ABSTRACT
Non-small cell lung cancer (NSCLC) is the most common malignant tumor of lung, which seriously threatens the life of people. It has been reported that lncRNA prostate cancer-associated transcript 6 (PCAT6) could facilitate the metastasis of NSCLC cells. However, whether lncRNA PCAT6 in NSCLC cells could affect the tumor microenvironment (TME) remains unclear. In the present study, the level of PCAT6 in NSCLC cells was detected using RT-qPCR. The effects of PCAT6 knockdown on the viability and apoptosis in NSCLC cells were detected with CCK-8 and flow cytometry assay. NSCLC cell-derived exosomes were isolated with ultracentrifugation. Next, transwell assay was conducted to assess the migration and invasion of NSCLC cells. Dual-luciferase reporter assay was performed to verify the relationship among PCAT6, miR-326, and KLF1 in A549 cells. In addition, nanoparticle tracking analysis (NTA) was applied to detect the particle size of isolated exosomes. Moreover, ELISA assay was performed to detect the levels of IL-1β and IL-10 in the supernatant of macrophage. We found knockdown of PCAT6 significantly inhibited the viability, migration, and invasion of NSCLC cells. In addition, dual-luciferase reporter assay illustrated that miR-326 was the target of PCAT6 and KLF1 was the target of miR-326 in NSCLC cells. Moreover, NSCLC cells-derived exosomes could promote macrophages M2 polarization by transporting PCAT6. Meanwhile, macrophages M2 polarization was able to promote the metastasis and epithelial-mesenchymal transition (EMT) process of NSCLC cells via regulating PCAT6/miR-326/KLF1 axis. Taken together, knockdown of lncRNA PCAT6 suppressed the growth of NSCLC cells by inhibiting macrophages M2 polarization via miR-326/KLF1 axis.
GRAPHICAL ABSTRACT

Highlights
PCAT6 was upregulated in non-small cell lung cancer.
Knockdown of PCAT6 significantly inhibited the tumorgenesis of lung cancer.
Knockdown of PCAT6 inhibited macrophages M2 polarization via regulating miR-326/KLF1 axis.
Introduction
It is well known that non-small cell lung cancer (NSCLC) seriously threatens the life of people [Citation1,Citation2]. It has been reported that the tumorigenesis of NSCLC is related to smoking, environmental pollution, radioactive substances, and genetic factors [Citation3–5]. The clinical manifestations of NSCLC are cough, chest pain, dyspnea, fever, and so on [Citation6–8]. Up to now, the main treatments for NSCLC include molecular targeted drug therapy, surgery [Citation9–12]. Although great progress has been made in the diagnosis and treatment of NSCLC, the prognosis of the patients with advance NSCLC remains poor.
As we know, macrophages play an extremely vital role in the development of the body and the balance of the internal environment [Citation13]. Macrophages are derived from peripheral blood mononuclear cells (PBMCs) [Citation14,Citation15]. In addition, macrophages can form two extreme phenotypes (M1/M2 phenotypes) respond to different stimulating factors [Citation16, Citation17]. M1 macrophages are thought to have a close relationship with inflammation [Citation17], while M2 macrophages are generally believed to have the functions of promoting tissue repair, vascular regeneration, fibrosis, and tumorigenesis [Citation18]. In addition, when a malignant tumor occurs in the body, macrophages gradually differentiate into tumor-associated macrophages (TAMs) under the action of the tumor microenvironment (TME) [Citation18–20]. On the other hand, the metastasis of tumor cells could be prevented when tumor-related macrophages M2 polarization was inhibited [Citation21,Citation22]. Strikingly, the macrophages polarization is closely related to Kruppel-like factor 1 (KLF1) [Citation23], since KLF1 is an important participator during the process of macrophages polarization [Citation24].
Long non-coding RNA (lncRNA) is a class of non-coding RNA with the length of more than 200 nucleotides [Citation25]. Recent studies have shown that lncRNA is able to indirectly regulate gene expression via interacting with microRNA (miRNA) as a competing endogenous RNAs (ceRNA) [Citation26,Citation27]. In addition, lncRNA plays an important role in the tumorigenesis of lung cancer [Citation28,Citation29]. For example, upregulation of lncRNA DANCR promotes the invasion of lung cancer cells via sequestering miR-216a [Citation28]. Overexpression of lncRNA TUC338 accelerates the tumorigenesis of lung cancer by regulating MAPK pathway [Citation30]. In addition, the expression of lncRNA PCAT6 is upregulated in a variety of tumors, including hepatocellular carcinoma, ovarian cancer, and NSCLC [Citation31–34]. Of note, lncRNA PCAT6 (prostate cancer-associated transcript 6) was reported to facilitate the proliferation and invasion of NSCLC cells by regulating miR-330-5p [Citation34]. However, whether lncRNA PCAT6 could regulate macrophages M2 polarization in NSCLC remains unclear. Therefore, we aimed to explore the relationship between lncRNA PCAT6 and TME in NSCLC in the current study. We hope the findings might shed new lights on investigating novel effective treatment for the patient with NSCLC.
Material and methods
Cell culture
The human normal lung epithelial cell lines BEAS-2B, human NSCLC cell lines (A549, NCI-H1299 and NCI-H1975) as well as human acute monocytic leukemia cell lines THP-1 were provided by ATCC (Manassas, VA, USA). RIPM1640 (Thermo Fisher) supplemented with 10% FBS was applied for culturing these cells. All these cells are placed at 37°C with 5% CO2 according to the previous literature [Citation35].
