ABSTRACT
Testicular germ cell tumor (TGCT) is a relatively rare entity tumor, accounting for only 1% of all male cancers. However, it is the most common solid tumor in young men between 15 and 34 years old. Long noncoding RNAs (lncRNAs) are involved in various physiological and pathological processes. However, the functions of lncRNAs in TGCT have only rarely been investigated. LncRNAs associated with TGCT were identified using Gene Expression Omnibus (GEO) database and UCSC XENA database data mining. The effects of LINC00313 on NCCIT cell migration and invasion were evaluated in transwell assays. The expression levels of epithelial-mesenchyme transition (EMT)-related proteins in cells knockdown of LINC00313 were analyzed by Western blot. Correlation analyses between lncRNA LINC00313 expression and copy number variation (CNV) and immune cell infiltration were carried out using The Cancer Genome Atl as (TCGA) data. The effect of Panobinostatin targeting LINC00313 in TGCT cells was investigated. We observed higher LINC00313 expression in TGCT. The migratory and invasive properties of TGCT cells were augmented by LINC00313, likely via its effects on modulating the expression of epithelial-mesenchyme transition (EMT) related proteins: CTNNB1, ZEB1, CDH2, Snail and VIM. Moreover, LINC00313 expression and CNV correlated negatively with the infiltration of immune cells. In addition, Panobinostat might be a possible candidate drug to target LINC00313 in TGCT. LINC00313 performs important pro-migration and invasion functions in the pathogenesis of TGCT. LINC00313 could be used as diagnostic, prognostic, immune marker and therapeutic target to develop effective treatment of TGCT.
Graphical Abstract
Highlights
LINC00313 could be a prognostic marker of TGCT.
LINC00313 promote the migration and invasion of TGCT.
LINC00313 influence the EMT signaling of TGCT.
Introduction
Testicular germ cell tumor (TGCT) is a relatively rare malignancy, representing only 1% of all cancers in men. However, it is the most common solid tumor in young men between the ages of 15 and 34 [Citation1]. TGCTs are classified broadly into seminomas, which resemble primordial germ cells (PGCs), and non-seminomas, which are either undifferentiated (embryonic carcinoma) or differentiated (teratoma, yolk sac tumor, choriocarcinomas) [Citation2]. The incidence of TGCTs has increased worldwide during recent years, particularly in men of European descent, although a decline in mortality rates has been reported in western countries. For the clinical management of TGCTs, increased levels of lactate dehydrogenase, alpha-fetoprotein, and human chorionic gonadotropin are essential tools for diagnosis, risk assessment, and patient prognosis [Citation3]. However, serum levels of these tumor markers are shown increased in only 60% of patients with TGCTs [Citation4]. Hence, alternative and more functional markers should be investigated and introduced for the diagnosis and prognosis of TGCTs.
Noncoding DNA covers 95% of DNA sequences in the human genome, most of which are transcribed into various non-coding RNAs (ncRNAs). Testes express large numbers of ncRNAs, mainly microRNAs (miRNAs), piwi-interacting RNAs (piRNAs) and long noncoding (lncRNAs), which play roles in the regulation of gene expression. LncRNAs are conventionally defined as transcripts with lengths exceeding 200 nucleotides that are not translated into protein. Functions of lncRNAs rely on their capacity to bind and regulate a molecular partner, either via base-pair interactions or through their secondary structure [Citation5]. Alterations in lncRNA expression might be involve in testicular germ cell tumorigenesis. This hypothesis is supported by the observation that many TGCT risk loci identified in a genome-wide association study (GWAS) are in the non-coding regions of the genome [Citation6]. PRY4-IT1, a lncRNA produced within the intronic region of SPRY4 (encoding sprouty RTK signaling antagonist 4), acts as an oncogene in TGCT development and its expression is silenced in TGCT cells [Citation7]. Previously, we found that the expression of lncRNA LINC00467 correlated positively with poor prognosis and the pathological grade of TGCT [Citation8]. However, lncRNAs in TGCT have only been investigated rarely. Recently, some studies reported that lncRNA LINC00313 promote tumorigenesis and metastasis in Human cancer. In this study, we explore the role of LINC00313 in TGCT progression to understand the importance of the LINC00313 in TCGT and provide insights into the role of LINC00313 in the progression of TGCT.
To the best of our knowledge, the relationship between LINC00313 and TGCT has not been previously reported. In this study, we firstly reported that LINC00313 was overexpressed in TGCT. Therefore, this study aims to address this research question with the hope of discovering an additional therapeutic target and molecular marker of clinical significance in TGCT treatment and diagnosis.
