miR-218-5p overexpression significantly repressed PC-9 and A549 cell migration (P < 0.05, ). Transwell assay indicated that miR-218-5p overexpression dramatically hampered cell invasion (P < 0.05, ). To sum up, miR-218-5p overexpression repressed LUAD cell progression.
Figure 2. MiR-218-5p represses LUAD cell progression. (a) Transfection efficiency of miR-218-5p mimics in LUAD cells (PC-9 and A549). (b) The effect of overexpressed miR-218-5p on LUAD cell proliferation. (c) The effect of miR-218-5p overexpression on LUAD cell migration (40×). (d) The impact of overexpressed miR-218-5p on LUAD cell invasion (100×). ***P < 0.001.
2.3 MiR-218-5p directly targets ERO1A in LUAD
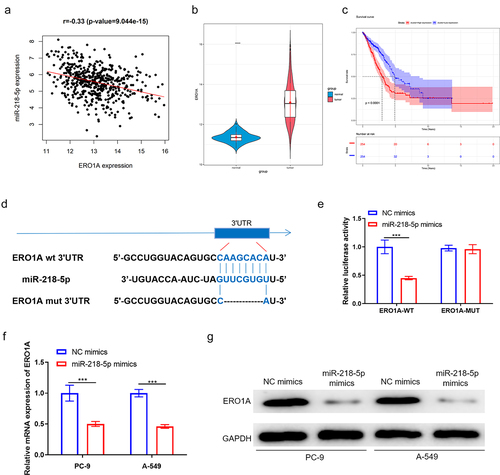
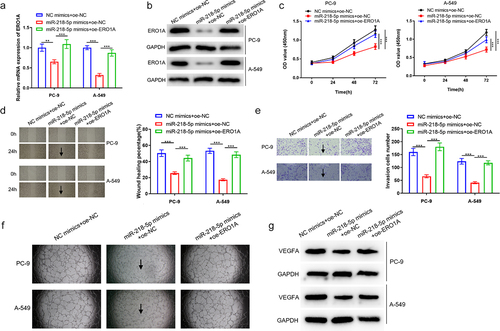
ERO1A has been reported as a tumor-promoting factor in lung cancer [Citation29,Citation30]. To identify whether miR-218-5p could specifically suppress ERO1A expression in LUAD, a correlation analysis was introduced (). Pearson analysis revealed the negative relevance of miR-218-5p to ERO1A, and ERO1A was thought as a downstream gene of miR-218-5p. Additionally, expression analysis showed that ERO1A was prominently stimulated in LUAD tissue based on TCGA-LUAD dataset (). Moreover, survival analysis revealed that ERO1A level was relevant to LUAD patient’s poor prognosis (). Furthermore, TargetScan database analyzed that miR-218-5p had targeted binding sites with ERO1A (). Hence, we posited ERO1A as a target of miR-218-5p.
Figure 3. ERO1A is a target of miR-218-5p. (a) Scatter diagram of correlation between ERO1A and miR-218-5p. (b) Violin plot of ERO1A expression level in normal (blue) and tumor (red) samples. (c) Survival analysis of ERO1A expression on patient’s prognosis. Blue: low expression, red: high expression. (d) Potential target sequences of miR-218-5p on ERO1A transcript. (e) Verification of the targeting between two genes. (f-g) The impacts of miR-218-5p high expression on ERO1A mRNA and protein expression. ***P < 0.001.
Thereafter, dual-luciferase reporter gene detection unveiled that the luciferase intensity markedly declined in LUAD cells with ERO1A-WT and miR-218-5p mimics (P < 0.05), which further indicated that miR-218-5p could target ERO1A 3’-UTR (). Furthermore, qRT-PCR and western blot discovered that miR-218-5p upregulation prominently restrained ERO1A expression in LUAD cells (P < 0.05, ). Above all, our data presented that ERO1A was a direct downstream target of miR-218-5p and was utilized for subsequent analyses.
