ABSTRACT
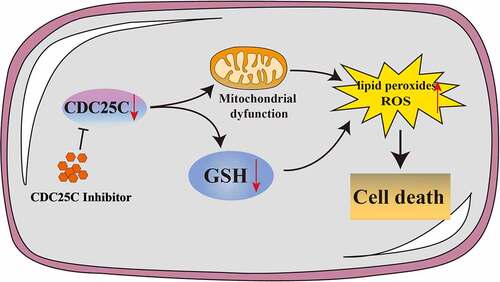
Pancreatic adenocarcinoma (PAAD) is a common digestive tract malignant tumor with an extremely poor prognosis. The survival and prognosis may significantly improve if it is diagnosed early. Therefore, identifying biomarkers for early diagnosis is still considered a great clinical challenge in PAAD. Cell Division Cycle 25C (CDC25C), a cardinal cell cycle regulatory protein, directly mediates the G2/M phase and is intimately implicated in tumor development. In the current study, we aim to explore the possible functions of CDC25C and determine the potential role of CDC25C in the early diagnosis and prognosis of PAAD. Expression analysis indicated that CDC25C was overexpressed in PAAD . In addition, survival analysis revealed a strong correlation between the enhanced expression of CDC25C and poor survival in PAAD. Furthermore, pathway analysis showed that CDC25C is related to TP53 signaling pathways, glutathione metabolism, and glycolysis. Mechanically, our in vitro experiments verified that CDC25C was capable of promoting cell viability and proliferation. CDC25C inhibition increases the accumulation of ROS, inhibits mitochondrial respiration, suppresses glycolysis metabolism and reduces GSH levels. To summarize, CDC25C may be involved in energy metabolism by maintaining mitochondrial homeostasis. Our results suggested that CDC25C is a potential biological marker and promising therapeutic target of PAAD.
GRAPHICAL ABSTRACT
Highlights
CDC25C was upregulated in PAAD tissues and cell lines which was correlated with PAAD prognosis.
CDC25C was involved in TP53 signaling pathways, glutathione metabolism, and glycolysis.
CDC25C inhibition triggered mitochondrial dysfunction and reduced glycolysis metabolism.
1. Introduction
The incidence of pancreatic adenocarcinoma (PAAD) is increasing by approximately 1% annually worldwide, and PAAD ranks third as the reason for cancer-related mortality [Citation1,Citation2]. A large part of PAAD patients was generally diagnosed at an advanced stage owing to insidious and atypical clinical symptoms. Despite the development of new medical technologies and improvements in treatment methods, the survival of PAAD patients has not greatly improved [Citation3]. Therefore, diagnostic biomarkers and therapeutic targets for PAAD are essential for preventing the occurrence and development of PAAD. Recent evidence suggests circulating tumor DNA (ctDNA) is a potential prognostic marker and KRAS mutant ctDNA was detected in 62% of pre-operative plasma samples [Citation4]. Moreover, tRNA-derived small RNAs and microRNAs were also identified as biomarkers for PAAD [Citation5–7]. Despite the increasing biomarkers regarding PAAD, the sensitivity and specificity of them remain unsatisfactory [Citation8]. Thus, developing novel effective early diagnostic biomarkers for PAAD is crucial.
Previous studies have shown that the CDC25 phosphatase family, composed of CDC25A, CDC25B, and CDC25C in humans, regulates the cell cycle progression by dephosphorylating the CDK-Cyclin complex [Citation9]. CDC25C, a tyrosine-protein phosphatase required for cell cycle progression, is located in the cytoplasm and can relocate from the cytoplasm to the nucleus during the interphase [Citation10]. Current studies have revealed that CDC25C has substantial potential applicability for early diagnosis and prognosis of tumors; for example, CDC25C can predict the response to radiotherapy in esophageal squamous cell carcinoma patients, and forecast the postsurgical survival rate of such patients [Citation11,Citation12]. Moreover, the over-expression of CDC25C showed a strong correlation with poor prognosis in 1,700 tumor samples of breast cancer [Citation13]. Nevertheless, only a few studies reported the effect of CDC25C in the early diagnosis and prognosis of PAAD.