Materials and methods
Online database data analysis
The Gene Set Cancer Analysis (GSCA; http://bioinfo.life.hust.edu.cn/GSCA/#/) database is an interactive web version tool developed by the research team of Huazhong University of Science and Technology [Citation9]. It was used to analyze the relationship between LINC00313 CNV and the infiltration of two immune cell types, CD8 + T cells and dendritic cells (DCs), in TGCT. The expression data of LINC00313 in normal samples, seminoma samples and non-seminoma samples are obtained from the GEO database (https://www.ncbi.nlm.nih.gov/geo/). These data are from the same dataset as GSE3218 [Citation10,Citation11]. Gene Expression Profiling Interactive Analysis (GEPIA, http://gepia.cancer-pku.cn/) was developed based on the TCGA and Genotype-Tissue Expression (GTEx) databases [Citation12]. We used the UCSC XENA (https://xena.ucsc.edu/) online tool [Citation13] to analyze the expression of LINC00313 and its correlation with the survival of patients with TGCT from TCGA. TGCT patients were divided into two groups according to the expression of a certain gene (high or low expression). In addition, the GEPIA database was used to analyze the correlation between LINC00313 and the expression of epithelial-mesenchymal transition (EMT) related genes, such as VIM (encoding vimentin, ZEB1 (encoding zinc finger E-box binding homeobox 1), SNAIL (encoding snail family transcriptional repressor 1), CTNNB1 (encoding catenin beta 1), and CDH2 (encoding N-cadherin). We have analyzed interacted miRNAs associated with the lncRNA and genes by using Targetscan (https://www.targetscan.org/vert_72/) and miRcode (http://mircode.org/index.php)[Citation14–16]. A model of EMT-related genes regulated by LINC00313 based on the prognostic classification of TGCT was constructed using the online tool ASSISTANT for Clinical Bioinformatics (https://www.aclbi.com/static/index.html#/) along with the R software package glmnet (v 4.1–1) and timeROC (v 0.4) based on TCGA TGCT cohort data. Sangerbox tools (http://sangerbox.com/ Index) were used to assess the relationships between LINC00313 expression levels and the immune score, stromal score, and immune cell enrichment score in TGCT. LINC00313 involvement in immune cell and immune-related pathway was analyzed using ImmuLnc [Citation17] tool (http://bio-bigdata. hrbmu.edu.cn/ImmLnc/index.jsp). Finally, LncMap [Citation18] tool was used to analyze the relationship between LINC00313 expression and the half-maximal inhibitory concentration (IC50) of various drugs.
Cell culture and siRNA transfection
The human TGCT cell line, NCCIT, used in this study, was donated by the Associate Researcher Su-Ren Chen. NCCIT cells were cultured in Roswell Park Memorial Institute (RPMI)-1640 medium (Gibco, Grand Island, NY, USA) containing 10% fetal bovine serum (FBS; Gibco) at 37°C in 95% air and 5% carbon dioxide (CO2). Confluent cells were digested and passaged with 0.1% trypsin (Gibco). Negative control small interfering RNA (siRNA) and siRNAs targeting LINC00313 were designed and synthesized by Tsingke Biotechnology Co., Ltd (Beijing, China). TGCT cells were prepared before transfection. Plasmid transfection was performed using Lipofectamine 2000 transfection reagent (Invitrogen, Waltham, MA, USA) according to the manufacturer’s instructions. All siRNAs were designed and synthesized by Ribobio (Guangzhou, China). LINC00313 siRNA-1: GCTTCCTGGATTGCATAAA; LINC00313 siRNA-2: GGCTCCTTCTCCTTACATA. Quantitative real-time reverse transcription PCR (qRT-PCR) with relative quantification was used to assess the knockdown efficiency of the siRNAs to identify the most effective molecule.
Transwell cell migration and invasion experiments
Transwell migration and invasion assays were performed using 8.0 μm Transwell Permeable Supports (Corning Inc., Corning, NY, USA). Cells at a density of 5 × 104 cells per well in 100 µl serum-free medium were seeded into the upper chamber pre-coated with Matrigel Matrix (BD Biosciences, San Jose, CA, USA), Then, 600 μl medium containing 15% FBS was added to the lower chamber. The time for the migration assay was 12 h and that for the invasion assay was 36 h, cells that did not invade through the membrane were mechanically removed with a cotton swab. Next, 4% paraformaldehyde was used to fix the cells on the bottom surface of the membrane for 10 min, which were then stained with crystal violet solution. The number of invaded cells was counted in five randomly selected fields.
