ABSTRACT
Breast cancer (BC) is one of the most prevalent gynecologic malignant tumors with a poor prognosis and the second leading cause of cancer-related deaths in women worldwide. In recent years, it has been shown that long non-coding RNA (lncRNA) plays an important role in the development of breast cancer (BC). An antisense lncRNA from the MCF2 cell line (MCF2L-AS1) has been discovered recently and has been shown to function in a variety of malignancies. However, its function as a regulator of BC development has yet to be determined. Herein, the bioinformatics study analysis showed that MCF2L-AS1 was frequently highly expressed in BC tumors, and this overexpression was associated with worse patient outcomes. BC cells’ proliferation, migration, and invasion are inhibited when MCF2L-AS1 is silenced, whereas the inverse is evident when MCF2L-AS1 is overexpressed. It was also observed that MCF2L-AS1 knockdown decreased carcinogenesis in xenograft tumor models. Furthermore, we discovered that MCF2L-AS1 could bind to and improve the transcription activity of the yes-associated protein (YAP). However, following YAP knockdown, this lncRNA’s ability to drive BC malignancy was considerably reduced. In conclusion, MCF2L-AS1 may represent a potential predictive biomarker in BC patients, as well as a key regulator of BC cell proliferation. It works through positive feedback processes involving direct YAP binding and subsequent modulation of intracellular gene expression. Our findings add to our understanding of MCF2L-AS1 regulation and its potential as a therapeutic target in patients with this fatal cancer type.
Graphical Abstract
KEYWORDS:
Highlights
LncRNA-MCF2L-AS1 expression is up-regulated in BC, correlated with poor BC patient prognosis.
Knocking down MCF2L-AS1 could reduce the proliferative, migratory, and invasive activity of BC cells,
YAP is a target gene of MCF2L-AS1.
MCF2L-AS1 could promote the transcriptional activity of YAP in BC cells.
Introduction
Breast cancer (BC) is the most common malignancy and the second deadliest cancer type among women worldwide [Citation1]. Despite substantial advances in the early diagnosis and treatment of BC patients over the past two decades, this disease remains a major cause of morbidity and mortality. Treatments for early-stage breast cancer are generally curative, however therapy for late breast cancer largely focuses on boosting the overall quality of life and extending life expectancy [Citation2,Citation3]. As a result, an improved understanding of BC’s underlying processes is essential to better guide the diagnosis and treatment of this lethal malignancy.
The emergence of high-throughput sequencing technologies has led to an increasing focus on the biological role of non-coding RNAs (ncRNAs). Protein-coding gene sequences make up just 2% of the genome, however, the other 75% is thought to be translated as noncoding RNAs (ncRNAs) [Citation4,Citation5]. Long ncRNAs (lncRNAs) are over 200 nucleotides in length and lack protein-coding potential [Citation6]. They are primarily involved in key biological processes that can regulate gene expression via chromatin remodeling and at the post-transcriptional and translational levels [Citation7]. Evidence shows that lncRNAs are closely associated with the development as well as the prognosis of patients with breast cancer (BC) [Citation8–10]. Studies based on sequencing have shown that deregulation of lncRNAs is a frequent finding in malignancies, underlining their importance as both regulators of oncogenic processes and prospective biomarkers that may guide patient assessment and therapy [Citation11,Citation12].
The 1173-bp lncRNA MCF.2 cell line-derived transforming sequence like antisense RNA 1 (MCF2L-AS1) was recently discovered to be dysregulated in the presence of some malignancies, including colorectal cancer and non-small cell lung cancer [Citation13,Citation14]. Mechanistically, several pathways have been identified whereby MCF2L-AS1 can drive enhanced tumor progression and/or metastasis [Citation13–16]. However, the clinical relevance and functional roles of this lncRNA in BC have yet to be explored. Thus, there is a dire need to explore the biological functions and regulatory networks associated with MCF2L-AS1 regarding BC progression.
The Hippo signaling pathway has been identified as a central regulator of many aspects of tumor development [Citation17–20]. Several transcription factors may interact with the downstream yes-associated protein (YAP) in the Hippo pathway to affect intracellular transcriptional responses and hence change physiology [Citation21]. In BC, YAP is commonly identified as an oncogene, with its dysregulation potentially favoring invasive and metastatic activity [Citation22,Citation23]. Phosphorylation of the LATS1/2 kinase protein by the Hippo pathway is the conventional method for controlling YAP activity, although other pathways may also regulate it [Citation24]. In BC, for example, there is evidence that glucocorticoid receptor signaling can activate YAP activity [Citation25]. Recent work also suggests that YAP can influence actin dynamics and overall cellular movement [Citation20,Citation26,Citation27]. However, the specific YAP binding partners linked to tumor growth are unknown, and whether lncRNAs may directly alter YAP activity in this pathological situation has yet to be determined.
Through bioinformatics approaches, MCF2L-AS1 was found to be significantly upregulated in BC tumors and correlated with poor patient prognosis. We hypothesized that the high expression of McF2l-as may be a pathogenic factor promoting the development of breast cancer. However, the role of MCF2L-AS1 in BC remains unclear. Therefore, the purpose of this work was to explore the processes by which MCF2L-AS1 regulates BC tumor cell proliferation and transcription to establish a strong theoretical foundation for future research into the pathophysiology of BC.
Materials and methods
Clinical samples
Using paired tumor and paracancerous tissue samples obtained from 130 BC patients who received mammectomy at Baoji Municipal Central Hospital between October 2017 and 2020, correlations between MCF2L-AS1 expression and patient clinicopathological features were investigated. This research was authorized by the Institutional Review Board of Baoji Municipal Central Hospital (permission number. F20170716), and signed informed consent was obtained from all patients. Prior to storage, liquid nitrogen was used to snap-freeze patient samples after mammectomy.