THP-1 monocytes were firstly induced into M0 macrophages by treating with PMA (200 nM) for 24 h. Then, M0 macrophages was induced to M2 macrophages M2 polarization by treating with IL4/IL13 (10 ng/mL) for 24 h.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNAs from BEAS-2B, A549, NCI-H1299, NCI-H1975, or exosomes derived from A549 were extracted using Trizol reagent (ELK Biotechnology, Wuhan, China). Next, total RNAs was reversely transcribed into corresponding cDNA through EntiLink™ 1st Strand cDNA Synthesis Kit (ELK Biotechnology). After that, the StepOne™ Real-Time PCR System (Thermo Fisher Scientific, Waltham, MA, USA) was applied to perform RT-qPCR. The primers were listed as follows: β-actin, forward, 5’-GTCCACCGCAAATGCTTCTA-3’, reverse, 5’-TGCTGTCACCTTCACCGTTC-3’; KLF1, forward, 5’-AGGATGACTTCCTCAAGTGGTG-3’, reverse, 5’-GAGAAGTTGGTGAGGAGGAGATC-3’; PCAT6, forward, 5’-TCCAACTCCCAGACCTCACG-3’, reverse, 5’-GAGGAGCGCCTCATCACCAG-3’. The method of 2−ΔΔCT was applied to calculate the expression of lncRNA PCAT6 according to the previous literature [Citation36]. β-actin was viewed as an internal control.
Cell transfection
SiRNAs against lncRNA PCAT6 and KLF1 were provided by RiboBio. The sequences of siRNAs were presented as following: PCAT6 siRNA1, 5’-ACATCCCTAGGTGTCTCCATCCTCA-3’; PCAT6 siRNA2, 5’-CATCCCTAGGTGTCTCCATCCTCAT-3’; PCAT6 siRNA3, 5’-TCTCCATCCTCATTCGGTCCATCCA-3’; siRNA control, 5’-ACAATCCGTGGCTCTCTACCCTTCA-3’; KIF1 siRNA1, 5’-AGTACCAAGGGCACTTCCAGCTCTT-3’; KIF1 siRNA2, 5’-GACTGCAGAGGATCCAGGTGTGATA-3’. PCAT6 siRNAs, KLF1 siRNAs, or negative control were transfected into NSCLC cells with Lipofectamine® 2000 according to the previous literature [Citation37].
Cell Counting Kit-8 (CCK-8) assay
CCK‐8 assay kit (Dojindo, Kumamoto, Japan) was conducted to evaluate the viability of A549 or NCI-H1299 cells. Firstly, A549 or NCI-H1299 cells (5 × 103 cells/well) were seeded into 96-well plates. After treatments, cells were incubated with 10 µL CCK-8 for another 2 h according to the previous literature [Citation36]. Then, the absorbance of cells at 450 nm was monitored using a microplate reader.
Flow cytometry assay
Annexin-V-FITC apoptosis detection kit (Tianjin Sanjian Biotechnology Co., Ltd., Tianjin, China) was used to detect the apoptosis of A549 or NCI-H1299 cells. Firstly, A549 or NCI-H1299 cells (5 × 104/mL) were seeded into 6-well plates. After treatments, cells were incubated with 10 µL Annexin V-FITC and PI solution for 30 min in the dark. After that, the apoptosis of A549 or NCI-H1299 cells was monitored using a flow cytometer according to the previous literature [Citation38].
Transwell assay
The migration and invasion of NSCLC cells were detected by transwell assay. Transwell or matrigel-coated transwell (8 μm pore size) was provided by Corning (New York, NY, USA). Firstly, A549 or NCI-H1299 cells were injected into the upper chambers with 200 μL serum-free RIPM1640. Meanwhile, the lower chamber was supplemented with 600 μL RIPM1640 containing 10% FBS. Then, cells were cultured at 37°C for 24 h. Next, 0.1% crystal violet was applied to stain the cells that in the lower chambers. After that, the number of migration or invasion cells was counted using a microscope according to the previous literature [Citation38].
Dual-luciferase reporter assay
The wild-type/mutated-type (WT/MT) sequence of PCAT6 or KLF1 was inserted into the pGL6-miRNA reporter vector. Then, the WT or MT recombinant plasmid along with the miR-326 mimics (CCUCUGGGCCCUUCCUCCAG) or mimics control (mimics-ctrl) were transfected into A549 cells using Lipofectamine 2000. After that, a dual-Luciferase reporter assay system (Beyotime) was used to monitor the luciferase activities of A549 cells according to the previous literature [Citation38]. MiR-326 mimics and mimics control were provided with RioBio (Guangzhou, China).
Exosomes isolation
Exosomes in medium were isolated using differential ultracentrifugation according to previous study [Citation39]. Firstly, A549 cells were maintained in RPMI1650 for 48 h. Then, culture supernatants were collected by centrifuging at 2,000 × g for 20 min. Next, the supernatants were centrifuged at 10,000 × g for 30 min to remove cellular debris. After that, the pellets in supernatants were collected by centrifuging at 100,000 × g for 60 min twice according to the previous literature [Citation40].