Western blotting
The cellular protein precipitates were extracted using radioimmunoprecipitation assay (RIPA) lysis buffer (Beyotime, Nanjing, China). Proteins were separated on 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels, electro-blotted onto a polyvinylidene difluoride (PVDF) membrane, and incubated with anti-VIM antibodies (Cell Signaling Technology, Danvers, MA, USA), anti-ZEB1 antibodies (Cell Signaling Technology, Danvers, MA, USA), anti-SNAIL antibodies (Cell Signaling Technology, Danvers, MA, USA), anti-CTNNB1 antibodies (Cell Signaling Technology, Danvers, MA, USA), anti-CDH2 antibodies (Cell Signaling Technology, Danvers, MA, USA), or anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) antibodies ((Millipore, Billerica, MA, USA). Then, the membranes were incubated with secondary anti-rabbit, anti-donkey, or anti-mouse horseradish peroxidase-conjugated antibodies (Santa Cruz Biotechnology). A gel imaging system was used to scan the protein bands using a chemiluminescent ECL reagent (Millipore). GAPDH was used as an internal reference.
Statistical analysis
Statistical analysis was performed using SPSS ® 18.0 package for windows (IBM Corp., Armonk, NY, USA). Data are expressed as percentages. A P-value < 0.05 was considered as statistically significant. Student’s t test was used to calculate the significance of the difference between the two groups. Significant differences between multiple data sets were calculated using one-way analysis of variance (ANOVA). disease free interval survival (DFI) and progression free interval survival (PFI) were calculated using a log-rank test.
Results
In the present study, we tried to explore the function of LINC00313 in the Testicular Germ Cell Tumors based on a series of bioinformation analyses and in vitro experiments. We found LINC00313 was higher expression in TGCT and associated significantly with prognosis of patients. Further analysis found that LINC00313 regulated expression of EMT-related proteins and associated with immune cell infiltration. Finally, we have explored the association of LINC00313 expression and drug activity in TGCT.
LINC00313 CNV is associated with immune cell infiltration
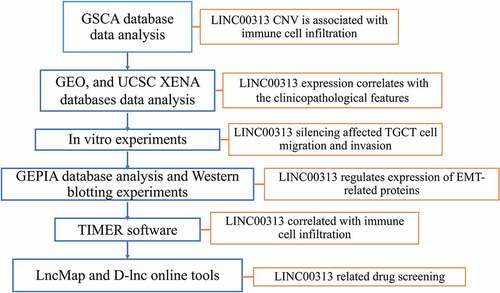
The relationship between LINC00313 CNV and TGCT was explored. As shown in ) amplification was the most common CNV type of LINC00313 in TGCT, and most of the amplifications were heterozygous CNVs. Further analysis of immune cell infiltration revealed that LINC00313 CNV correlated negatively and significantly with infiltrate scores, along with CD4 + T cell and DC infiltration ()). Moreover, we found that patients with a high copy number of LINC00313 had lower DFI and PFI ()).
Figure 1. Copy number variation of LINC00313 correlates with tumor immune invasion in TGCT.
(A) Stacked plot showing the proportional distribution of copy number variation of LINC00313 in TGCT; (B) Bubble plot of heterozygous copy number variation of LINC00313 in TGCT; (C) Bubble plot of homozygosis copy number variation of LINC00313 in TGCT; (D) Correlation between LINC00313 and the immune infiltration score, CD8 + T cell infiltration, and DC cell infiltration in TGCT. (E&F) The correlation between the copy number of LINC00313 and the DFI and PFI of patients with TGCT. CNV: Copy Number Variation; TGCT: Testicular Germ Cell Tumor; Homo: Homozygous; Amp: Amplification; Del: Deletion; cor: correlation; FDR: False Discovery Rates; DC: dendritic cells; TCGA: The Cancer Genome Atlas; DFI: Disease free interval; PFI: Progression free interval.
Higher LINC00313 Expression in TGCT
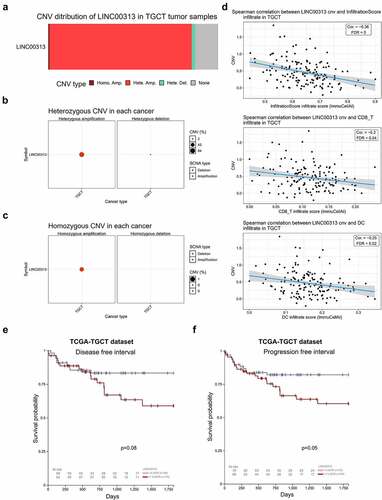
To determine the expression of LINC00313 in TGCT, TGCT data in the GEO database was used to analyze the expression of LINC00313 statistically, which showed that LINC00313 expression was upregulated significantly in TGCT tissues ()). Meanwhile, as shown in ) LINC00313 expression was upregulated significantly in different types of TGCT (seminoma and non- seminoma). As show in , the expression of LINC00313 has high sensitivity and specificity for distinguishing normal samples from TGCT samples, seminoma samples, non- seminoma samples. Moreover, LINC00313 expression was also associated significantly with DFI and PFI survival in patients with TGCTs (). Thus, LINC00313 could be used as an independent diagnosis and prognostic indicator for patients with TGCT.