2.4 MiR-218-5p restrains LUAD cell malignant progression via binding ERO1A
To determine whether miR-218-5p-mediated ERO1A expression inhibition represses LUAD progression, we constructed the following groups: NC mimics+oe-NC, miR-218-5p mimics+oe-NC, and miR-218-5p mimics+oe-ERO1A. testified transfection efficiency of miR-218-5p and ERO1A, and it was discovered that ERO1A level was notably lower in miR-218-5p mimics+oe-NC group than that in miR-218-5p mimics+oe-ERO1A group (P < 0.05). Afterward, CCK-8 assay revealed that oe-ERO1A reduced the inhibition of miR-218-5p overexpression on LUAD cell proliferative ability (P < 0.05, ). Similarly, ERO1A overexpression partially reversed the suppression of miR-218-5p stimulation on LUAD cell migration (P < 0.05, ). Transwell assay presented that miR-218-5p overexpression restrained LUAD cell invasion, while was reversed by ERO1A overexpression (P < 0.05, ). Angiogenesis abilities and VEGFA protein expression were evaluated subsequently, where it was observed that angiogenesis ability and VEGFA protein level could be inhibited by miR-218-5p upregulation while the inhibiting effects could be reversed by the ERO1A overexpression (). Collectively, miR-218-5p/ERO1A axis may affect LUAD cell processes, and angiogenesis through mediating ERO1A.
Figure 4. MiR-218-5p represses LUAD cell progression, and angiogenesis through targeting ERO1A. (a) ERO1A mRNA level in NC mimics+oe-NC, miR-218-5p mimics+oe-NC and miR-218-5p mimics+oe-ERO1A groups. (b) ERO1A protein expression level in each treatment group. (c) Changes in proliferation of LUAD cells. (d) Changes in migration of LUAD cells (40×). (e) Changes in invasion of LUAD cells (100×). (f) Angiogenesis status in the differently treated groups. (g) VEGFA protein expression level in the differently treated groups. **P < 0.01 vsPvs
3. Discussion
LUAD faces a huge challenge for clinical treatment owing to its difficult diagnosis in the early stage, easy recurrence and bad prognosis [Citation31]. Major development of gene sequencing technology accelerates targeted therapy for LUAD, contributing to patients’ survival improvement [Citation32]. MiRNAs have been attested to induce tumor suppression or acceleration, and exert the function of prognosis-prediction biomarkers [Citation33]. Differential-expression miRNAs in LUAD were determined utilizing bioinformatics method, from which downregulated miR-218-5p was uncovered [Citation12]. Likewise, this work confirmed this downregulation in LUAD tissue. Moreover, miR-218-5p upregulation repressed LUAD cell functions. In concordance with these findings, Zhu et al. [Citation11]. described that dramatic miR-218-5p downregulation in non-small cell lung cancer correlates with suppression in cell migration and proliferation through binding to EGFR. Hence, we hypothesized that miR-218-5p was able to constrain LUAD cell invasion and migration as an inhibitor.
Additionally, miRNAs have wide targets to affect tumor progression, like cell proliferation, death, angiogenesis, metastasis and drug resistance [Citation34–36]. Despite empirical evidence relating to functions of miR-218-5p in LUAD development, investigation on the exact mechanism is warranted. This work uncovered the negative relevance of miR-218-5p to ERO1A utilizing correlation analysis, due to which ERO1A was chosen for study. ERO1A was a target of miR-218-5p discovered by bioinformatics method. Dual-luciferase method also verified this targeting relationship. It is well known that ERO1A oxidates PDI to facilitate disulfide bond [Citation37,Citation38] that is a requisite for protein functions [Citation39]. Moreover, high-expression ERO1A in cancers pertains to tumor cell migration and proliferation [Citation18,Citation22,Citation40]. Meanwhile, ERO1A overexpression was associated with poor prognosis [Citation19,Citation37] that coincides exactly with our data. Afterward, functional experiment denoted that miR-218-5p constrained LUAD cell functions through inhibiting ERO1A expression.