Cell culture
The BT-549 and MDA-MB-231 BC cell lines were obtained from the American Type Culture Collection (VA, USA), while control MCF-10A cells were received from the Shanghai Institutes for Biological Sciences of the Chinese Academy of Sciences (Shanghai, China). The cell lines were cultured as previously described [Citation10]. BC cell lines were cultured in high-glucose DMEM (Gibco, NY, USA) supplemented with 10% FBS (Biological Industries, Kibbutz Beit Haemek, Israel) as per recommended protocols, whereas MCF-10A cells were grown in F12/DMEM (Gibco) supplemented with 10% FBS. Cells were cultured in a 37°C humidified 5% CO2 incubator. The siRNA sequence is shown in Supplementary Table S1.
Cell transfection
The cell transfection was conducted using a slightly modified version of a previously published protocol [Citation28]. For the knockdown of MCF2L-AS1 and YAP, specific siRNA sequences targeting MCF2LAS1 (si-MCF2L-AS1) and YAP (si-YAP) were provided by GenePharma (Shanghai, China). Nonspecific siRNAs were used as negative control (NC). To overexpress MCF2L-AS1, the whole sequence of MCF2L-AS1 was subcloned into pcDNA3.0 vector with empty vectors being utilized as NC. The constructs were transfected into BC cells that were 60–70% confluent using Lipofectamine 3000 (Invitrogen, CA, USA) based on provided instructions. At 48 h post-transfection, cells were collected for downstream analysis.
Quantitative real-time polymerase chain reaction (qPCR)
The qPCR was conducted using a slightly modified version of a previously published protocol [Citation29]. RNA was extracted from samples using TRIzol (15596026, Invitrogen), followed by a PrimeScript RT reagent (RR047, Takara Bio Inc., Shiga, Japan) to prepare cDNA based on provided directions. Hieff™ qPCR SYBR® Green Master Mix (No Rox) (Yeasen) and a StepOne-Plus instrument (Applied Biosciences) were then used for triplicate analyses of samples via qPCR. The GAPDH served as a normalization control. Melt curves were generated to ensure the specificity of amplification products. The 2−ΔΔCt method was used to assess relative gene expression.
MTT assay
A cell proliferation assay was performed with an MTT kit (Sigma) according to the manufacturer’s instructions. Cells were plated in 96-well plates (5000/well). At appropriate time points, 100 uL of MTT reagent was added to each well (0.5 mg/ml, Sigma), and cells were then incubated for 4 h at 37°C. Media was then aspirated and 150 uL of DMSO (Sigma) was added per well, with absorbance at 570 nm then being measured.
Transwell assay
A previously reported methodology [Citation29] was significantly changed to accomplish the Transwell detection. 24-well Transwell inserts (Corning) coated or uncoated with Matrigel (Dongcheng, Ningbo, Zhejiang, China) were used to examine cellular invasive and migratory activities. At 24 h post-transfection, appropriate cells were added into the upper chamber of a Transwell insert, while 600 uL of DMEM containing 10% FBS was added to the lower chamber. Matrigel-coated inserts (25 mg/50 mL, 60 μL) were employed for invasion assays. Moreover, migratory/invasive cells were incubated for 24 hours, subsequently fixed with methanol, stained with Giemsa for 2 hours, and scanned under a light microscope.
Western blotting
A significantly modified version of a previously reported Western blotting procedure was used [Citation30]. In brief, an SDS lysis buffer (2 M thiourea, 2% (w/v) DTT, 7 M urea) supplemented with 1% (v/w) protease inhibitors (Pierce Biotechnology) was used to extract proteins from individual samples. Proteins were then separated via 10% SDS-PAGE and transferred onto PVDF membranes (Millipore, IPVH00010). Blots were blocked with 5% nonfat milk for 2 h, after which they were incubated at 4°C with anti-YAP (Biorbyt, orb193680), anti-E-cadherin (Cell Signaling Technology, 3195), anti-Vimentin (Abcam, ab92547), or anti-N-cadherin (Abcam, ab18203) overnight, with all antibodies being diluted 1:1000. Blots were then washed three times and probed with HRP-conjugated secondary antibodies (1:2000) for 2 h at room temperature. Furthermore, protein bands were visualized using the SuperSignalWest Femto Chemiluminescent Substrate Western Blotting detection reagent (Thermo Scientific).
Xenograft model assay
Based on a previously published study, the xenograft model assay was developed by making minor alterations to that research [Citation31]. The Yan’an University Institutional Animal Care and Use Committee authorized all animal research, and the Animal Ethical and Welfare Committee approved all procedures (Approval No. 2018DWLS101). Nude athymic BALB/c mice (male, 4–6 weeks old) from the Model Animal Research Center of Nanjing University were housed in a specific pathogen-free environment. Ten female mice (4 weeks old) were divided into two groups (shNC and shMCF2L-AS1). Appropriate MDA-MB-231 cells transfected with sh-MCF2L-AS1 and pENTR vector (EV) were harvested and washed two times using cold PBS, combined with Matrigel (BD Biosciences) at a 1:1 ratio, and subcutaneously injected into the right flank of individual mice (2 × 105 cells/mouse). Upon euthanasia of mice with tumors that had grown to a diameter of 10–15 nm after 5 weeks, tumors were removed, scanned, and weighed before being stained with immunofluorescent dye.
Immunofluorescent staining
The Immunofluorescent staining was based on a previously published work [Citation32], with slight adjustments. Tissue slices were deparaffinized, rehydrated, and washed three times in PBS for 10 min. Antigen retrieval was performed by boiling samples in a microwave for 10 min in a 10 mM citrate buffer (pH 6.0). Samples were then washed three times with PBST for 10 min each, blocked using 5% BSA for 1 h, and stained with anti-Ki-67 (Novus biology, NBP2-54791) at 4°C overnight. Slides were then washed again with PBST, stained for 2 h with secondary antibodies at room temperature, counterstained with DAPI for 5 min, rinsed using PBS, mounted using Immu-Moun or VectaShield, and visualized with an LSM700 confocal microscope (Carl Zeiss AG).