Nanoparticle Tracking Analysis (NTA)
NTA was applied to identify whether pellets were exosomes by detecting the particle size. Firstly, pellets were washed using deionized water. Then, polystyrene microspheres were applied to calibrate ZetaView analyzer (Particle Metrix, Meerbusch, Germany). Next, pellets were diluted with 1X PBS buffer (Biological Industries, Israel). After that, ZetaView analyzer was conducted to detect whether these pellets conformed to the structure and particle size of the exosomes according to the previous literature [Citation40,Citation41].
Exosome uptake
A549 Exo (exosomes isolated from A549 cells) or A549PCAT [Citation6] siRNA [Citation1] Exo (exosomes derived from PCAT6 siRNA1-treated A549) were labeled with PKH26 staining dye. Then, A549 Exo or A549PCAT [Citation6] siRNA [Citation1] Exo were co-incubated with macrophage for 24 h. Besides, cytoskeleton or nuclei was stained by phalloidin or DAPI, respectively. Next, the internalization of A549 Exo or A549PCAT [Citation6] siRNA [Citation1] Exo was monitored using a fluorescence microscope according to the previous literature [Citation41,Citation42].
ELISA assay
Human IL1β (Interleukin 1 Beta) ELISA Kit or Human IL10 (Interleukin 10) ELISA Kit was used to detect the expressions of IL-1β or IL-10 in the supernatant of macrophage according to the manufacturer’s instructions. Human IL1β (Interleukin 1 Beta) ELISA Kit and Human IL10 (Interleukin 10) ELISA Kits were purchased from ELK Biotechnology according to the previous literature [Citation43].
Western blot assay
Total proteins from A549 cells were isolated using RIPA lysis buffer (Aspen Biotechnology, Wuhan, China). BCA kit was performed to quantify the concentration of proteins. After that, proteins (10 μg/lane) were separated by 10% SDS-PAGE and then transferred onto PVDF membranes. Next, PVDF membranes were incubated with primary antibodies anti-KLF1 (Abcam, Cambridge, MA, USA; 1:1000; Rabbit), anti-E-cadherin (1:1000; Rabbit), anti-N-cadherin (1:1000; Rabbit), anti-β-catenin (1:1000; Rabbit), anti-cleaved caspase 3 (1:1000; Rabbit), anti-Bcl-2 (1:1000; Rabbit), or anti-β-actin (1:1000; Rabbit) at 4°C overnight. Subsequently, PVDF membranes were incubated with the secondary antibodies (1:5000; Goat Anti-Rabbit) for 1 h. Then, the protein bands were observed using an efficient chemiluminescence (ECL) kit according to the previous literature [Citation35]. β-actin was viewed as an internal control.
Statistical analysis
These experimental data were presented as mean ± SD. One-way analysis of variance (ANOVA) and Tukey’s test were used to determine statistical differences. GraphPad Prism (La Jolla, CA, USA) was applied to analyze these data. All experiments were repeated three times. P < 0.05 was regarded as significant difference according to the previous literature [Citation43].
Results
Knockdown of PCAT6 inhibits the viability of NSCLC cells by inducing apoptosis
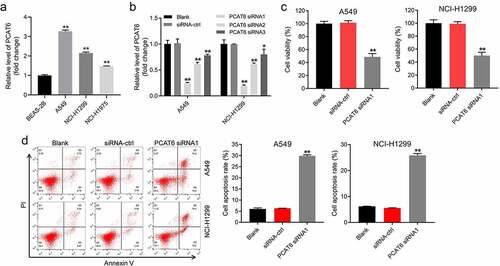
In order to explore the role of lncRNA PCAT6 in NSCLC, we first knockdown the expression of PCAT6 in NSCLC cells. Then, the effects of PCAT6 siRNA on the viability and apoptosis were detected. As shown in , the expression of PCAT6 in A549, NCI-H1299, or NCI-H1975 was significantly higher than that in BEAS-2B. As the expression of PCAT6 in A549 and NCI-H1299 was higher than that in NCI-H1975, A549, and NCI-H1299 cells were selected of use for the following experiments. In addition, PCAT6 siRNAs notably inhibited the expression of PCAT6 (). Since NSCLC cells were more sensitive to PCAT6 siRNA1, PCAT6 siRNA1 was used in the following experiments (). The result of CCK8 suggested PCAT6 siRNA1 markedly inhibited the viability of NSCLC cells (). Meanwhile, PCAT6 siRNA1 remarkably induced the apoptosis of NSCLC cells via increasing the expression of cleaved caspase 3 ( and supplementary Fig. 1A, 1B). All in all, knockdown of PCAT6 inhibited the viability of NSCLC cells by inducing apoptosis.
Figure 1. Knockdown of PCAT6 inhibited the viability of NSCLC cells by inducing apoptosis. (a) RT-qPCR was conducted to detect the expression of PCAT6 in BEAS-2B, A549, NCI-H1299 or NCI-H1975 cells. (b) NSCLC cells were transfected with PCAT6 siRNA1, PCAT6 siRNA2, PCAT6 siRNA3 or siRNA-ctrl using Lipofectamine® 2000. RT-qPCR was conducted to detect the level of PCAT6 in NSCLC cells. (c) CCK-8 assay was conducted to assess the viability of A549 and NCI-H1299 cells. (d) Flow cytometry was conducted to measure the apoptosis of A549 and NCI-H1299 cells. *P < 0.05, **P < 0.01 compared with control group; n = 3.