In this study, we aim to explore the possible functions of CDC25C and determine the potential role of CDC25C in the early diagnosis and prognosis of PAAD. By analyzing Gene Expression Profiling Interactive Analysis (GEPIA) and Oncomine databases, the expression abundance of CDC25C was dramatically elevated in PAAD and negatively associated with the PAAD prognosis. Moreover, we discovered that CDC25C was related to glutathione metabolism and glycolysis in PAAD. Finally, the effects of CDC25C were examined in vitro and the results revealed that CDC25C inhibitors could hinder cell proliferation, trigger mitochondrial dysfunction, and suppress glycolysis metabolism in PAAD cells. In summary, this study provides evidence that CDC25C is involved in regulating mitochondrial respiration and glycolysis metabolism in PAAD cells. These findings demonstrated that CDC25C may be a promising diagnostic and prognostic predictive biomarker.
2. Materials and methods
2.1. Gene expression analysis
GEO database was used to analyze the expression of CDC25C in PAAD and normal pancreatic tissues [Citation14,Citation15]. We selected 4 GEO chip data sets (GSE71729, GSE106189, GSE16515, GSE77435) and divided them into two groups to compare the expression of CDC25C, each of which contained pancreatic tumor and pancreatic normal tissues.
UALCAN is an online tool to analyze the gene expression and prognosis based on TCGA database [Citation16,Citation17]. We utilized UALCAN to analyze CDC25C expression by different aspects of sample types, including tumor grade, patient’s race, age, methylation level and TP53 mutation status. Besides, the expression of CDC25C was also confirmed by GEPIA which can process and visualize data based on TCGA and GTEx databases [Citation18].
Oncomine is a comprehensive online analysis web designed to analyze cancer genetic information including differential gene analysis, gene expression and clinical correlation analysis. The differential expression of CDC25C between carcinoma and adjacent tissues was explored by the Oncomine database.
2.2. Survival analysis
GEPIA aimed to investigate the relationship between CDC25C expression and survival of PAAD patients. According to the expression level of CDC25C, 178 PAAD patients were divided into two groups (high CDC25C expression group and low CDC25Cexpression group), and then analyzed the difference of disease-free survival and overall survival between the two groups.
2.3. CDC25C-Related gene enrichment analysis
STRING is a search tool for retrieving protein interaction relationships [Citation19,Citation20]. To elucidate the role and signaling pathways of differentially expressed genes of PAAD tissues compared with non-cancerous tissues, we obtained the top 50 genes that interact with CDC25C, and then constructed the protein-protein interaction networks (PPI) through the STRING tool. Moreover, pathway analysis of the enriched genes was performed using Metascape which is a gene function annotation tool based on many metabolic pathway databases [Citation21]. Meanwhile, the GSEA of CDC25C function in PAAD was also carried out in R software.
2.4. Immune infiltration analysis
CIBERSORT is an online analysis database that analyzes the immune cell infiltration in samples [Citation22]. In this study, 134 samples that met the criteria for immune infiltration analysis were included to analyze the distribution of 22 different immune cells in PAAD. Tumor Immune Estimation Resource (TIMER) is an online tumor immune research tool to reveal the infiltration of immunocytes in the tumor microenvironment [Citation23]. Immune infiltration analysis of CDC25C was conducted by CIBERSORT and TIMER databases.
2.5. Chemicals, reagents, and antibodies
Enhanced Cell Counting Kit-8 (CCK8 kit), Hoechst 33,258 was acquired from Beyotime (Shanghai, China). NSC95397 and NSC663284 were purchased from MedChemExpress (New Jersey, USA). DCF-DA, C11-BODIPY (581/591) and MitoSOX were purchased from Thermo Fisher Scientific (Waltham, MA). The antibodies to CDC25C (ab32444), GAPDH (ab8245), β-Actin (ab8226), GRD1 (ab124995), PKM2 (ab89364), c-Myc (ab32072), NDUFS3 (ab177471), SDHB (ab175225), NDUFV2 (ab183715), COX IV (ab202554) were purchased from Abcam. GAPDH and β-Actin antibodies were used with 5000-fold dilution and other antibodies were used with 1000-fold dilution.
2.6. Cell culture
SW1990, BXPC3 and HPDE6-C7 cells were subcultured and preserved in our laboratory. The cell lines were cultivated in DMEM high glucose medium (Hyclone, USA) supplemented with 10% Fetal Bovine Serum (Gibco, USA) and 1% penicillin and streptomycin, (Beyotime, China). Cells were then cultured at 37°C and 5% CO2.