TUNEL assays
TUNEL staining was performed with a TUNEL Bright-Green Apoptosis Detection kit (Vazyme, A112-03) as per recently published protocols [Citation33].
RNA immunoprecipitation
The RNA immunoprecipitation procedure was adapted from a previously published study with minor modifications 30. Two 15 cm cell culture plates were collected and rinsed with PBS before being crosslinked with 1% formaldehyde. Following this, the crosslinking process was halted by the addition of glycine to the solution (0.125 M). Cells were then collected via centrifugation and resuspended in IP lysis buffer (50 mM HEPES at pH 7.5, 1 mM EDTA, 0.4 M NaCl, 0.5% Triton X-100, 10% glycerol, 1 mM DTT) containing RNase inhibitor (Ambion), 1 mM PMSF, and protease inhibitors (GenDEPOT). Samples were then subjected to sonication, and pre-rinsed protein G agarose (Millipore) was employed to pre-clear the lysates. Lysates were then incubated overnight with anti-YAPa (Biorbyt, orb193680) or control IgG (Santa Cruz, sc-2025) (2 μg each) at 4°C, followed by the addition of rinsed protein G agarose and incubation for 1 h at 4°C. Furthermore, agarose beads were washed with IP lysis buffer, after which samples were centrifuged, and RNA in the collected supernatants was isolated using an RNeasy Mini Kit (Qiagen) with DNase I. Experimental primers are listed in Table S1, after which qPCR analyses were conducted, with data being expressed based on fold enrichment over control IgG. To avoid identifying the main antibody’s heavy chain, a secondary antibody (Abmart, M21006) was utilized for Western blot validation of the YAP protein.
RNA pull-down
The RNA pull-down was based on a previously published work with minimal modifications [Citation34]. MDA-MB-231 cells were transfected with 3’-biotinylated MCF2L-AS1 or scramble control constructs and incubated for 48 h, after which they were UV-irradiated (400 mJ/cm2). Cells were then homogenized using CLIP lysis buffer supplemented with 0.2 U/ul of recombinant RNase Inhibitor (Promega) and a Complete protease inhibitor cocktail (EDTA-free; Roche). Streptavidin-agarose beads (Pierce) were then utilized to precipitate proteins and bound biotin-conjugated RNAs at 4°C. Beads were then rinsed twice with lysis buffer, high-salt buffer, and PNK buffer respectively. After precipitation, Western blotting and qPCR were used to assess protein and RNA levels within these precipitate samples.
Luciferase reporter assay
YAP transcriptional activity was monitored via a luciferase reporter assay approach [Citation35]. Briefly, Lipofectamine 3000 was used to transfect MDA-MB-231 cells with the 8xGTIIC-luciferase (Plasmid #34615) and β-gal plasmids (Ambion, USA). A Luciferase Reporter assay kit (BioVision, Inc., K801-200) was then used to monitor luciferase activity within these cells, while a β-Galactosidase Enzyme Assay System with Reporter Lysis Buffer (Promega Corporation, E2000) was used based upon provided instructions to assess β-gal activity.
Chromatin immunoprecipitation assay (CHIP)
A ChIP assay kit from Millipore (17–371) was used according to the manufacturer’s protocol. MDA-MB-231 cells were transfected with si-MCF2L-AS1 or nonspecific siRNAs. After cross-linking, 5 μg of the antibody against YAP (Biorbyt, orb193680) was added to immunoprecipitate endogenous YAP. After immunoprecipitation, protein–DNA cross-links were reversed and DNA was purified to remove the chromatin proteins to be used for qPCR. Primer sequences are listed in Supplementary Table S1.
Results
In this study, we explored the role of MCF2L-AS1 in BC. Our findings demonstrated that MCF2L-AS1 was increased in BC and that silencing MCF2L-AS1 inhibited BC cell proliferation, migration, and invasive activity both in vitro and in vivo, as well as invasive activity. Further, MCF2L-AS1 can directly bind to YAP within BC cells, thereby enhancing YAP transcriptional activity and stimulating malignant activity. The carcinogenic effects of MCF2L-AS1 in these BC cells were abated when YAP was knocked down. Our findings highlight the functional roles of MCF2L-AS1 in BC, providing new insights into BC pathogenesis.
MCF2L-AS1 upregulation in BC tumors is correlated with poor patient outcomes
In our study, we proposed to screen a lncRNA that regulates the progression of breast cancer. We assessed differentially expressed lncRNAs between BC tumor and healthy tissue samples in the published GSE145144 microarray dataset from the GEO database. MCF2L-As1 was upregulated in BC tumors relative to normal tissues as determined based on Transcripts Per Million (TPM) score values in this dataset ()). The TANRIC tool (https://bioinformatics.mdanderson.org/public-software/tanric/) was used to evaluate lncRNA expression patterns in the TCGA data set, which allowed us to further refine our findings. This analysis similarly confirmed MCF2L-AS1 to be upregulated in BC tumors relative to paired and unpaired healthy tissues ()). Higher levels of MCF2L-AS1 were also found to be associated with poorer disease-free survival (DFS) ()).
Figure 1. BC patients exhibit MCF2L-AS1 upregulation that is correlated with a worse prognosis. (a, b) A GEO dataset was queried to compare the expression of MCF2L-AS1 in BC tumors and paracancerous normal tissue. (c) MCF2L-AS1 expression levels were compared in BC patient tumors and unpaired healthy tissue samples present within the TCGA database (normal = 105, tumor = 837). (d) MCF2L-AS1 levels were assessed in 105 pairs of BC tumors and paracancerous healthy tissue samples. (e) DFS curves for BC patients (n = 1068) were established using the Kaplan-Meier plotter database, revealing higher levels of MCF2L-AS1 to be correlated with worse patient OS. Data were generated based on quartile cutoffs for the expression of MCF2L-AS1. (f) MCF2L-AS1 levels in BC patient tumor tissues and paracancerous samples (n = 130) were assessed, with GAPDH to normalize these levels. (g) Median MCF2L-AS1 expression was used to classify patients into those expressing low or high levels of this lncRNA (each n = 65), revealing MCF2L-AS1-high patients to have poorer survival outcomes as compared to patients expressing lower levels thereof. (**p < 0.01, ***p < 0.001).