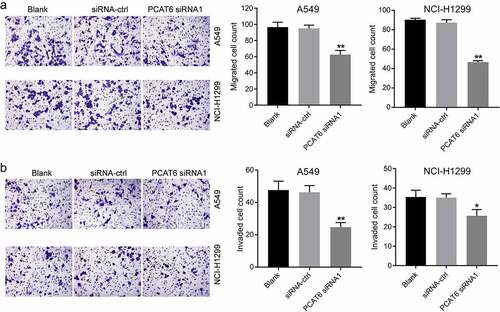
Knockdown of PCAT6 inhibits the migration and invasion of NSCLC cells
Next, to investigate the effect of PCAT6 siRNA1 on the metastasis of NSCLC cells, transwell assay was performed. As indicated in ), PCAT6 siRNA1 notably inhibited the metastasis of NSCLC. To sum up, knockdown of PCAT6 was able to inhibit the metastasis of NSCLC cells.
The interaction among miR-326, PCAT6, and KLF1 in NSCLC cells
With the aim of exploring the mechanism by which PCAT6 mediated the tumorigenesis of NSCLC, starBase was applied to predict the corresponding binding of PCAT6. The result of database displayed that miR-326 might be a candidate target of PCAT6 (). And it has been reported that miR-326 plays an important role in NSCLC [Citation44,Citation45]. Therefore, the relationship between miR-326 and PCAT6 was focused. In addition, the result of dual-luciferase reporter assay illustrated that miR-326 mimics markedly inhibited the luciferase activity of A549 cells transfected with PCAT6 wild-type sequence; however, miR-326 mimics had no effect on A549 cells transfected with PCAT6 mutant-type sequence ().
Figure 3. MiR-326 was the target of PCAT6 and KLF1 was the target of miR-326 in NSCLC cells. (a) StarBase was applied to predict the putative binding sites between miR-326 and PCAT6. (b) Dual-luciferase reporter assay was performed to verify the relationship between miR-326 and PCAT6 in A549 cells. (c) Targetscan, miRWalk and miRDB bioinformatics databases were applied to predict the putative binding sites between miR-326 and KLF1. (d) Dual-luciferase reporter assay was performed to identify the relationship between miR-326 and KLF1 in A549 cells. (e) The level of KLF1 in A549 cells was evaluated using RT-qPCR. **P < 0.01 compared with control group; n = 3.
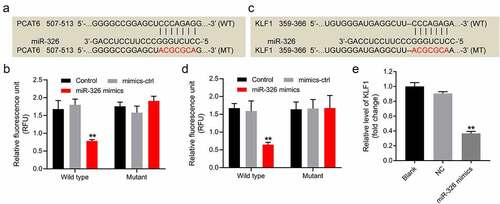
Next, the target of miR-326 was investigated using Targetscan, miRWalk, and miRDB bioinformatics databases. These databases commonly implied that KLF1 might be a direct target of miR-326 (). In addition, KLF1 is closely related to the development of lung-related diseases [Citation46]. Meanwhile, miR-326 mimics remarkably decreased the luciferase activity of A549 cells harboring KLF1 WT; whereas there was no change on the luciferase activity of A549 cells harboring KLF1 MT (). And miR-326 mimics significantly downregulated the expression of KLF1 in A549 cells (). Moreover, downregulation of KLF1 inhibited the migration of NSCLC cells and induced the apoptosis (Supplementary Fig. 2A, B, and C). All these data indicated that miR-326 was the target of PCAT6 and KLF1 was the target of miR-326 in NSCLC cells.
NSCLC cells-derived exosomes transported PCAT6 can be internalized by macrophage
With the aim to explore whether PCAT6 could regulate macrophages M2 polarization, NSCLC cell-derived exosomes were isolated. The result of NTA illuminated that the diameter of pellets derived from NSCLC cells ranges from 100 to 150 nm (). Besides, pellets expressed specific protein markers TSG101 and CD9 (). Next, in an attempt to investigate whether PCAT6 could be transport from NSCLC cells into macrophages through exosomes, A549 Exo, or A549PCAT [Citation6] siRNA [Citation1] Exo was co-incubated with macrophage. As indicated in , PKH26 could be observed in macrophages suggesting A549 Exo and A549PCAT [Citation6] siRNA [Citation1] Exo was internalized by macrophage. Meanwhile, the expression of PCAT6 in macrophage that were treated with A549PCAT [Citation6] siRNA [Citation1] Exo was much lower than that treated with A549 Exo (). All these data indicated NSCLC cells-derived exosomes could be internalized by macrophage.
Figure 4. NSCLC cells-derived exosomes transported PCAT6 could be internalized by macrophage. (a) NTA was applied to identify whether pellets were exosomes by detecting the particle size. (b) Western blot assay was applied to detect the level of specific protein markers TSG101 and CD9. (c) A549/A549 PCAT [Citation6] siRNA [Citation1] cells were co-incubated with macrophage for 24 h. (d) RT-qPCR was conducted to detect the expression of PCAT6 in macrophage. **P < 0.01 compared with A549 Exo group, n = 3.