2.7. Cell viability assay
CCK8 kit was used to detect the cell viability according to the product instruction. The seeding density of 96-well plates (NEST, China) was 2 × 104 cells/well. After designed treatments, CCK8 was added for 2 h incubation. Finally, the microplate reader (Thermo Fisher Scientific, Inc) was utilized to measure the absorbance of each well at 450 nm.
2.8. Growth curve assay
The growth curve of cells was assayed by the CCK8 kit. The cell density was 0.5 × 104 cells/well, and the cells were grown in 96-well plates. The cell proliferation was tested at 0, 24, 48 and 72 h after predesigned treatment. Then add 10 µL CCK-8 into each well, and incubate at 37°C in a 5% CO2 incubator. Finally, the absorbance of each well was detected using a microplate reader.
2.9. Cell cloning assay
The clonogenic ability of cells was carried out according to our previous published paper [Citation24]. The cells were collected and seeded on a 12-well plate with a density of 2,000 cells/well. After the cells grew in the well plate for 14 days, the cells were fixed with 4% paraformaldehyde (Leagene, China) for 15 min and stained with 0.5% crystal violet for 30 minutes. The colonies were imaged using a digital camera and counted by the Image J program.
2.10. EdU assay
The 5-ethynyl-2-deoxyuridine (EdU) incorporation assay was performed using EdU assay kit (Beyotime, Shanghai, China). The cells (1 × 105) were seeded in a confocal dish (Cellvis, USA) at 37°C away from light overnight. Next, cells were exposed to NSC663284 or NSC95397 (inhibitors of CDC25C) for 24 h and incubated with EdU (10 µM) for 2 h at 37°C. Subsequently, 4% paraformaldehyde was used for fixing cells and cells were permeabilized by 0.3% TritonX-100 in PBS for 15 min. Finally, Cells were washed 3 times in PBS followed by 30 min incubation with 1.72 mL Click Reaction Buffer, 80 µL CuSO4, 4 µL Azide 594, and 200 µL Click Additive Solution. Nucleic was co-stained with Hoechst 33,258 for 10 min. Images were obtained with laser confocal microscopy (Leica TCS SP5, German).
2.11. Measurement of ROS productions and lipid peroxide
The SW1990 cells were seeded on 6-well plates and each well contained 8 × 105 cells. The cells were incubated with NSC663284 and NSC95397 for 4 h and then stained with DCF-DA (5 µM) and C11-BODIPY (4 µM) for 30 min. Next, the fluorescence intensity was determined by CytoFLEX SRT flow cytometry (Beckman Coulter, USA).
2.12. Confocal microscopy assay
SW1990 cells were cultured in the confocal dish overnight at 37°C. Following indicated treatments for 4 h, the cells were stained with DCF-DA (5 µM) and MitoSOX (3 µM) in an incubator at 37°C for 30 min. Meanwhile, Hoechst 33,258 (10 µg/mL) was applied to the visualization of nucleus. Lastly, a laser confocal microscope was used to photograph the confocal dish.
2.13. Determinations of OCR and ECAR
The SW1990 cells were cultured in an XFe 24 seahorse cell culture microplate (Agilent, USA). In the concurrent phase, the sensor cartridge (Agilent, USA) was hydrated overnight at 37°C in an incubator without CO2. Cells were subjected to the treatments with NSC663284 and NSC95397 for 4 h, respectively. Then the microplate was replaced with a Seahorse base medium and maintained in a CO2-free incubator for 1 h. The oxygen consumption rate (OCR) represents the basic oxygen consumption of cells, including mitochondrial oxidative phosphorylation and proton leakage oxygen consumption. OCR was monitored continuously in real-time by serial addition of oligomycin (1.5 μM), carbonyl cyanide 4-(trifluoromethoxy) phenylhydrazone (FCCP) (2 μM), and Antimycin A/Rotenone (0.5 μM). And the ability of the extracellular acidification rate (ECAR) indirectly shows the glycolytic ability of cells. ECAR was monitored on a real-time basis by serial addition of glucose (10 mM), oligomycin (1 μM), and 2-Deoxyglucose (2-DG) (50 mM). Finally, the OCR and ECAR were analyzed by the Seahorse XFe 24 Bioanalyzer (Agilent, USA).