In the early stage of cancer, distant metastasis usually occurs through blood vessels, while tumor cells are capable of stimulating the formation of novel blood vessels by directly releasing VEGF [Citation41]. Additionally, VEGF contributes to directly target tumor cell growth and metastasis [Citation42]. Thus, angiogenesis has been a hopeful target for lung cancer management and the corresponding anti-angiogenesis drugs are developed in clinical trials [Citation43], but the mechanism of action still needs to be further studied. MiRNA dysregulation might result in abnormal angiogenesis and cancer development [Citation44]. MiR-141 upregulation in serum exosomes in lung cancer might work on angiogenesis and tumorigenesis through targeting GAX [Citation45]. MiR-126 expression repair downregulates VEGF to represses lung cancer cell growth [Citation46]. Our experimental data suggested that miR-218-5p upregulation restrains angiogenesis and VEGFA protein expression to hamper LUAD cell invasion and migration, which is probably due to the role of miRNA in angiogenesis [Citation45].
Furthermore, this research observed that ERO1A overexpression reversed LUAD cell functions and angiogenesis induced by miR-218-5p. Yang et al. [Citation20] stated that ERO1A high-expression in metastatic hepatocellular carcinoma tissue accelerates cancer cell angiogenesis and metastasis through SIPR1/STAT3/VEGF-A pathway. Transcription factor nuclear factor IB overexpression increases ERO1A expression to stimulate breast cancer angiogenesis and aggravate prognosis [Citation22]. Little has been uncovered with regard to miR-218-5p-ERO1A targeting, we therefore summarized a novel mechanism in which miR-128-5p regulating LUAD cell functions and angiogenesis by directly targeting ERO1A. Nonetheless, the research on ERO1A downstream pathway and its impacts on oxidative folding awaits to be deepened.
Conclusion
This work indicated the targeting relationship of miR-218-5p and ERO1A, and clarified their exact roles in LUAD progression. Data unveiled that miR-218-5p restrained LUAD cell proliferation, invasion, migration, and angiogenesis through modulating ERO1A. The results help us to figure out the pathogenesis of LUAD and to develop novel targeted therapeutic regimens in the future. Additionally, current studies lack evidence to confirm the regulation of miR-218-5p/ERO1A axis on in vivo tumor metastasis and lack of studies on miR-218-5p regulation-related pathways, which encourage us to explore the molecular mechanisms and signaling pathways in the progression of LUAD.
Contributors’ statement:
(I) Conception and design: Gang Chen
(II) Administrative support: Kunyu Wang
(III) Provision of study materials or patients: Gang Chen
(IV) Collection and assembly of data: Qihao Wang
(V) Data analysis and interpretation: Gang Chen
(VI) Manuscript writing: All authors
(VII) Final approval of manuscript: All authors
Ethics approval and consent to participate
Not applicable.
Supplemental Material
Download Zip (1.6 MB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data Availability Statement:
The data and materials in the current study are available from the corresponding author on reasonable request.
Supplementary material:
Supplemental data for this article can be accessed here
Additional information
Funding
References
- Weber M, McWilliams A, Canfell K. Prospects for cost-effective lung cancer screening using risk calculators. Transl Cancer Res. 2019;8(S2):S141–s144.
- Ferlay J, Colombet M, Soerjomataram I, et al. Cancer statistics for the year 2020: an overview. Int J Cancer. 2021;149(4):778–789.
- Patel MI, Cheng I, Gomez SL. US lung cancer trends by histologic type. Cancer. 2015;121:1150–1152.
- Peters S, Adjei AA, Gridelli C, et al. Metastatic non-small-cell lung cancer (NSCLC): ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2012;23(Suppl 7):vii56–64.
- Hirsch FR, Scagliotti GV, Mulshine JL, et al. Lung cancer: current therapies and new targeted treatments. Lancet. 2017;389:299–311.
- Blandin Knight S, Crosbie PA, Balata H, et al. Progress and prospects of early detection in lung cancer. Open Biol. 2017;7(9):170070.
- Saliminejad K, Khorram Khorshid HR, Soleymani Fard S, et al. An overview of microRNAs: biology, functions, therapeutics, and analysis methods. J Cell Physiol. 2019;234(5):5451–5465.
- Chen L, Heikkinen L, Wang C, et al. Trends in the development of miRNA bioinformatics tools. Brief Bioinform. 2019;20(1836–1852):1836–1852.
- Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269.
- Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16(3):203–222.
- Zhu K, Ding H, Wang W, et al. Tumor-suppressive miR-218-5p inhibits cancer cell proliferation and migration via EGFR in non-small cell lung cancer. Oncotarget. 2016;7(19):28075–28085.