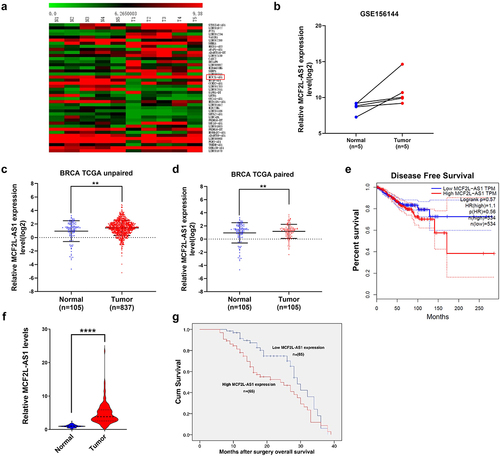
Following, MCF2L-AS1 expression was compared in 130 paired BC patient tumor and paracancerous tissue samples via qPCR (), revealing MCF2L-AS1 in tumors consistent with that observed in the TCGA dataset ()). Similarly, we found higher MCF2L-AS1 expression to be associated with a more advanced TNM stage (III/IV), whereas patients with a less advanced TNM stage tended to express lower levels of this lncRNA (). MCF2L-AS1 was also correlated with poorer prognostic outcomes among BC patients (p < 0.01; )). These findings imply that high levels of MCF2L-AS1 overexpression in BC tumors are associated with a poor prognosis for patients.
Table 1. The relationship between MCF2L-AS1 expression and breast cancer patient clinicopathological characteristics
MCF2L-AS1 regulates breast cancer cell proliferation
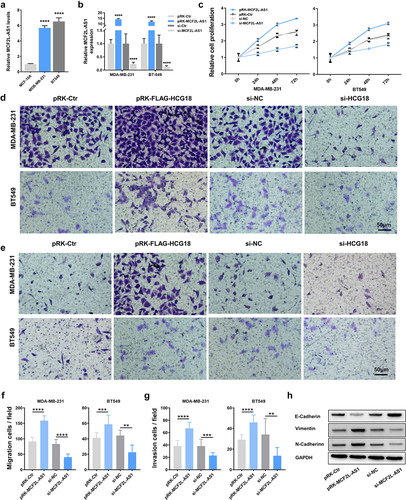
Moreover, we wanted to explore the MCF2L-AS1 effect in BC cell proliferation because of its role in regulating tumor cell oncogenicity. To accomplish this, the levels of MCF2L-AS1, a lncRNA, in the MDA-MB-231 and BT-549 BC cell lines were compared to those in the human MCF-10 mammary epithelial cell line as a reference, and both cell lines showed substantial increases ()). To examine the link between MCF2L-AS1 and cell growth, an MTT assay was conducted to evaluate MDA-MB-231 and BT-549 cells transfected using MCF2L-AS1, inhibitor, or control constructs. lncRNA levels were decreased in cells transfected with the MCF2L-AS1 inhibitor but increased in cells transfected with the pRK-MCF2L-AS1 vector ()). Inhibiting MCF2L-AS1 suppressed MDA-MB-231 and BT-549 cell proliferation. However, this proliferative activity was enhanced relative to that in control cells following pRK-MCF2L-AS1 transfection ()). As a consequence of these findings, MCF2L-AS1 has been identified as a key regulator of increased BC cell proliferation.
Figure 2. MCF2L-AS1 enhances the migratory, proliferative, and invasive activity of BC cells. (a, b) MCF2L-AS1 knockdown or overexpression in MDA-MB-231 and BT549 cells was confirmed via qPCR. (c). The impact of overexpressing or inhibiting MCF2L-AS1 expression on the proliferation of MDA-MB-231 and BT549 cells was determined via MTT assay. (d-g) The impact of overexpressing or inhibiting MCF2L-AS1 expression on the migration (d, f) and invasion (e, g) of MDA-MB-231 and BT549 cells was determined via transwell assay. (h) EMT-related protein levels were assessed via Western blotting in the indicated treatment groups. (**p < 0.01, ***p < 0.001, ****P < 0.0001).
MCF2L-AS1 promotes the migratory and invasive activity of BC cells
Cell invasion is a significant aspect of cancer progression that involves the migration of tumor cells into contiguous tissues and the dissolution of extracellular matrix proteins. Here, transwell assays were further performed to explore the MCF2L-AS1 ability to influence BC cell migratory and invasive activity. In these assays, inhibiting MCF2L-AS1 significantly decreased the numbers of migratory and invasive MDA-MB-231 and BT-549 cells, whereas the overexpression of this lncRNA yielded the opposite phenotype ()). These results suggested that MCF2L-AS1 functions as a promoter of BC cell metastatic growth.
Cell adhesion molecules are implicated in invasion and metastasis in various cancers [Citation36,Citation37]. Therefore, we performed qPCR to detect the expression of cell adhesion molecules that have been confirmed to be involved in tumor invasion and metastasis (such as E-cadherin, N-cadherin, and Vimentin). In our BC cell lines, we found that MCF2L-AS1 was able to influence the expression of these markers ()), corroborating our finding that MCF2L-AS1 could promote BC metastasis.