2.14. Western blot
Cells were treated as described earlier. Briefly, the collected cells were lysed via RIPA lysis buffer containing protease and phosphatase inhibitors (Beyotime) and the supernatant was quantified by BCA Protein Quantification Kit (Boxbio Science & Technology, Beijing). Next, the proteins in these samples were separated by 10% SDS-PAGE and transferred to PVDF membranes (Bio-Rad). Membranes were blocked in 5% skimmed milk for 1 h and then incubated with the primary antibodies on a shaker for 12 h in a chromatography cabinet. On the following day, the blots were washed with TBST and incubated with horseradish peroxidase (HRP) labeled secondary antibodies for 1 h. The HRP signals were detected by a Fdbio-Plco ECL kit (Fdbio science). Finally, the blots were scanned by Gel Imager (Bio-Rad).
2.15. Statistical analysis
All data obtained were processed and statistically calculated with GraphPad Prism (version 5.0). Data obtained above were presented as the mean ± standard deviation (SD). The differences between two assay groups were performed by the Student’s t-test. One-way ANOVA was used for analyzing comparisons among multiple groups. p < 0.05 was considered to be the statistically significant level.
3. Results
3.1. CDC25C expression is high in PAAD
CDC25C was reported to be a potential biomarker in cancers, but the role of CDC25C in PAAD remains unknown [Citation11]. In the current study, we explore the regulatory role of CDC25C and found that CDC25C may promote cell proliferation and regulate energy metabolism by maintaining mitochondrial homeostasis in PAAD.
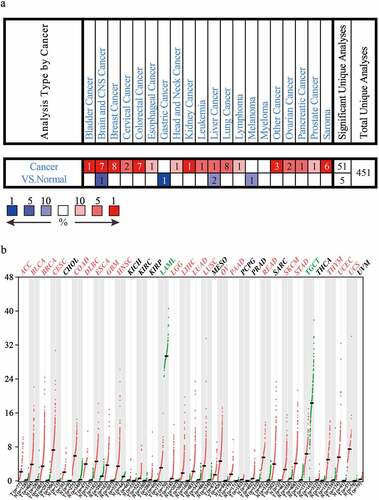
Firstly, to explore the expression characteristics of CDC25C in different cancer types, we analyzed CDC25C expression using the Oncomine database. As displayed in ), CDC25C expression was significantly elevated in various tumors, including PAAD tissues, compared with its expression levels in normal samples. Next, the CDC25C expression level was also analyzed using the GEPIA online database. Consistent with the data obtained from the Oncomine database, CDC25C was highly expressed in 21 out of 33 cancers ()). The results of the two bioinformatics databases suggested CDC25C is overexpressed in multiple cancers.
Figure 1. CDC25C is overexpressed in multiple cancers.
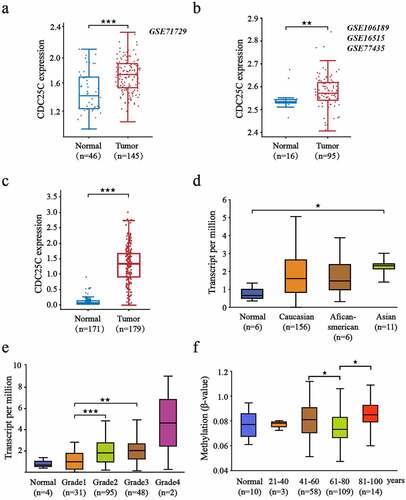
Subsequently, to further investigate the expression of CDC25C in PAAD, four datasets acquired from the GEO database were selected and then divided into two groups according to tissue types, each of which contained pancreatic tumor and healthy pancreatic tissues ()). The results of group 1 and group 2 showed enhanced expression of CDC25C in pancreatic carcinoma compared with adjacent tissues (p = 5.9e-06 and p = 0.0069, respectively). Similarly, the analysis from GEPIA also revealed CDC25C was upregulated in PAAD (n = 179) compared with that in the normal group (n = 171) (p = 5.60e-56) ()). Additionally, further results indicated that CDC25C expression was the highest in Asians among all races ()). CDC25C expression increased continuously from grade 1 to grade 4, suggesting that CDC25C was significantly associated with the malignancy of pancreatic cancer, especially from grade 1 to grade 2 (p = 1.83e-05) ()). Compared with patients in the 61–80 age group, those in the 41–60 and >80 age groups were discovered to have higher CDC25C methylation levels ()).
Figure 2. CDC25C is highly expressed in PAAD.