- Peng Z, Pan L, Niu Z, et al. Identification of microRNAs as potential biomarkers for lung adenocarcinoma using integrating genomics analysis. Oncotarget. 2017;8:64143–64156.
- Yang Q, Li J, Hu Y, et al. MiR-218-5p suppresses the killing effect of natural killer cell to lung adenocarcinoma by targeting SHMT1. Yonsei Med J. 2019;60:500.
- Zhu J, Wang M, Hu D. Development of an autophagy-related gene prognostic signature in lung adenocarcinoma and lung squamous cell carcinoma. PeerJ. 2020;8(e8288):e8288.
- Tanaka T, Kutomi G, Kajiwara T, et al. Cancer-associated oxidoreductase ERO1-α promotes immune escape through up-regulation of PD-L1 in human breast cancer. Oncotarget. 2017;8:24706–24718.
- Kutomi G, Tamura Y, Tanaka T, et al. Human endoplasmic reticulum oxidoreductin 1-α is a novel predictor for poor prognosis of breast cancer. Cancer Sci. 2013;104:1091–1096.
- Seol SY, Kim C, Lim JY, et al. Overexpression of endoplasmic reticulum oxidoreductin 1-α (ERO1L) is associated with poor prognosis of gastric cancer. Cancer Res Treat. 2016;48:1196–1209.
- Zhang J, Yang, J., Lin, C., Liu, W., Huo, Y., Yang, M., Jiang, S. H., Sun, Y., et al. Endoplasmic Reticulum stress-dependent expression of ERO1L promotes aerobic glycolysis in Pancreatic Cancer. Theranostics. 2020;10:8400–8414. DOI:10.7150/thno.45124
- Zhou J, Liu B, Li Z, et al. Proteomic analyses identify differentially expressed proteins and pathways between low-risk and high-risk subtypes of early-stage lung adenocarcinoma and their prognostic impacts. Mol Cell Proteomics. 2021;20:100015.
- Yang S, Yang C, Yu F, et al. Endoplasmic reticulum resident oxidase ERO1-Lalpha promotes hepatocellular carcinoma metastasis and angiogenesis through the S1PR1/STAT3/VEGF-A pathway. Cell Death Dis. 2018;9(1105). DOI:10.1038/s41419-018-1134-4
- Luo W, He D, Zhang J, et al. Knockdown of PPARδ induces vegfa-mediated angiogenesis via interaction with ERO1A in human colorectal cancer. Front Oncol. 2021;11(713892). DOI:10.3389/fonc.2021.713892
- Zilli F, Marques Ramos P, Auf der Maur P, et al. The NFIB-ERO1A axis promotes breast cancer metastatic colonization of disseminated tumour cells. EMBO Mol Med. 2021;13(e13162). DOI:10.15252/emmm.202013162
- Gan TQ, Chen, W. J., Qin, H., Huang, S. N., Yang, L. H., Fang, Y. Y., Pan., et al. Clinical value and prospective pathway signaling of microRNA-375 in lung adenocarcinoma: a study based on the Cancer Genome Atlas (TCGA), Gene Expression Omnibus (GEO) and bioinformatics analysis. Med Sci Monit. 2017;23:2453–2464.
- Jiang N, Zou C, Zhu Y, et al. HIF-1ɑ-regulated miR-1275 maintains stem cell-like phenotypes and promotes the progression of LUAD by simultaneously activating Wnt/β-catenin and Notch signaling. Theranostics. 2020;10:2553–2570.
- Huang D, Li C. circ-ACACA promotes proliferation, invasion, migration and glycolysis of cervical cancer cells by targeting the miR-582-5p/ERO1A signaling axis. Oncol Lett. 2021;22(795). DOI:10.3892/ol.2021.13056
- Wang G, Han J, Wang G, et al. ERO1α mediates endoplasmic reticulum stress-induced apoptosis via microRNA-101/EZH2 axis in colon cancer RKO and HT-29 cells. Hum Cell. 2021;34:932–944.
- Yang J, Xu Y, Huo Y, et al. ERO1L promotes hepatic metastasis through activating epithelial-mesenchymal transition (EMT) in pancreatic cancer. J Immunol Res. 2021;2021(5553425):1–10.