![Figure 4. NSCLC cells-derived exosomes transported PCAT6 could be internalized by macrophage. (a) NTA was applied to identify whether pellets were exosomes by detecting the particle size. (b) Western blot assay was applied to detect the level of specific protein markers TSG101 and CD9. (c) A549/A549 PCAT [Citation6] siRNA [Citation1] cells were co-incubated with macrophage for 24 h. (d) RT-qPCR was conducted to detect the expression of PCAT6 in macrophage. **P < 0.01 compared with A549 Exo group, n = 3.](/cms/asset/98caedf9-b856-4452-86e1-757e9b32db18/kbie_a_2076388_f0004_oc.jpg)
A549 Exo is able to promote macrophages M2 polarization
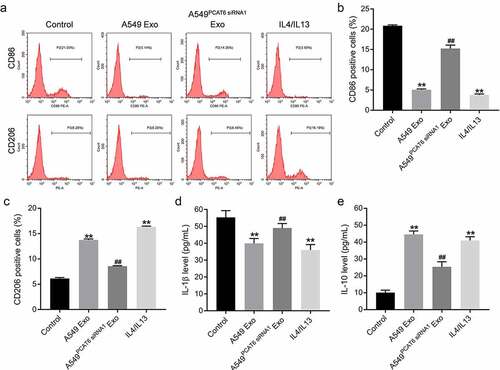
To detect the effect of NSCLC cell-derived exosomes on macrophage polarization, macrophage-related markers were detected. The results of flow cytometry and ELISA assay indicated that A549 Exo or IL4/IL13 significantly downregulated the expression of CD86 and IL-1β (M1 macrophage makers) and increased the level of CD206 and IL-10 (M2 macrophage makers) in macrophage; however, the effects of A549 Exo was abolished when PCAT6 was eliminated (). All these results suggested that A549 Exo was able to promote macrophages M2 polarization by transporting PCAT6.
Figure 5. A549 Exo was able to promote macrophages M2 polarization. (A, B and C) The expressions of CD86 and CD206 in macrophage were detected by flow cytometry assay. (d and e) ELISA assay was performed to detect the expressions of IL-1β and IL-10 in the supernatant of macrophage. **P < 0.01 compared with control group; ##P < 0.01 compared with A549 Exo group; n = 3.
Macrophages M2 polarization significantly promote the migration and invasion of NSCLC cells
With the aim to further explore the influence of macrophages M2 polarization on NSCLC cells, A549 cells were co-cultured with macrophage (PMA-treated THP-1), MExo (A549 Exo-treated macrophage), (A549PCAT [Citation6] siRNA [Citation1] Exo-treated macrophage MPCAT [Citation6] siRNA [Citation1]) or MIL [Citation4]/IL [Citation13] (IL4/IL13 treated macrophage) (). The outcome of co-culture illustrated MExo or MIL [Citation4]/IL [Citation13] significantly promoted the metastasis of A549 cells, while the effects of MExo on cell migration and invasion were neutralized when PCAT6 was eliminated. Taken together, macrophages M2 polarization was able to promote the metastasis of NSCLC cells.
Figure 6. Macrophages M2 polarization was able to promote the migration and invasion of NSCLC cells. (a) THP-1 monocytes were induced into M0 macrophages by treating with PMA for 24 h. Then, M0 macrophages were treated with A549 Exo or A549PCAT [Citation6] siRNA [Citation1] Exo for 24 h. Next, A549 were co-cultured with macrophage. (b) Transwell assay was conducted to assess the metastasis of A549 cells. **P < 0.01 compared with A549 group. ##P < 0.01 compared with the A549 + MExo group; n = 3. M: M0 macrophages; MExo: M0 macrophages incubated with A549 Exo; MIL [Citation4]/IL [Citation13]: M0 macrophages treated with IL4/IL13 to differentiate into M2 macrophages; MPCAT [Citation6] siRNA [Citation1]: M0 macrophages were incubated with A549PCAT [Citation6] siRNA [Citation1] Exo.
![Figure 6. Macrophages M2 polarization was able to promote the migration and invasion of NSCLC cells. (a) THP-1 monocytes were induced into M0 macrophages by treating with PMA for 24 h. Then, M0 macrophages were treated with A549 Exo or A549PCAT [Citation6] siRNA [Citation1] Exo for 24 h. Next, A549 were co-cultured with macrophage. (b) Transwell assay was conducted to assess the metastasis of A549 cells. **P < 0.01 compared with A549 group. ##P < 0.01 compared with the A549 + MExo group; n = 3. M: M0 macrophages; MExo: M0 macrophages incubated with A549 Exo; MIL [Citation4]/IL [Citation13]: M0 macrophages treated with IL4/IL13 to differentiate into M2 macrophages; MPCAT [Citation6] siRNA [Citation1]: M0 macrophages were incubated with A549PCAT [Citation6] siRNA [Citation1] Exo.](/cms/asset/1e2ab574-c609-48bd-9424-983ab1d58201/kbie_a_2076388_f0006_oc.jpg)
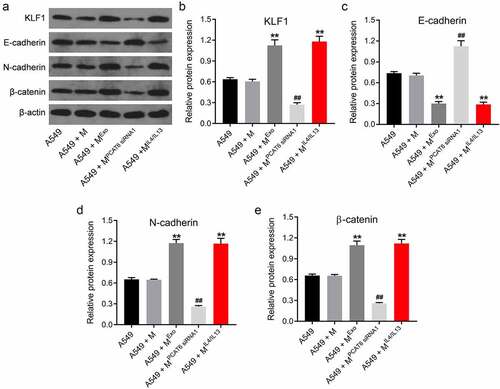
Macrophages M2 polarization promotes EMT process of NSCLC cells via regulating PCAT6/miR-326/KLF1 axis
In order to further explore the mechanism by which macrophages M2 polarization regulated the progression of NSCLC cells, western blot was applied. As indicated in ), MExo markedly increased the level of KLF1 in A549 cells, while these increases were reversed when PCAT6 was eliminated. Furthermore, MExo or MIL [Citation4]/IL [Citation13] notably downregulated the expression of E-cadherin and upregulated the levels of N-cadherin and β-catenin A549 cells; however, the effects of MExo on these proteins were abolished when PCAT6 was eliminated (). All these results illustrated that macrophages M2 polarization was able to promote EMT process of NSCLC cells via regulating PCAT6/miR-326/KLF1 axis.