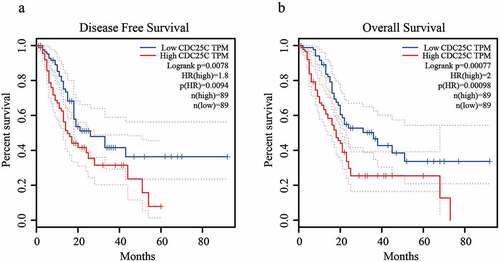
3.2. CDC25C expression is negatively associated with the prognosis of PAAD
To confirm the association between CDC25C overexpression and PAAD prognosis, we divided all PAAD samples into CDC25C high and low expression groups according to the average CDC25C expression for prognostic analyses. It was observed that patients with PAAD in the CDC25C low expression group possessed a relatively better disease-free and overall survival than those in the high expression group (). The result of the survival analysis indicated that the enhanced CDC25C expression showed a strong correlation with poor PAAD prognosis, suggesting the potential of CDC25C as a biomarker to predict the prognosis of PAAD.
3.3. Functional enrichment analyses of CDC25C related genes
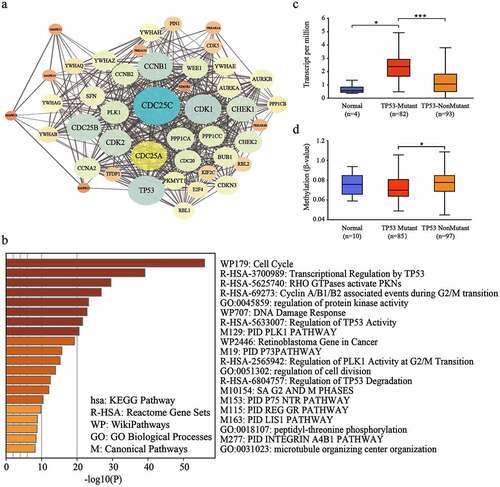
Because of the prognostic value of CDC25C, we want to further probe the biological function of CDC25C in PAAD. PPI network was constructed through correlation analysis, and 50 genes related to CDC25C were obtained from the STRING database ()). Consequently, the enrichment analysis was carried out by a Metascape online database, and the result exhibited that these genes were markedly enriched in cell cycle, TP53 signaling pathways, DNA damage response, protein kinase activity regulation, and immunomodulatory pathways ()). Notably, three TP53-related pathways were enriched, particularly transcriptional regulation by TP53, regulation of TP53 activity, and the regulation of TP53 degradation ()). Additionally, CDC25C was overexpressed in TP53-mutants patients compared with that TP53-nonmutants (p = 1.10e-07) ()). Furthermore, methylation levels were estimated and CDC25C DNA methylation level was discovered to be elevated in patients with TP53-non mutant compared with that in the TP53-mutant ()). These results illustrated that TP53 signaling pathways might have some connection between CDC25C and PAAD.
Figure 4. Gene enrichment analysis of CDC25C.
3.4. CDC25C acts as an immune-related gene in PAAD
We then explored the distribution of 22 immune cells in PAAD and the correlation analysis was displayed in ). The correlation heatmap reflects a higher correlation within the subgroups of the different tumor-infiltrating immune cells ()). To further verify the interaction between CDC25C and the 22 tumor-infiltrating immune cells in PAAD, the relationship between CDC25C and immune cell infiltration was explored. Five types of immune cells exhibited a strong association with the CDC25C expression ()). The result revealed that two cells had a positive relationship with CDC25C, particularly macrophage M0 (R = 0.18, p = 0.039) and follicular helper T cells (R = 0.19, p = 0.028), and three cell types had a negative relationship with CDC25C, particularly naive B cells (R = −0.23, p = 0.0078), monocytes (R = −0.28, p = 0.0011), and CD4+ memory resting T cells (R = −0.17, p = 0.044). Then a differential analysis was performed and discovered that the significantly different immune cells between the high and low CDC25C expression groups were naive B cells, memory B cells, CD8+ T cells, monocytes, and activated dendritic cells (p < 0.001, p = 0.011, p = 0.020, p = 0.012, and p = 0.036, respectively) ()). Intersection analysis revealed that naive B cells and monocytes harbored the strongest correlation with CDC25C expression in PAAD ()).
Figure 5. Immune infiltration signature analysis of PAAD with different CDC25C expressions.