Figure 2. Expression of LINC00313 correlates with the clinicopathological features of patients with TGCT.
(A) GEO database analysis shows that LINC00313 was notably upregulated in TGCT. (B & C) GEO database showing that LINC00313 was notably upregulated across different subtypes of TGCT. (D, E & F) The expression of LINC00313 has high sensitivity and specificity for distinguishing normal samples from TGCT samples, seminoma samples, non-seminoma samples. (G & H) The correlation between LINC00313 and the DFI and PFI of patients with TGCT. GEO: Gene Expression Omnibus; AUC: Area Under The Curve.
Silencing of LINC00313 inhibited the migration and invasion of TGCT cells
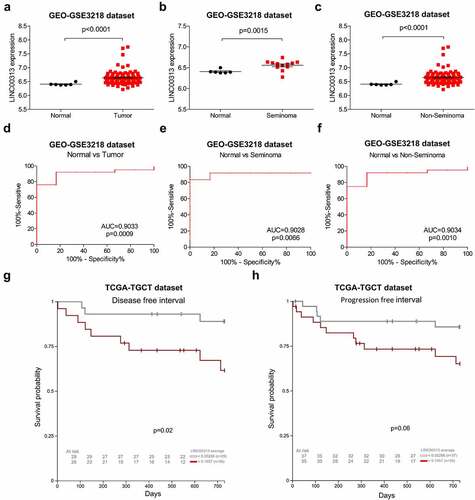
To determine whether LINC00313 can promote the migration and invasion of TGCT, we selected NCCIT cells for further in vitro experiments. The expression levels of LINC00313 in NCCIT was determined using qRT-PCR to verify the effects of the silencing of LINC00313 (). The migration and invasion abilities of TGCT cells was investigated using the Transwell assay(). LINC00313 silencing decreased the number of invaded and migrated cells significantly. This indicates that LINC00313 plays an important role in the migration and invasion of TGCT, which warrants further investigation.
Figure 3. The impact of LINC00313 on the migratory and invasive properties of TGCT. (A) The silencing efficiency of LINC00313 siRNA on the TGCA cell line was assessed using qRT-PCR. (B&C) Transwell cell migration assays enabled an assessment of how LINC00313 silencing affected TGCT cell migration (D&E) Transwell cell invasion assays enabled an assessment of how LINC00313 silencing affected TGCT cell invasion. NC: Negative Control; siRNA: small interfering RNA.
LINC00313 regulates expression of EMT-related proteins
The EMT signaling pathway has been implicated in several types of cancers. Notably, LINC00313 has previously been found to be linked to the EMT process in thyroid cancer cells [Citation19]. Based on this, we analyzed the relationship between LINC00313 and EMT markers (VIM, ZEB1, SNAIL, CTNNB1, and CDH2) using the GEPIA database and found that LINC00313 expression correlated significantly and positively with these EMT markers (). Therefore, we speculated that LINC00313 might be involved in process of TGCT by regulating EMT signals. Therefore the expression of EMT-related proteins after silencing of LINC00313 was assessed using western blotting. The levels of VIM, ZEB1, SNAIL, CTNNB1, and CDH2 proteins were downregulated significantly (), suggesting that the LINC00313-mediated EMT might be an important mechanism involved in TGCT migration and invasion. Then, we have analyzed interacted miRNAs associated with the lncRNA and genes, and drawn a lncRNA-miRNA-gene interaction network (). LINC00313 might mediate EMT through miR-138-5p, miR-150-5p, miR-204-5p and miR-205-5p.
Figure 4. Effects of silencing of LINC00313 on EMT-related proteins were investigated using Western blotting experiments.
(A–E) The association between LINC00313 and EMT markers were analyzed with the GEPIA database (F) After silencing of LINC00313, the levels of VIM, ZEB1, SNAIL, CTNNB1, and CDH2 were downregulated significantly. (G) LINC00313-miRNA-gene interaction network. TPM: Transcripts Per Kilobase Million.
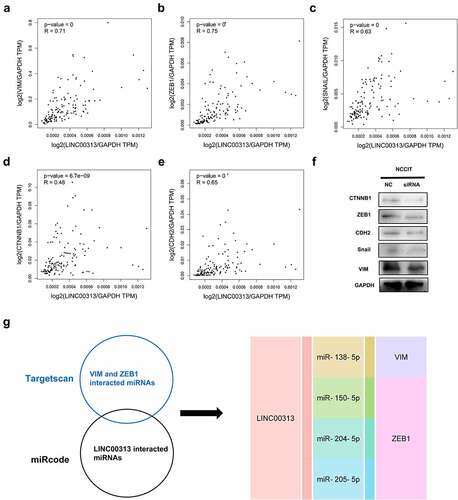
We proceeded to build a risk model through Lasso regression with reference to the EMT-related genes regulated by LINC00313 (). The combination of 4 genes out of 5 input genes yielded the highest C-index (), Risk score = (0.3675)*CTNNB1+(−0.5661)*ZEB1+(0.5392)*VIM+(0.4163)*SNAI1, lambda.min = 0.0056. Based on our model, we stratified patients according to low- and high-risk groups (). We noted that as the risk score increased, the patient’s mortality rate also gradually increased (). Patients in the high-risk group possessed a poorer DFS (). Finally, we plotted a time-dependent ROC curve and found that the AUCs at 1, 3, and 5 years were 0.75, 0.698, and 0.584, respectively().