Figure 7. Macrophages M2 polarization was able to promote EMT process of NSCLC cells via regulating PCAT6/miR-326/KLF1 axis. (A, B, C, D and E) Western blot assay was applied to detect the level of KLF1, E-cadherin, N-cadherin and β-catenin in A549 cells. **P < 0.01 compared with A549 group. ##P < 0.01 compared with the A549 + MExo group; n = 3.
Discussion
The biological function of PCAT6 in lung cancer has received extensive attention [Citation47]. Su et al. reported that sevoflurane could restrain the viability and proliferation of lung cancer cells through regulating PCAT6/miR-326/axis [Citation47]. In addition, Cui et al. indicated that PCAT6 was able to facilitate the proliferation and invasion of NSCLC cells by regulating miR-330-5p 34. Moreover, Shi et al. suggested that knockdown of PCAT6 could inhibit the growth of NSCLC cells in vitro and in vivo via inhibiting LATS2 [Citation48]. This current study showed that knockdown of PCAT6 inhibited the viability, metastasis of NSCLC cells by via PCAT6/miR-326/KLF1 axis. Our research results were consistent with previous studies. In addition, we first explore the connection between PCAT6 and TME in NSCLC.
In recent years, the TME has received great attention as well. TME are recognized that the metastasis of tumors are related to the environment of tumor cells [Citation49]. It is worth noting that most of the macrophages in tumors are of M2 type [Citation50]. For example, lncRNA XIST was highly expressed in macrophages [Citation51]. Knockdown of lncRNA XIST was able to inhibit macrophages M2 polarization, which in turn alleviated the occurrence and development of lung cancer [Citation51]. Besides, it has been reported that PCAT6 was able to promote macrophages M2 polarization in cholangiocarcinoma [Citation52]. Meanwhile, PCAT6 could boost the viability, metastasis and EMT process of triple-negative breast cancer [Citation53]. In turn, the level of PCAT6 was increased by M2 macrophages in breast cancer [Citation53]. And M2 macrophages-induced PCAT6 increase promoted the development of triple-negative breast cancer [Citation53]. In the present study, A549 Exo could promote macrophages M2 polarization via regulating PCAT6/miR-326/KLF1 axis. Meanwhile, macrophages M2 polarization was able to promote the metastasis and EMT process of NSCLC cells by regulating PCAT6/miR-326/KLF1 axis. These studies commonly showed that PCAT6-induced macrophages M2 polarization, which in turn promoted the tumorgenesis.
In addition, it has been reported that KLF1 is closely related to macrophages polarization [Citation24]. In the present study, we found that knockdown of lncRNA PCAT6 suppressed the growth of NSCLC cells by inhibiting macrophages M2 polarization via miR-326/KLF1 axis. These phenomena commonly indicated that KLF1 acts as a vital role during macrophages polarization. Besides, KLF1 acts as a crucial role in the process of EMT [Citation54]. For instant, Li et al showed that knockdown of KLF1 was able to decrease the viability, metastasis, and EMT process of gastric cancer cells [Citation54]. In this study, knockdown of PCAT6 was able to inhibit the metastasis and EMT process of NSCLC cells. In addition, knockdown of KLF1 inhibited the migration of NSCLC cells via inducing apoptosis. These data are consistent with previous study, suggesting that KLF1 plays an important role in regulating the process of EMT.
It is worth noting that lncRNA PCAT6 regulates A549 metastasis not only through regulating EMT. We deduced PCAT6 may affect the metastasis of A549 via regulating different miRNAs. In addition, we found PCAT6 regulated macrophages M2 polarization, which in turn affected the metastasis of A549 as well. All in all, the detailed mechanism by which PCAT6 regulated the metastasis of NSCLC remains unclear and this is the limitation of current study.
Conclusion
In conclusion, NSCLC cells-derived exosomes transported PCAT6 significantly promote macrophages M2 polarization. Meanwhile, macrophages M2 polarization was able to promote the metastasis and EMT process of NSCLC cells via regulating PCAT6/miR-326/KLF1 axis. This study might point out that the tumorigenesis and development of NSCLC could be alleviated by inhibiting macrophages M2 polarization.
Supplemental Material
Download Zip (10.5 MB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/21655979.2022.2076388
Additional information
Funding
References
- Herbst RS, Morgensztern D, Boshoff C. The biology and management of non-small cell lung cancer. Nature. 2018;553:446–454.
- Pennell NA, Arcila ME, Gandara DR, et al. Biomarker testing for patients with advanced non-small cell lung cancer: real-world issues and tough choices. American Society of Clinical Oncology educational book American Society of Clinical Oncology Annual Meeting, America. 2019; 39:531–542.
- Loeb LA, Ernster VL, Warner KE, et al. Smoking and lung cancer: an overview. Cancer Res. 1984;44(12 Pt 1):5940–5958.