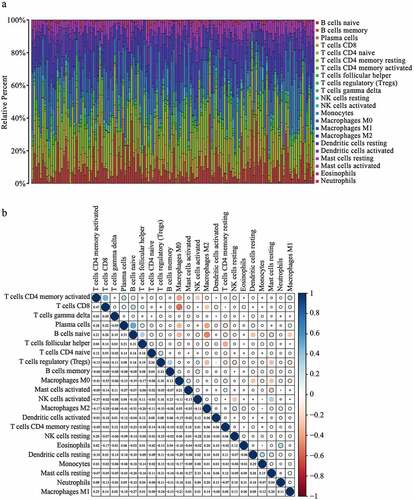
Figure 6. The correlation analysis of CDC25C and immune cells in PAAD.
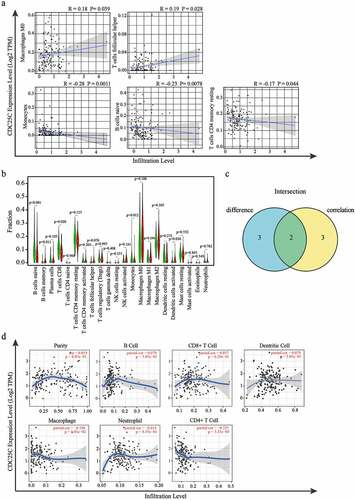
Furthermore, the relationship between CDC25C and immune cells or tumor purity was examined by the TIMER database. The result displayed in ) showed the negative correlation between CDC25C and CD4+ T cells (r = −0.225, p = 3.3e-03) in PAAD. Cumulatively, our data suggested that CDC25C expression significantly influenced the immune activity in PAAD.
3.5. CDC25C promotes the viability and proliferation of PAAD cell lines
Previous studies showed that NSC663284 and NSC95397 could specifically inhibit CDC25 expression [Citation25]. In this study, the inhibition effect of NSC663284 and NSC95397 on PAAD cell lines (SW1990 and BXPC3) and normal pancreatic epithelial cells (HPDE6-C7) were validated. Treatment with NSC663284 and NSC95397 dramatically inhibited CDC25C protein expression ()). Meanwhile, HPDE6-C7 cells exhibited lower CDC25C protein expression compared with that of the SW1990 and BXPC3 cells. This result is similar to our bioinformatics analysis demonstrated that CDC25C had a higher expression in PAAD than normal tissue.
Figure 7. CDC25C promotes the viability of PAAD cell lines.
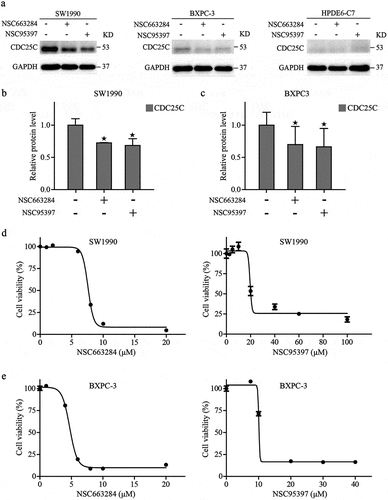
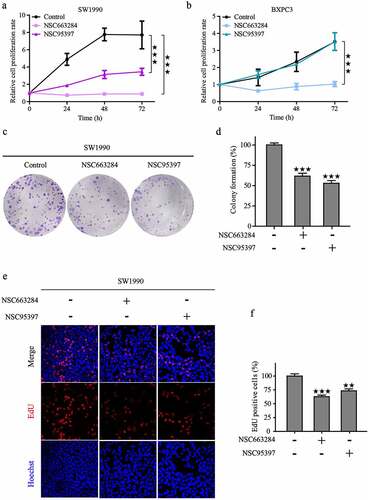
To explore the impact of CDC25C on the viability and proliferation of PAAD cells, SW1990 and BXPC3 cells were incubated with various concentrations of NSC95397 and NSC663284 for 24 h, and the viability of PAAD cells was assessed using CCK8 kit. As displayed in , NSC95397 and NSC663284 inhibited the viability of SW1990 and BXPC3 cells in a dose-dependent manner. Next, the growth and colony-forming ability of PAAD cells under the administration of NSC663284 and NSC95397 were assessed. The results revealed that the proliferative and clonal capacity of PAAD cells was suppressed after the inhibition of CDC25C ()). To further confirm this observation, we also performed an EdU assay to test the effects of the two inhibitors on DNA synthesis and found that the treated cells had a reduced proportion of EdU-positive nuclei compared to the control ()). Collectively, these results demonstrated that CDC25C could promote the viability and proliferation of PAAD cells.
Figure 8. CDC25C promotes the proliferation of PAAD cell lines.