Figure 5. Establishing a prognostic EMT-related gene risk signature for TGCT patients in the TCGA TGCT cohort. (A)LASSO Cox regression of the 5 EMT-related genes regulated by LINC00313. (B) Screening of the parameter in the LASSO Cox regression. (C) After dividing patients into high- and low-risk groups, the risk distribution, survival, and expression of 4 related genes for each patient was displayed. (D) Significant differences of DFS were observed between high-and low-risk TCGA TGCT patients (E) ROC curves exhibits the predictive sensitivity and specificity of the risk score at 1, 3, and 5 years. HR: hazard ratio.
LINC00313 is associated with immune cell infiltration
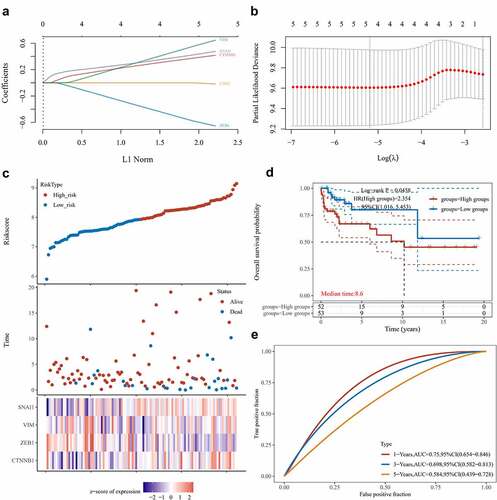
The relationship between the TGCT immune and stromal scores and LINC00313 was explored using the TIMER software. Our analysis found that LINC00313 expression correlated significantly and negatively with the estimated score and the immune score but not with the stromal score ( and ). Meanwhile, the quantities of CD8 + T cells, neutrophils, and dendritic cells were significantly lower in TGCT samples with high LINC00313 expression (). Furthermore, Spearman correlation analysis revealed that CD8 + T cells and DCs infiltration negatively correlated with LINC00313 expression (). The lower the expression of LINC00313, the higher the degree of CD8 + T cells and DCs tumor infiltration. Additionally, we also noted a significantly suppressed expression of immune checkpoint genes LAG3, PDCD1, and TIGIT in TGCT samples with high LINC00313 expression (). Candidate immune signaling pathways influenced by LINC00313 were investigated using the ImmuLnc tool. We found that levels of cytokines, transforming growth factor-beta (TGF-β) family members, chemokines, tumor necrosis factor (TNF) family members receptors, and cytokine receptors were raised when LINC00313 was overexpressed, while the T cell receptor (TCR) signaling pathway and antigen processing and presentation were reduced after LINC00313 overexpression (). Based on the above findings, we hypothesize that the TGCT tumor immune response and immune microenvironment were likely modulated by LINC00313.
Figure 6. Relationship between LINC00313 and immune cells and immune pathways in the TGCT immune microenvironment. (A) Correlation between LINC00313 and immune score. (B) Correlation between LINC00313 and stromal score. (C) Difference in immune cell score between high and low expression of LINC00313 in TGCT patients. (D) Correlation between LINC00313 and immune cell enrichment scores. (E) Differential expression of immune checkpoint genes between high and low expression of LINC00313 in TGCT patients. (F) Correlation between LINC00313 and the enrichment scores of various immune-related pathways. G1: LINC00313 high group; G2: LINC00313 low group. Est: Estimation; ES: Enrichment score.
Potential drug targeting LINC00313 in TGCTs
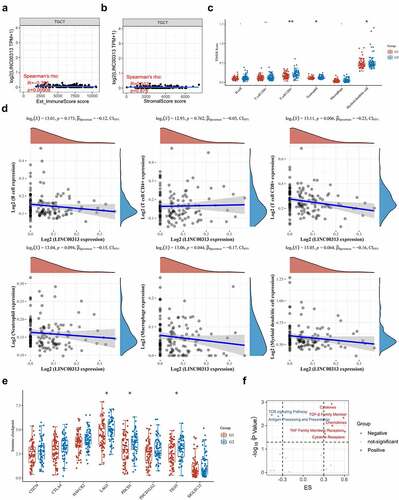
Our previous results indicated that LINC00313 potentially affects the migration and invasion of TGCTs, thus highlighting LINC00313 as a novel therapeutic target. Therefore, we explored the association of LINC00313 expression and drug activity in TGCTs. We used the LncMap tool to calculate the Spearman correlation coefficient between LINC00313 expression levels and the IC50 values of drugs. We observed that the LINC00313 expression was influenced by 15 drugs, among which panobinostat was found to be the best candidate drug to target LINC00313 in TGCT (). In addition, we also performed the same analysis using another prediction tool D-lnc. Interestingly, panobinostat was also found to be a drug that was highly compatible and had multiple potential binding sites ().