MCF2L-AS1 knockdown suppresses the in vivo growth of breast tumors
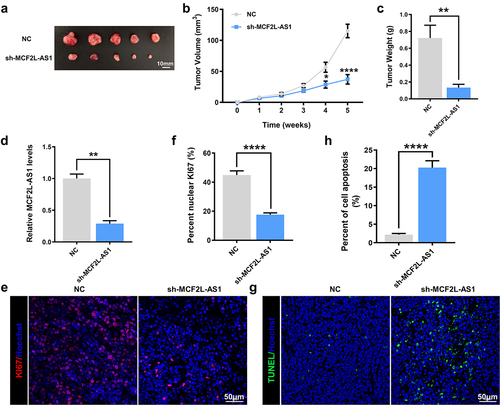
Following up on the in vitro findings, we investigated the effect of knocking down MCF2L-AS1 on breast cancers in vivo. MDA-MB-231 cells transduced with the sh- MCF2L-AS1/pENTR vector (EV) were used in a nude mice xenograft model. Up to 35 days after the knockdown of MCF2L-AS1, there was a dramatic decrease in tumor volume (67.7%) in the sh-MCF2L-AS1 group compared with controls ()). Similarly, knocking down this lncRNA decreased the average weights of tumors by 86% relative to control tumors ()). Successful MCF2L-AS1 silencing in these tumors was confirmed via qPCR ()). Additionally, immunostaining analysis of the proliferation marker KI-67 was performed in resected tumor tissues. It was observed that MCF2L-AS1 silencing significantly suppressed the proliferation of xenograft tumor cells ()). The TUNEL staining further revealed an increase in intratumoral apoptosis in tumors in which MCF2L-AS1 had been knocked down ()). consequently, these data suggested that knocking down MCF2L-AS1 was sufficient to inhibit in vivo breast tumor growth.
Figure 3. MCF2L-AS1 enhances the growth of BC tumors in vivo. (a). Images of tumors collected at 5 weeks post-implantation. (b) Tumor volumes were measured at the indicated time points. (c) Tumor weight values were calculated in the indicated groups at 5 weeks post-injection. (d) MCF2L-AS1 knockdown was confirmed in MDA-MB-231 tumors via qPCR. (e, f) MCF2L-AS1-silenced MDA-MB-231 tumors exhibited decreased Ki-67 staining, consistent with reduced proliferative activity. ****P < 0.0001. (g, h). MCF2L-AS1-silenced MDA-MB-231 tumors were assessed via TUNEL staining, revealing an increase in intratumoral apoptotic cell death. (*p < 0.05, **p < 0.01, ****P < 0.0001).
MCF2L-AS1 directly interacts with YAP in BC cells
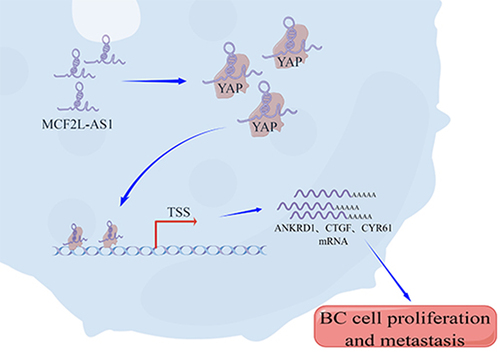
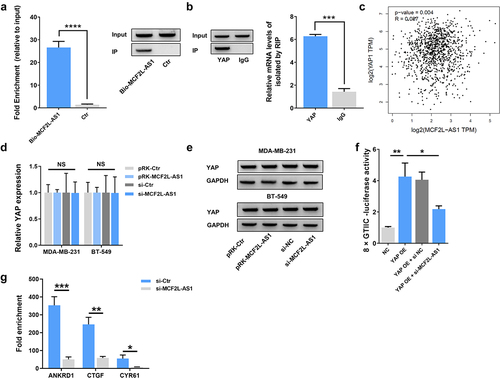
Additionally, we endeavored to understand further completely the molecular pathways by which MCF2L-AS1 influences the malignant behaviors of BC cells. Reportedly, YAP is an essential regulator of the growth of BC tumors. We performed RIP and RNA pull down assays and detected a direct binding interaction between YAP and MCF2L-AS1 ()). However, according to the the data from TCGA dataset by calculating the Pearson correlation coefficient, MCF2L-AS1 was found uncorrelated with YAP ()). Further, we found that altering MCF2L-AS1 expression in BC cells had no impact on YAP mRNA or protein levels therein ()).
Figure 4. MCF2L-AS1 interacts with YAP and thereby modulates its transcriptional regulatory activity. (a). YAP co-precipitation was achieved using 3`-biotinylated synthetic oligonucleotides, with YAP being detected in the resultant eluates via Western blotting. (b). RIP-qPCR was performed to detect the interacting ability of YAP and MCF2L-AS1 among one another. (c). The expression of MCF2L-AS1 was not correlated with YAP in the tumor tissues of BC patients. (d-e). The mRNA and protein level expression of YAP in BC cells was measured following MCF2L-AS1 knockdown. (f). 8 × GTIIC-luciferase activity was assessed in BC cell lines. YAP OE: YAP overexpression; si MCF2L-AS1: MCF2L-AS1 knockdown. (*p < 0.05, **p < 0.01, ***p < 0.001, ****P < 0.0001). (g). ChIP-qPCR analysis shows the occupancy of ANKRD1, CTGF, and CYR61 promoters YAP. Endogenous YAP was immunoprecipitated from control or MCF2L-AS1 silencing BC cells. n = 3 technical replicates per group.
MCF2L-AS1 promotes BC development through the transcriptional activity of YAP protein
As YAP proteins interact with TEAD and TAZ to actively regulate the transcription of target genes, we explored the regulatory function of MCF2L-AS1 on the transcriptional activity of YAP using a luciferase assay. We utilized the 8× GTIIC-luciferase YAP1/TAZ reporter construct containing three YAP1/TAZ/TEAD DNA binding sites prepared from the −200/+27 fragment of the human CTGF gene promoter to clarify the functional relevance of this regulatory interaction [Citation38]. Knocking down MCF2L-AS1 significantly decreased YAP transcriptional activity in this luciferase reporter assay system ()). Moreover, we analyzed YAP target gene promoters by ChIP assays. MCF2L-AS1 silencing significantly decreased the occupancy of target gene promoters by endogenous YAP ()). Therefore, our data demonstrated that MCF2L-AS1 can bind to YAP and regulate the transcriptional regulatory activity of YAP.