3.6. CDC25C regulates mitochondrial function and ROS generation
Mitochondria play a crucial role in energy metabolism and regulate the processes of cell proliferation and apoptosis. We then monitored mitochondrial homeostasis using DCF-DA and MitoSOX probes to further explore the role of CDC25C in PAAD cells [Citation26]. First, we investigated the production of cellular and mitochondrial ROS after NSC663284 and NSC95397 treatment and the results showed that the fluorescence intensity of DCF-DA and MitoSOX enhanced remarkably ()).
Figure 9. CDC25C regulates mitochondrial function and ROS generation.
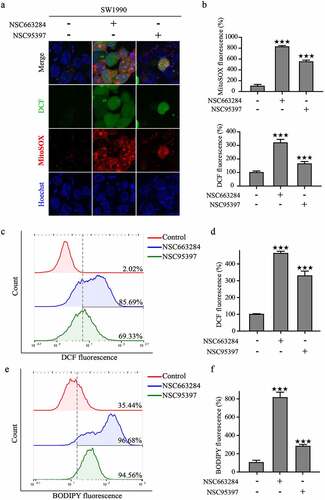
Notably, our recent studies have reported that mitochondrial dysfunction would induce excessive ROS and lipid peroxides [Citation27,Citation28]. Therefore, we then detected the formation of lipid peroxide in SW1990 cells using the fluorescent probe of C11-BODIPY. The inhibition of CDC25C significantly increased lipid peroxide production ()). These findings indicated that CDC25C may participate in the regulation of mitochondrial function.
3.7. CDC25C regulates mitochondrial respiration and glycolysis metabolism.
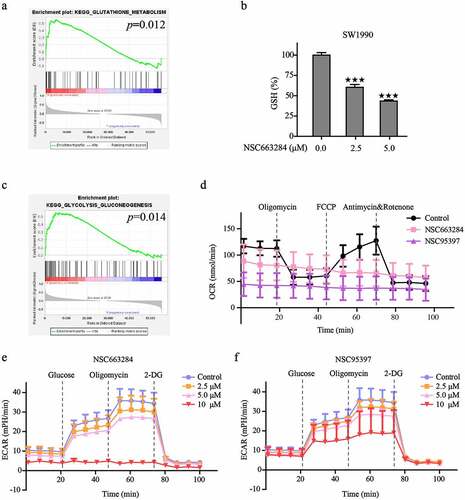
Our previous studies have revealed that the decreased level of glutathione (GSH) was the reason for the excess accumulation of lipid peroxides and oxidative stress [Citation29,Citation30]. Concomitantly, GSEA analysis suggested that CDC25C might enhance the GSH metabolism and activity of glycolysis (p = 0.012, p = 0.014, respectively), which indicated that other important functions of CDC25C may be associated ()). Consequently, the level of GSH was detected and the result showed that CDC25C inhibition could dramatically reduce the GSH level ()). Mitochondrial is the main organelle of ROS production for the electron leakage from the electron transport chain [Citation26,Citation31]. To check whether the increase of oxidative stress and exhaustion of GSH is due to the suppression of CDC25C, OCR and ECAR were analyzed by the Seahorse XFe 24 Bioanalyzer. Results showed that the basal respiration, ATP production, maximal respiration, and coupling efficiency were all substantially suppressed after the inhibition of CDC25C by NSC663284 and NSC95397 () and S1). In addition, CDC25C inhibitors resulted in lower ECAR compared with the control, illustrating that inhibition of CDC25C was capable of blocking mitochondrial oxidative respiratory and glycolysis metabolism. ()). In conclusion, these data suggested that CDC25C exerts a regulatory role in mitochondrial respiration and glycolysis metabolism.
Figure 10. CDC25C regulates mitochondrial respiration and glycolysis metabolism.
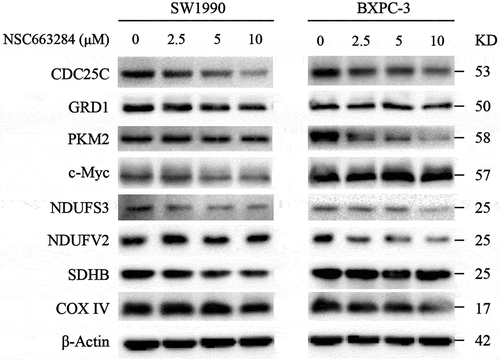
To further investigate the functions of CDC25C in PAAD, the expression level of proteins associated with GSH, glycolysis, and mitochondrion metabolism were evaluated by western blot analysis. CDC25C inhibitors could reduce the expression levels of oxidative respiratory chain (NDUFS3, NDUFV2, SDHB and COX IV), glycolytic regulatory proteins (PKM2 and c-Myc), and glutathione reductase (GRD1) (). Collectively, these findings together with the bioinformatics analysis support the notion that CDC25C plays a potential role in mitochondrial homeostasis and could be a potential biomarker in PAAD.