Figure 7. LINC00313 related drug screening. (A) LncMap analysis of the correlation between LINC00313 expression and the IC50 of various drugs. (B) D-lnc online tool predicts the score and binding site of drugs that may interact with LINC00313. Hit represents the position where the drug acts on LINC00313.
Discussion
The most frequent cancer in the young male population is TGCT. Radical orchiectomy is typically the first treatment for all testicular cancers, which has a high cure rate [Citation20]. However, in the patients with refractory disease, there are few alternative treatments. Meanwhile, although therapeutic approaches have improved, approximately 25% of the patients had relapse [Citation21]. Hence, the metastasis and recurrence of TGCT are one of the most intractable problems in the clinical setting. Previous studies have confirmed that AKT3 expression was higher in TGCT patients and might be a potential therapeutic target and a novel molecular marker of TGCT [Citation22]. Wu etc. have reported that Toll-like receptor 2 (TLR2) might affect the prognosis of TGCT, as well as be an indicator of immune function in the tumor microenvironment [Citation23]. However, the roles of lncRNAs in TGCT have been less studied before. In recent years, research on lncRNAs has received increased attention and has become a hotspot of tumor research [Citation24]. LncRNAs plays a crucial role in tumorigenesis, invasion, metastasis, and recurrence. There was a study reported that H19, a Long Non-coding RNA, played an oncogenic role in pan-cancer including TGCT and showed that miRNA-mediated lncRNA-TF-gene co-regulation is complicated yet important in cancers. And suggested that co-regulation-based approach is promising to identify TFs or genes related to cancer [Citation25]. Here, we especially report the role of LINC00313 in TGCT, while also referring to the valuable experience of previous studies.
In previous studies, LINC00313 was shown to be upregulated and correlated with poor prognosis in some cancers. For example, LINC00313 was reported to be highly expressed in lung cancer and to indicate shorter overall survival (OS) [Citation26]. High expression of LINC00313 was associated with shorter OS in osteosarcoma [Citation27]. To the best of our knowledge, the expression and role of LINC00313 in TGCT has not been reported. In the present study, we performed data mining using public databases to show that LINC00313 expression was upregulated significantly in TGCT tissue and that there was significantly association between DFI survival and LINC00313 expression. This is similar to the findings of another research team in osteosarcoma [Citation27] and cervical carcinoma [Citation28], who found that patients with high LINC00313 expression had poorer overall and disease-free survival. Moreover, the expression of LINC00313 has a good diagnostic value for TGCT. These results indicate that LINC00313 may be a potential diagnostic and prognostic marker for TGCT.
We also found significant associations between LINC00313 and tumorigenesis and metastasis. LINC00313 knockdown suppressed cell migration and invasion in the TGCT cell line. These data support the view that LINC00313 plays a very important role in the migration and invasion of TGCT cells. Previous studies demonstrated that LINC00313 participates in the progression of various tumors [Citation29,Citation30], which was consistent with our findings. The mechanism of LINC00313 dysregulation in TGCT requires further research, although our results showed that LINC00313 promote the migration and invasion abilities of TGCT cell lines. To our knowledge, our study is the first to characterize the functional role of LINC00313 in TGCTs.
EMT is critical for local invasion and cell dissemination, which in cancer, are associated with tumor initiation and progression, stemness, survival, and resistance to therapy [Citation31]. In our study, the western blotting showed that LINC00313 might exert its tumor-promotive role by modulating EMT signaling, which was consistent with the results of cell invasion and migration. EMT plays an important role in carcinogenesis, especial for local invasion and cell dissemination. Previous studies have also indicated that LINC00313 accelerated the progression, migration, and EMT of cancer cells, including cervical carcinoma [Citation28] and papillary thyroid carcinoma [Citation19,Citation30]. However, whether LINC00313 participates in the EMT process in TGCTs remains to be further confirmed.