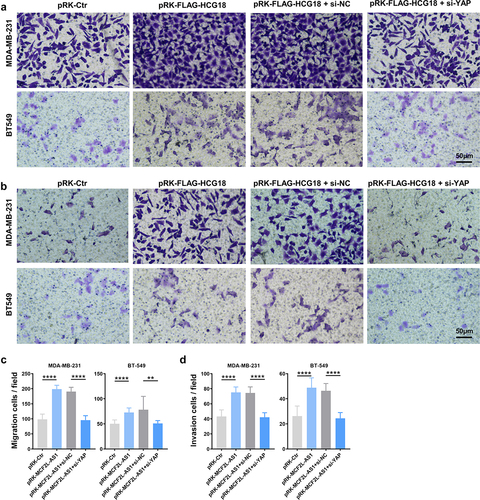
We further explored the functional correlation between MCF2L-AS1 and YAP in the pathology of BC. It was found that knocking down YAP also sufficiently ablates the ability of MCF2L-AS1 to promote the metastatic growth of BC cells ()). Collectively, MCF2L-AS1 could directly bind to YAP, and promote its transcriptional activity in BC cells, and the function of MCF2L-AS1 in promoting BC development depends on the expression of YAP.
Figure 5. MCF2L-AS1 interacts with YAP to control the migratory and invasive activity of BC cells. (a, b) YAP knockdown was sufficient to rescue the observed enhancement of BC cell migratory activity observed following MCF2L-AS1 overexpression. (c, d). The ability of MCF2L-AS1 to promote the invasive activity of BC cells can be reversed by knocking down YAP. ****P < 0.0001.
Discussion
Breast cancer is one of the most prevalent and severe cancers globally, The number of cases worldwide keeps increasing annually [Citation39,Citation40]. A key challenge remains the identification of novel predictors of BC progression and the identification of new targets for interventional drug development efforts. Based on their length, non-coding RNAs can be classified as small non-coding RNAs (< 1200 nucleotides) or lncRNAs (> 200 nucleotides) [Citation4,Citation7]. Many recent studies have documented essential roles for lncRNAs as regulators of gene expression at the transcriptional, translational, and epigenetic levels in both physiological and pathogenic situations [Citation41]. Dysregulation of lncRNA expression patterns is now recognized as a prevalent finding in a variety of malignancies, with these ncRNAs having the ability to control oncogenic processes such as tumor cell motility, proliferation, survival, invasion, stemness, microenvironmental remodeling, and metastasis [Citation42–44]. Several studies have examined the functions of lncRNAs in BC [Citation9,Citation10,Citation31]. MCF2L-AS1 is a lncRNA that was recently identified and found to be dysregulated in multiple human cancer types in which it regulates progression-related disease processes [Citation11]. Cai et al. showed MCF2L-AS1/miR-105/ IL-1β Axis Regulates Colorectal Cancer Cell Oxaliplatin Resistance [Citation45]. Zhang et al. showed MCF2L-AS1 aggravates proliferation, invasion, and glycolysis of colorectal cancer cells via the crosstalk with miR-874-3p/FOXM1 signaling axis [Citation15]. Li et al. showed MCF2L-AS1 promotes the cancer stem cell-like traits in non-small cell lung cancer cells through regulating miR-873-5p level [Citation13]. Nevertheless, our research was the first to highlight the importance of MCF2L-AS1 for breast cancer prevention and treatment. In this study, we examined MCF2L-AS1 expression profiles using sequencing data from the TCGA and GEO databases and found that it was significantly upregulated in BC tumors and cell lines compared to normal control samples and that this upregulation was associated with a poor prognosis for BC patients.
To explore the role of MCF2L-AS1 in BC, we knocked down the MCF2L-AS1 gene and found that this reduced the proliferative, migratory, and invasive activity of BC cells, whereas overexpressing this lncRNA had the opposite effect. Cell adhesion molecules are well known to play essential roles in invasion and metastasis in human carcinomas [Citation46]. Significantly, we found that overexpressing MCF2L-AS1 resulted in the upregulation of Vimentin and N-cadherin together with the downregulation of E-cadherin. As a consequence of these findings, it is clear that MCF2L-AS1 is a tumor enhancer in BC.
YAP plays a central role in oncogenic processes, functioning as a proto-oncogene that is overexpressed in many tumors [Citation47–49]. In addition, patients with BC displaying high YAP activity appeared to have a higher overall positive prognosis, leading to the belief that YAP might act as a tumor suppressor in breast cancer [Citation50–53]. Given its ability to regulate oncogenesis, tumor progression, and neovascularization, YAP represents a promising target for anticancer treatment. Further, YAP has been recognized as a transcription factor in promoting tumor progression and metastasis, regulated by several upstream mechanisms [Citation54–56]. We herein found that MCF2L-AS1 was able to bind to YAP and activate its transcriptional activity. Knocking down YAP was sufficient to reverse the effects of overexpressing MCF2L-AS1, thereby negatively regulating BC cell progression. Overall, our data suggest that the MCF2L-AS1/YAP axis may represent a promising target for suppressing BC growth. We additionally found that the MCF2L-AS1/YAP axis was able to promote BC tumor development and metastasis, suggesting that the Hippo pathway may regulate MCF2L-AS1, although further work is required to test this hypothesis.
Nonetheless, there are still a few limitations in this work. Apart from the roles of the factors we revealed, more unknown biological functions still need to be investigated about MCF2L-AS1. Whether MCF2L-AS1 regulates other molecules such as enzymes or kinases, which participate in tumor progression, is also worth studying.
Conclusion
Precisely, the study demonstrated that lncRNA MCF2L-AS1 is highly expressed in BC tissues and cells. MCF2L-AS1 knockdown may inhibit BC cells proliferation and metastasis via inhibiting YAP transcriptional activity. It may provide a theoretical and experimental basis for MCF2L-AS1 to be a potential target for BC treatment therapies.