- Zeng F, Wang Q, Wang S, et al. Linc00173 promotes chemoresistance and progression of small cell lung cancer by sponging miR-218 to regulate Etk expression. Oncogene. 2020;39:293–307.
- Lei Y, Zang, R., Lu, Z., Zhang, G., Huang, J., Liu, C., Wang, Z., Mao, S., Che, Y., Wang., et al. ERO1L promotes IL6/sIL6R signaling and regulates MUC16 expression to promote CA125 secretion and the metastasis of lung cancer cells. Cell Death Dis. 2020;11(853). DOI:10.1038/s41419-020-03067-8
- Shi X, Wu J, Liu Y, et al. ERO1L promotes NSCLC development by modulating cell cycle-related molecules. Cell Biol Int. 2020;44:2473–2484.
- Liu LP, Lu, L., Zhao, Q. Q., Kou, Q. J., Jiang, Z. Z., Gui, R., Luo, Y. W., et al. Identification and validation of the pyroptosis-related molecular subtypes of lung adenocarcinoma by bioinformatics and machine learning. Front Cell Dev Biol. 2021;9(756340). DOI:10.3389/fcell.2021.756340
- Arbour KC, Riely GJ. Systemic therapy for locally advanced and metastatic non-small cell lung cancer: a review. JAMA. 2019;322:764–774.
- Nelson KM, Weiss GJ. MicroRNAs and cancer: past, present, and potential future. Mol Cancer Ther. 2008;7:3655–3660.
- Wang B, Wang D, Yan T, et al. MiR-138-5p promotes TNF-alpha-induced apoptosis in human intervertebral disc degeneration by targeting SIRT1 through PTEN/PI3K/Akt signaling. Exp Cell Res. 2016;345:199–205.
- Mao Y, Wu S, Zhao R, et al. MiR-205 promotes proliferation, migration and invasion of nasopharyngeal carcinoma cells by activation of AKT signalling. J Int Med Res. 2016;44:231–240.
- Li S, Zeng A, Hu Q, et al. miR-423-5p contributes to a malignant phenotype and temozolomide chemoresistance in glioblastomas. Neuro Oncol. 2017;19:55–65.
- Shergalis AG, Hu S, Bankhead A 3rd, et al. Role of the ERO1-PDI interaction in oxidative protein folding and disease. Pharmacol Ther. 2020;210(107525). DOI:10.1016/j.pharmthera.2020.107525
- Araki K, Nagata K. Functional in vitro analysis of the ERO1 protein and protein-disulfide isomerase pathway. J Biol Chem. 2011;286:32705–32712.
- Cabibbo A, Pagani M, Fabbri M, et al. ERO1-L, a human protein that favors disulfide bond formation in the endoplasmic reticulum. J Biol Chem. 2000;275:4827–4833.
- Yan W, Wang X, Liu T, et al. Expression of endoplasmic reticulum oxidoreductase 1-α in cholangiocarcinoma tissues and its effects on the proliferation and migration of cholangiocarcinoma cells. Cancer Manag Res. 2019;11:6727–6739.
- Popper HH. Progression and metastasis of lung cancer. Cancer Metastasis Rev. 2016;35:75–91.
- Frezzetti D, Gallo M, Maiello MR, et al. VEGF as a potential target in lung cancer. Expert Opin Ther Targets. 2017;21:959–966.
- Shepherd FA. Angiogenesis inhibitors in the treatment of lung cancer. Lung Cancer. 2001;34(Suppl 3):S81–89.
- Sun LL, Li WD, Lei FR, et al. The regulatory role of micro RNA s in angiogenesis-related diseases. J Cell Mol Med. 2018;22:4568–4587.
- Wang W, Hong G, Wang S, et al. Tumor-derived exosomal miRNA-141 promote angiogenesis and malignant progression of lung cancer by targeting growth arrest-specific homeobox gene (GAX). Bioengineered. 2021;12:821–831. DOI:10.1080/21655979.2021.1886771
- Liu B, Peng XC, Zheng XL, et al. MiR-126 restoration down-regulate VEGF and inhibit the growth of lung cancer cell lines in vitro and in vivo. Lung Cancer. 2009;66:169–175.