- Rivera GA, Wakelee H. Lung cancer in never smokers. Adv Exp Med Biol. 2016;893:43–57.
- Molina JR, Yang P, Cassivi SD, et al. Non-small cell lung cancer: epidemiology, risk factors, treatment, and survivorship. 2008; 83:584–594.
- Smith JA, Harle A, Dockry R, et al. Aprepitant for cough in lung cancer. a randomized placebo-controlled trial and mechanistic insights. Am J Respir Crit Care Med. 2021;203(6):737–745.
- Malinowska K. The relationship between chest pain and level of perioperative anxiety in patients with lung cancer. Pol Przegl Chir. 2018;90(2):23–27.
- Ha D, Ries AL. Characterization of dyspnea in veteran lung cancer survivors following curative-intent therapy. J Cardiopulm Rehabil Prev. 2020;40(2):120–127.
- Hoy H, Lynch T, Beck M. Surgical Treatment of Lung Cancer. Crit Care Nurs Clin North Am. 2019;31(3):303–313.
- Rossi A, Di Maio M. Platinum-based chemotherapy in advanced non-small-cell lung cancer: optimal number of treatment cycles. Expert Rev Anticancer Ther. 2016;16(6):653–660.
- Imyanitov EN, Iyevleva AG, Levchenko EV. Molecular testing and targeted therapy for non-small cell lung cancer: current status and perspectives. Crit Rev Oncol Hematol. 2021;157:103194.
- Jonna S, Subramaniam DS. Molecular diagnostics and targeted therapies in non-small cell lung cancer (NSCLC): an update. Discov Med. 2019;27(148):167–170.
- Hirayama D, Iida T, Nakase H. The phagocytic function of macrophage-enforcing innate immunity and tissue homeostasis. Int J Mol Sci. 2017;19(1):92.
- Chanput W, Mes JJ, Wichers HJ. THP-1 cell line: an in vitro cell model for immune modulation approach. Int Immunopharmacol. 2014;23(1):37–45.
- Biriken D, Yazıhan N, Ş Y. [Investigation of cytokine and midkine responses of human THP-1 leukemia cells induced by phorbol-12-Myristate-13-Acetate (PMA) at different concentrations and times]. Mikrobiyoloji bulteni. 2018;52(2):147–155.
- Funes SC, Rios M, Escobar-Vera J, et al. Implications of macrophage polarization in autoimmunity. Immunology. 2018;154(2):186–195.
- Zhou D, Huang C, Lin Z, et al. Macrophage polarization and function with emphasis on the evolving roles of coordinated regulation of cellular signaling pathways. Cell Signal. 2014;26(2):192–197.
- Martinez FO, Sica A, Mantovani A, et al. Macrophage activation and polarization. Front Biosci. 2008;13(13):453–461.
- Genin M, Clement F, Fattaccioli A, et al. M1 and M2 macrophages derived from THP-1 cells differentially modulate the response of cancer cells to etoposide. BMC Cancer. 2015;15(1):577.
- Pan Y, Yu Y, Wang X, et al. Tumor-associated macrophages in tumor immunity. Front Immunol. 2020;11:583084.
- He Y, Du J, Dong Z. Myeloid deletion of phosphoinositide-dependent kinase-1 enhances NK cell-mediated antitumor immunity by mediating macrophage polarization. Oncoimmunology. 2020;9(1):1774281.
- Zeng XY, Xie H, Yuan J, et al. M2-like tumor-associated macrophages-secreted EGF promotes epithelial ovarian cancer metastasis via activating EGFR-ERK signaling and suppressing lncRNA LIMT expression. Cancer Biol Ther. 2019;20(7):956–966.
- Mukherjee K, Bieker JJ. Transcriptional control of gene expression and the heterogeneous cellular identity of erythroblastic island macrophages. Front Genet. 2021;12:756028.
- Mukherjee K, Xue L, Planutis A, et al. EKLF/KLF1 expression defines a unique macrophage subset during mouse erythropoiesis. eLife. 2021;10. DOI:10.7554/eLife.61070
- Wang J, Su Z, Lu S, et al. LncRNA HOXA-AS2 and its molecular mechanisms in human cancer. Clin Chim Acta. 2018;485:229–233.
- Zhu J, Zhang X, Gao W, et al. lncRNA/circRNA‑miRNA‑mRNA ceRNA network in lumbar intervertebral disc degeneration. Mol Med Rep. 2019;20(4):3160–3174.
- Wu X, Sui Z, Zhang H, et al. Integrated analysis of lncRNA-mediated cerna network in lung adenocarcinoma. Front Oncol. 2020;10:554759.
- Zhen Q, Gao LN, Wang RF, et al. LncRNA DANCR Promotes Lung Cancer by Sequestering miR-216a. Cancer control: J Moffitt Cancer Center. 2018;25(1):1073274818769849.
- Sun J, Zhang Z, Bao S, et al. Identification of tumor immune infiltration-associated lncRNAs for improving prognosis and immunotherapy response of patients with non-small cell lung cancer. J Immunother Cancer. 2020;e000110:8.
- Zhang YX, Yuan J, Gao ZM, et al. LncRNA TUC338 promotes invasion of lung cancer by activating MAPK pathway. Eur Rev Med Pharmacol Sci. 2018;22(2):443–449.
- Wang S, Chen Z, Gu J, et al. The role of lncRNA PCAT6 in cancers. Front Oncol. 2021;11:701495.