Author Contributions
Qing She performed most of the experiments, analyzed the data, prepared all figures and/or tables, and authored the draft of the paper.
Yuanyuan Chen, Hong Liu, and Jichao Tan performed some of the experiments, analyzed the data, and prepared .
Youhuai Li conceived and designed the experiments, reviewed the draft of the paper, and approved the final draft.
Ethics approval
The Institutional Review Board of Baoji Municipal Central Hospital approved this study (approval no. F20170716), with all patients having provided written informed consent.
All animal studies were approved by the Institutional Animal Care and Use Committee of Yan’an University, with all protocols having been approved by the Animal Ethical and Welfare Committee (Approval No. 2018DWLS101).
Supplemental Material
Download Zip (620 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The analyzed datasets generated during the study are available from the corresponding author on reasonable request.
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/21655979.2022.2074108
Additional information
Funding
References
- DeSantis C, Ma J, Goding Sauer A, et al. Breast cancer statistics, 2017, racial disparity in mortality by state. CA Cancer J Clin. 2017;67:439–448.
- Buchholz T, Wazer D. Molecular biology and genetics of breast cancer development: a clinical perspective. Semin Radiat Oncol. 2002;12:285–295.
- Harbeck N, Gnant M. Breast cancer. Lancet (London. England). 2017;389:1134–1150.
- Das T, Das TK, Khodarkovskaya A, et al. Non-coding RNAs and their bioengineering applications for neurological diseases. Bioengineered. 2021;12:11675–11698.
- Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21(11):1253–1261.
- Ponting C, Oliver P, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641.
- Perry R, Ulitsky I. The functions of long noncoding RNAs in development and stem cells. Development (Cambridge. England). 2016;143:3882–3894.
- Wang G, Duan P, Liu F, et al. Long non-coding RNA CASC7 suppresses malignant behaviors of breast cancer by regulating miR-21-5p/FASLG axis. Bioengineered. 2021;12:11555–11566.
- Chang J, Zhang Y, Ye X, et al. Long non-coding RNA (LncRNA) CASC9/microRNA(miR)-590-3p/sine oculis homeobox 1 (SIX1)/NF-κB axis promotes proliferation and migration in breast cancer. Bioengineered. 2021;12:8709–8723.
- Zhang M, Yang L, Hou L, et al. LncRNA SNHG1 promotes tumor progression and cisplatin resistance through epigenetically silencing miR-381 in breast cancer. Bioengineered. 2021;12(2):9239–9250.
- Arantes L, De Carvalho A, Melendez M, et al. Serum, plasma and saliva biomarkers for head and neck cancer. Expert Rev Mol Diagn. 2018;18(1):85–112.
- Wang L, Xu T, Cui X, et al. Downregulation of lncRNA SNHG7 inhibits proliferation and invasion of nasopharyngeal carcinoma cells through repressing ROCK1. Eur Rev Med Pharmacol Sci. 2020;24:7917.
- Li S, Lin L. Long noncoding RNA MCF2L-AS1 promotes the cancer stem cell-like traits in non-small cell lung cancer cells through regulating miR-873-5p level. Environ Toxicol. 2021;36:1457–1465.
- Kong W, Li H, Xie L, et al. LncRNA MCF2L-AS1 aggravates the malignant development of colorectal cancer via targeting miR-105-5p/RAB22A axis. BMC Cancer. 2021;21:1069.
- Zhang Z, Yang W, Li N, et al. LncRNA MCF2L-AS1 aggravates proliferation, invasion and glycolysis of colorectal cancer cells via the crosstalk with miR-874-3p/FOXM1 signaling axis. Carcinogenesis. 2021;42:263–271.
- Huang FK, Zheng CY, Huang LK, et al. Long non-coding RNA MCF2L-AS1 promotes the aggressiveness of colorectal cancer by sponging miR-874-3p and thereby up-regulating CCNE1. J Gene Med. 2021;23:e3285.
- Wang H, Di X, Bi Y, et al. Long non-coding RNA LINC00649 regulates YES-associated protein 1 (YAP1)/Hippo pathway to accelerate gastric cancer (GC) progression via sequestering miR-16-5p. Bioengineered. 2021;12:1791–1802.
- Zeng Y, Xu Q, Xu N. Long non-coding RNA LOC107985656 represses the proliferation of hepatocellular carcinoma cells through activation of the tumor-suppressive Hippo pathway. Bioengineered. 2021;12(1):7964–7974.
- Zanconato F, Cordenonsi M, Piccolo S. YAP/TAZ at the roots of cancer. Cancer Cell. 2016;29:783–803.
- Jiang N, Zhao L, Zong D, et al. Long non-coding RNA LUADT1 promotes nasopharyngeal carcinoma cell proliferation and invasion by downregulating miR-1207-5p. Bioengineered. 2021;12:10716–10728.
- Ji X, Zhong G, Zhao B. Molecular mechanisms of the mammalian Hippo signaling pathway. Yi Chuan. 2017;39:546–567.
- Pegoraro S, Ros G, Ciani Y, et al. A novel HMGA1-CCNE2-YAP axis regulates breast cancer aggressiveness. Oncotarget. 2015;6:19087–19101.
- Kim H, Jung W, Koo J. Expression of Yes-associated protein (YAP) in metastatic breast cancer. Int J Clin Exp Pathol. 2015;8:11248–11257.
- Zheng L, Xiang C, Li X, et al. STARD13-correlated ceRNA network-directed inhibition on YAP/TAZ activity suppresses stemness of breast cancer via co-regulating Hippo and Rho-GTPase/F-actin signaling. J Hematol Oncol. 2018;11:72.
- Sorrentino G, Ruggeri N, Zannini A, et al. Glucocorticoid receptor signalling activates YAP in breast cancer. Nat Commun. 2017;8:14073.
- Qiao Y, Chen J, Lim Y, et al. YAP regulates actin dynamics through ARHGAP29 and promotes metastasis. Cell Rep. 2017;19:1495–1502.