The tumor microenvironment (TME) has been widely implicated in tumorigenesis because it harbors tumor cells that interact with surrounding cells through the circulatory and lymphatic systems to influence the development and progression of cancer [Citation32]. Infiltrating inflammatory cells in the TME play an important role in determining tumor survival and patient prognosis [Citation33]. In recent years, the study of the involvement of lncRNAs in immune regulation has become a research hot spot. In the present study, we found that there was a close relationship between LINC00313 and immunity in tumor tissues. LINC00313 expression correlated significantly and negatively with estimate score and immune score and was correlated significantly and negatively with CD8 + T cells and dendritic cells in the TME. Meanwhile, LINC00313 CNV also correlated significantly and negatively with CD4 + T cells and dendritic cells. Our results suggested that LINC00313 might be involved in the tumor immune process and the expression level of LINC00313 might serve as a predictor of the response to tumor immunotherapy. However, this hypothesis should be confirmed by further in vivo experiments in animals. This is also a limitation of the study.
The cost of developing drugs has always been high; therefore, most potential drugs are tested through drug activity screening in vitro with lower cost and shorter time. Based on the drug activity screen data, we revealed the associations among LINC00313 and cancer drugs. Panobinostat was identified as the best possible candidate drugs to target LINC00313 in TGCTs. However, the evaluation of lncRNAs for drug therapy is still in its early stages [Citation34].
Conclusion
In summary, we report the role of LINC00313 in TGCTs. We identified that LINC00313 performs important pro-migration and invasion functions in TGCT pathogenesis. Our results indicated that LINC00313 might be a diagnostic, prognostic, immune marker and therapeutic target for TGCTs, and will facilitate accurate and effective treatment of TGCTs.
Abbreviations
| TGCT | = | Testicular germ cell tumor; |
| lncRNAs | = | Long noncoding RNAs |
| GEO | = | Gene Expression Omnibus |
| CNV | = | copy number variation |
| TCGA | = | The Cancer Genome Atlas |
| PGCs | = | primordial germ cells |
| ncRNAs | = | non-coding RNAs |
| miRNAs | = | microRNAs |
| piRNAs | = | piwi-interacting RNAs |
| GSCA | = | Gene Set Cancer Analysis |
| DCs | = | dendritic cells |
| GEPIA | = | Gene Expression Profiling Interactive Analysis |
| GTEx | = | Genotype-Tissue Expression |
| EMT | = | epithelial-mesenchymal transition |
| RPMI | = | Roswell Park Memorial Institute |
| FBS | = | fetal bovine serum |
| siRNA | = | small interfering RNA |
| qRT-PCR | = | Quantitative real-time reverse transcription PCR |
| RIPA | = | radioimmunoprecipitation assay |
| SDS-PAGE | = | sodium dodecyl sulfate-polyacrylamide gel electrophoresis |
| PVDF | = | polyvinylidene difluoride |
| ANOVA | = | one-way analysis of variance |
| DFI | = | disease free interval survival |
| PFI | = | progression free interval survival |
| TGF-β | = | transforming growth factor beta |
| TNF | = | tumor necrosis factor |
| TCR | = | T cell receptor |
| TME | = | The tumor microenvironment |
| OS | = | overall survival. |
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Mannuel HD, Mitikiri N, Khan M, et al. Testicular germ cell tumors: biology and clinical update. Curr Opin Oncol. 2012;24(3):266–271.
- Batool A, Liu XM, Zhang CL, et al. Recent advances in the regulation of testicular germ cell tumors by microRNAs. Front Biosci (Landmark Ed). 2019;24(4):765–776.
- Deng C, Li Y, Zhou L, et al. HoxBlinc RNA Recruits Set1/MLL Complexes to Activate Hox Gene Expression Patterns and Mesoderm Lineage Development. Cell Rep. 2016;14(1):103–114.
- Lam MT, Cho H, Lesch HP, et al. Rev-Erbs repress macrophage gene expression by inhibiting enhancer-directed transcription. Nature. 2013;498(7455):511–515.
- Hong SH, Kwon JT, Kim J, et al. Profiling of testis-specific long noncoding RNAs in mice. BMC Genomics. 2018;19(1):539.
- Pyle LC, Nathanson KL. Genetic changes associated with testicular cancer susceptibility. Semin Oncol. 2016;43(5):575–581.
- Das MK, Furu K, Evensen HF, et al. Knockdown of SPRY4 and SPRY4-IT1 inhibits cell growth and phosphorylation of Akt in human testicular germ cell tumours. Sci Rep. 2018;8(1):2462.
- Bo H, Zhu F, Liu Z, et al. Integrated analysis of high-throughput sequencing data reveals the key role of LINC00467 in the invasion and metastasis of testicular germ cell tumors. Cell Death Discov. 2021;7(1):206.
- Liu CJ, Hu FF, Xia MX, et al. GSCALite: a web server for gene set cancer analysis. Bioinformatics. 2018;34(21):3771–3772.
- Kushwaha R, Jagadish N, Kustagi M, et al. Interrogation of a context-specific transcription factor network identifies novel regulators of pluripotency. Stem Cells. 2015;33(2):367–377.