- Luo Y, Lin J, Zhang Y, et al. LncRNA PCAT6 predicts poor prognosis in hepatocellular carcinoma and promotes proliferation through the regulation of cell cycle arrest and apoptosis. Cell Biochem Funct. 2020;38(7):895–904.
- Kong FR, Lv YH, Yao HM, et al. LncRNA PCAT6 promotes occurrence and development of ovarian cancer by inhibiting PTEN. Eur Rev Med Pharmacol Sci. 2019;23(19):8230–8238.
- Cui LH, Xu HR, Yang W, et al. lncRNA PCAT6 promotes non-small cell lung cancer cell proliferation, migration and invasion through regulating miR-330-5p. Onco Targets Ther. 2018;11:7715–7724.
- Lai F, Zhang H, Xu B, et al. Long non-coding RNA NBR2 suppresses the progress of colorectal cancer in vitro and in vivo by regulating the polarization of TAM. Bioengineered. 2021;12(1):5462–5475.
- Liu Y, Liu J, Cui J, et al. Role of lncRNA LINC01194 in hepatocellular carcinoma via the miR-655-3p/SMAD family member 5 axis. Bioengineered. 2022;13(1):1115–1125.
- Tan D, Li G, Zhang P, et al. LncRNA SNHG12 in extracellular vesicles derived from carcinoma-associated fibroblasts promotes cisplatin resistance in non-small cell lung cancer cells. Bioengineered. 2022;13(1):1838–1857.
- Fu D, Zang L, Li Z, et al. Long non-coding RNA CRNDE regulates the growth and migration of prostate cancer cells by targeting microRNA-146a-5p. Bioengineered. 2021;12(1):2469–2479.
- Han M, Gu Y, Lu P, et al. Exosome-mediated lncRNA AFAP1-AS1 promotes trastuzumab resistance through binding with AUF1 and activating ERBB2 translation. Mol Cancer. 2020;19(1):26.
- Shou Y, Wang X, Liang Y, et al. Exosomes-derived miR-154-5p attenuates esophageal squamous cell carcinoma progression and angiogenesis by targeting kinesin family member 14. Bioengineered. 2022;13(2):4610–4620.
- Huang H, Zhong P, Zhang J, et al. Human umbilical cord-mesenchymal stem cells-derived exosomes carrying microRNA-15a-5p possess therapeutic effects on wilms tumor via regulating septin 2. Bioengineered. 2022;13(3):6136–6149.
- Ni Q, Zhang H, Shi X, et al. Exosomal microRNA-23a-3p contributes to the progression of cholangiocarcinoma by interaction with dynamin3. Bioengineered. 2022;13(3):6208–6221.
- Che Z, Xueqin J, Zhang Z. LncRNA OIP5-AS1 accelerates intervertebral disc degeneration by targeting miR-25-3p. Bioengineered. 2021;12(2):11201–11212.
- Wang R, Xu J, Xu J, et al. MiR-326/Sp1/KLF3: a novel regulatory axis in lung cancer progression. Cell Prolif. 2019;52(2):e12551.
- Sun C, Huang C, Li S, et al. Hsa-miR-326 targets CCND1 and inhibits non-small cell lung cancer development. Oncotarget. 2016;7(7):8341–8359.
- Bao H, Wang J, Zhou D, et al. Retracted article: protein–Protein interaction network analysis in chronic obstructive pulmonary disease. Lung. 2014;192(1):87–93.
- Su G, Yan Z, Deng M. Sevoflurane inhibits proliferation, invasion, but enhances apoptosis of lung cancer cells by Wnt/β-catenin signaling via regulating lncRNA PCAT6/miR-326 axis. Open Life Sci. 2020;15(1):159–172.
- Shi X, Liu Z, Liu Z, et al. Long noncoding RNA PCAT6 functions as an oncogene by binding to EZH2 and suppressing LATS2 in non-small-cell lung cancer. EBioMedicine. 2018;37:177–187.
- Lee SS, Cheah YK. The interplay between micrornas and cellular components of Tumour Microenvironment (TME) on Non-Small-Cell Lung Cancer (NSCLC) progression. J Immunol Res. 2019;2019:3046379.
- Weng YS, Tseng HY, Chen YA, et al. MCT-1/miR-34a/IL-6/IL-6R signaling axis promotes EMT progression, cancer stemness and M2 macrophage polarization in triple-negative breast cancer. Mol Cancer. 2019;18(1):42.
- Sun Y, Xu J. TCF-4 regulated lncRNA-XIST promotes M2 polarization of macrophages and is associated with lung cancer. Onco Targets Ther. 2019;12:8055–8062.
- Tu J, Wu F, Chen L, et al. Long non-coding RNA PCAT6 induces M2 polarization of macrophages in cholangiocarcinoma via modulating miR-326 and RhoA-ROCK signaling pathway. Front Oncol. 2020;10:605877.
- Dong F, Ruan S, Wang J, et al. M2 macrophage-induced lncRNA PCAT6 facilitates tumorigenesis and angiogenesis of triple-negative breast cancer through modulation of VEGFR2. Cell Death Dis. 2020;11(9):728.
- Li S, Li Y, Tan B, et al. Krüppel-like factor 1 serves as a facilitator in gastric cancer progression via activating the Wnt/β-catenin pathway. Acta Biochim Pol. 2021;68(4):765–774.