- Wang Z, Wu Y, Wang H, et al. Interplay of mevalonate and Hippo pathways regulates RHAMM transcription via YAP to modulate breast cancer cell motility. Proc Natl Acad Sci U S A. 2014;111:E89–98.
- Xu TP, Huang MD, Xia R, et al. Decreased expression of the long non-coding RNA FENDRR is associated with poor prognosis in gastric cancer and FENDRR regulates gastric cancer cell metastasis by affecting fibronectin1 expression. J Hematol Oncol. 2014;7:63.
- Zhang Q, Wang Z, Cheng X, et al. lncRNA DANCR promotes the migration an invasion and of trophoblast cells through microRNA-214-5p in preeclampsia. Bioengineered. 2021;12:9424–9434.
- Liu XH, Sun M, Nie FQ, et al. Lnc RNA HOTAIR functions as a competing endogenous RNA to regulate HER2 expression by sponging miR-331-3p in gastric cancer. Mol Cancer. 2014;13:92.
- Qin S, Ning M, Liu Q, et al. Knockdown of long non-coding RNA CDKN2B-AS1 suppresses the progression of breast cancer by miR-122-5p/STK39 axis. Bioengineered. 2021;12:5125–5137.
- Hua R, Wei H, Liu C, et al. FBXO47 regulates telomere-inner nuclear envelope integration by stabilizing TRF2 during meiosis. Nucleic Acids Res. 2019;47:11755–11770.
- Liu Y, Liu C, Chang Z, et al. Degradation of the separase-cleaved Rec8, a meiotic cohesin subunit, by the N-end rule pathway. J Biol Chem. 2016;291:7426–7438.
- Goodarzi H, Liu X, Nguyen HC, et al. Endogenous tRNA-derived fragments suppress breast cancer progression via YBX1 displacement. Cell. 2015;161:790–802.
- Kim J, Piao HL, Kim BJ, et al. Long noncoding RNA MALAT1 suppresses breast cancer metastasis. Nat Genet. 2018;50:1705–1715.
- Thiery JP, Acloque H, Huang RY, et al. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890.
- Acloque H, Adams MS, Fishwick K, et al. Epithelial-mesenchymal transitions: the importance of changing cell state in development and disease. J Clin Invest. 2009;119:1438–1449.
- Zhao B, Ye X, Yu J, et al. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 2008;22:1962–1971.
- Siegel R, Miller K, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7–30.
- Kwantwi LB, Wang S, Sheng Y, et al. Multifaceted roles of CCL20 (C-C motif chemokine ligand 20): mechanisms and communication networks in breast cancer progression. Bioengineered. 2021;12:6923–6934.
- Li Q, Li Z, Fan Z, et al. Involvement of non‑coding RNAs in the pathogenesis of myocardial ischemia/reperfusion injury (Review). Int J Mol Med. 2021;47:47.
- Zheng Y, Zeng J, Xia H, et al. Upregulated lncRNA Cyclin-dependent kinase inhibitor 2B antisense RNA 1 induces the proliferation and migration of colorectal cancer by miR-378b/CAPRIN2 axis. Bioengineered. 2021;12(1):5476–5490.
- Zhang C, Ren X, Zhang W, et al. Prognostic and clinical significance of long non-coding RNA SNHG12 expression in various cancers. Bioengineered. 2020;11(1):1112–1123.
- Liu SJ, Dang HX, Lim DA, et al. Long noncoding RNAs in cancer metastasis. Nat Rev Cancer. 2021;21(7):446–460.
- Cai M, Hu W, Huang C, et al. lncRNA MCF2L-AS1/miR-105/ IL-1β axis regulates colorectal cancer cell oxaliplatin resistance. Cancer Manag Res. 2021;13:8685–8694.
- Pastushenko I, Blanpain C. EMT transition states during tumor progression and metastasis. Trends Cell Biol. 2019;29(3):212–226.
- Yan J, Shi L, Lin S, et al. MicroRNA-624-mediated ARRDC3/YAP/HIF1α axis enhances esophageal squamous cell carcinoma cell resistance to cisplatin and paclitaxel. Bioengineered. 2021;12(1):5334–5347.
- Wang G, Yu X, Xia J, et al. MicroRNA-9 restrains the sharp increase and boost apoptosis of human acute myeloid leukemia cells by adjusting the Hippo/YAP signaling pathway. Bioengineered. 2021;12(1):2906–2914.
- Li Z, Lian Z, Ma J, et al. Integrin β3 overexpression contributes to podocyte injury through inhibiting RhoA/YAP signaling pathway. Bioengineered. 2021;12(1):1138–1149.
- von Eyss B, Jaenicke LA, Kortlever RM, et al. A MYC-driven change in mitochondrial dynamics limits YAP/TAZ function in mammary epithelial cells and breast cancer. Cancer Cell. 2015;28(6):743–757.
- Elster D, Tollot M, Schlegelmilch K, et al. TRPS1 shapes YAP/TEAD-dependent transcription in breast cancer cells. Nat Commun. 2018;9:3115.
- Li L, Ma T, Ma Y, et al. LncRNA HCG18 contributes to nasopharyngeal carcinoma development by modulating miR-140/CCND1 and Hedgehog signaling pathway. Eur Rev Med Pharmacol Sci. 2019;23:10387–10399.
- Maugeri-Saccà M, Barba M, Pizzuti L, et al. The Hippo transducers TAZ and YAP in breast cancer: oncogenic activities and clinical implications. Expert Rev Mol Med. 2015;17:e14.
- Harvey KF, Zhang X, Thomas DM. The Hippo pathway and human cancer. Nat Rev Cancer. 2013;13:246–257.
- Pobbati AV, Hong W. Emerging roles of TEAD transcription factors and its coactivators in cancers. Cancer Biol Ther. 2013;14:390–398.
- Quinn HM, Vogel R, Popp O, et al. YAP and β-catenin cooperate to drive oncogenesis in basal breast cancer. Cancer Res. 2021;81:2116–2127.