- Korkola JE, Heck S, Olshen AB, et al. Development and Validation of a Gene-Based Model for Outcome Prediction in Germ Cell Tumors Using a Combined Genomic and Expression Profiling Approach. PLoS One. 2015;10(12):e0142846.
- Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98–W102.
- Goldman MJ, Craft B, Hastie M, et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat Biotechnol. 2020;38(6):675–678.
- Lewis BP, Shih IH, Jones-Rhoades MW, et al. Prediction of mammalian microRNA targets. Cell. 2003;115(7):787–798.
- Riffo-Campos AL, Riquelme I, Brebi-Mieville P. Tools for Sequence-Based miRNA Target Prediction: what to Choose? Int J Mol Sci. 2016 ;17(12):1987.
- Xiao F, Zuo Z, Cai G, et al. miRecords: an integrated resource for microRNA-target interactions. Nucleic Acids Res. 2009;37(Database):D105–110.
- Li Y, Jiang T, Zhou W, et al. Pan-cancer characterization of immune-related lncRNAs identifies potential oncogenic biomarkers. Nat Commun. 2020;11(1):1000.
- Li Y, Li L, Wang Z, et al. LncMAP: pan-cancer atlas of long noncoding RNA-mediated transcriptional network perturbations. Nucleic Acids Res. 2018;46(3):1113–1123.
- Zhao X, Hu X. Downregulated long noncoding RNA LINC00313 inhibits the epithelial-mesenchymal transition, invasion, and migration of thyroid cancer cells through inhibiting the methylation of ALX4. J Cell Physiol. 2019;234(11):20992–21004.
- Facchini G, Rossetti S, Berretta M, et al. Prognostic and predictive factors in testicular cancer. Eur Rev Med Pharmacol Sci. 2019;23(9):3885–3891.
- Chieffi P, De Martino M, Esposito F. Further insights into testicular germ cell tumor oncogenesis: potential therapeutic targets. Expert Rev Anticancer Ther. 2020;20(3):189–195.
- Luo Y, Zhou Q, Zhu F, et al. Hypomethylation-driven AKT Serine/Threonine Kinase 3 promotes testicular germ cell tumors proliferation and negatively correlates to immune infiltration. Bioengineered. 2021;12(2):11288–11302.
- Wu H, Zhang Z, Xiao XY, et al. Toll-like receptor 2 (TLR2) is a candidate prognostic factor in testicular germ cell tumors as well as an indicator of immune function in the tumor microenvironment. Bioengineered. 2021;12(1):1939–1951.
- Dong J, Su M, Chang W, et al. Long non-coding RNAs on the stage of cervical cancer (Review). Oncol Rep. 2017;38(4):1923–1931.
- Li A, Mallik S, Luo H, et al. H19, a Long Non-coding RNA, Mediates Transcription Factors and Target Genes through Interference of MicroRNAs in Pan-Cancer. Mol Ther Nucleic Acids. 2020;21:180–191.
- Li M, Qiu M, Xu Y, et al. Differentially expressed protein-coding genes and long noncoding RNA in early-stage lung cancer. Tumour Biol. 2015;36(12):9969–9978.
- Chen H, Wahafu P, Wang L, et al. LncRNA LINC00313 Knockdown Inhibits Tumorigenesis and Metastasis in Human Osteosarcoma by Upregulating FOSL2 through Sponging miR-342-3p. Yonsei Med J. 2020;61(5):359–370.
- Zhai Y, Liu Y, Wang Z, et al. Long Non-Coding RNA LINC00313 Accelerates Cervical Carcinoma Progression by miR-4677-3p/CDK6 Axis. Onco Targets Ther. 2021;14:2213–2226.
- Wu WJ, Yin H, Hu JJ, et al. Long noncoding RNA LINC00313 modulates papillary thyroid cancer tumorigenesis via sponging miR-4429. Neoplasma. 2018;65(6):933–942.
- Yan DG, Liu N, Chao M, et al. SP1-induced upregulation of long noncoding RNA LINC00313 contributes to papillary thyroid cancer progression via the miR-422a. Eur Rev Med Pharmacol Sci. 2019;23(3):1134–1144.
- Mitschke J, Burk UC, Reinheckel T. The role of proteases in epithelial-to-mesenchymal cell transitions in cancer. Cancer Metastasis Rev. 2019;38(3):431–444.
- Arneth B. Tumor Microenvironment. Medicina (Kaunas). 2019;56(1):15.
- Luo Y, Sun Y, Li L, et al. METTL3 May Regulate Testicular Germ Cell Tumors Through EMT and Immune Pathways. Cell Transplant. 2020;29:963689720946653.
- Wang Y, Wang Z, Xu J, et al. Systematic identification of non-coding pharmacogenomic landscape in cancer. Nat Commun. 2018;9(1):3192.