4. Discussion
PAAD is a highly aggressive type of cancer with a poor prognosis resulting in an average loss in life expectancy of 14.7 years [Citation32]. Due to the nonspecific clinical manifestations and a dearth of reliable biomarkers for the early detection of PAAD, new predictors for its diagnosis and prognosis are urgently needed. This study aimed to analyze the potential application of CDC25C in PAAD using multiple public databases and cellular experiments to obtain a better understanding of the role and effect of CDC25C in PAAD.
CDC25C is a phosphatase with dual specificity. Studies have revealed that CDC25C is a key regulator in cell division, especially in the G2/M phase of cells. In breast cancer cells, CDC25C was involved in the G2/M transition process by over-activating the mitotic target cyclin B-Cdk1 complex based on the recruiting of multiple phosphorylations [Citation33]. Further, the expression of CDC25C can be inhibited by P53, leading to the arrest of the cell cycle in the G2/M phase [Citation34]. Therefore, CDC25C is very promising as a new target for tumor therapy.
To investigate the role of CDC25C in PAAD, we studied the expression levels and prognostic characteristics of CDC25C in PAAD. Bioinformatics data analysis suggested that CDC25C expression is remarkably elevated in PAAD, and high expression of CDC25C is closely associated with the poor prognosis in PAAD patients. In addition, UALCAN online analysis showed the inseparable correlation of CDC25C expression and TP53 mutation in PAAD. This conclusion is consistent with the results reported in the literature, which revealed that different TP53 genotypes can regulate multiple phosphorylation sites of CDC25C [Citation35]. Moreover, in this study, the result of western blot assay also demonstrated that CDC25C expression is enhanced in PAAD cells compared with HPDE6-C7 cells. Concomitantly, our results revealed that when CDC25C expression was suppressed, SW1990 and BXPC3 cell viability and proliferation were decreased.
To identify the specific function of CDC25C in PAAD, we then conducted a pathway enrichment analysis. The result revealed that CDC25C expression was significantly enriched in the cell cycle, TP53 signaling pathways, DNA damage response, regulation of protein kinase activity, immunomodulatory pathways, glutathione metabolism, and glycolysis in PAAD. Tumor occurrence and development are closely associated with the interactions between the tumor and immune cells [Citation36,Citation37]. Immune infiltration analysis showed that CDC25C overexpression can significantly inhibit the number of naive B cells in the tumor. B cells change the direction of the myeloid response through the humoral immune characteristics and significantly affect tumor progression [Citation38]. Since the specific mechanism is not clear, the inhibition of naive B cells by CDC25C may partly expound the poor prognosis of PAAD patients with high CDC25C expression.
The occurrence of PAAD is commonly initiated by the acquisition of activating KRAS mutations, which can lead to mitochondrial dysfunction and increase the level of ROS [Citation39]. KRAS also maintains PAAD cells by inducing glucose uptake and enhancing glycolysis [Citation40]. Notably, ROS predominantly arises from the mitochondria, and excessive ROS accumulation could further contribute to mitochondrial dysfunction. The survival of tumor cells requires a high level of glycolysis [Citation41,Citation42]. In the current study, the inhibition of CDC25C facilitated the accumulation of mitochondrial ROS, cytoplasmic ROS and lipid peroxidation, and caused the downregulation of mitochondrial proteins. These results prove that CDC25C takes part in the modulation of mitochondrial homeostasis. Based on this, the real-time OCR, a sign of mitochondrial stress, was monitored, and our present research revealed that CDC25C inhibition dramatically diminished basal respiration, maximal respiration, and ATP production. Next, we also investigated the glycolytic metabolism using a Seahorse XFe 24 Extracellular Flux Analyzer and discovered that CDC25C inhibition not only reduced the OCR of SW1990 cells, but also inhibited glycolytic capability.
In addition to glycolysis, our GSEA analysis also revealed that CDC25C was related to glutathione metabolism. GSH, as a crucial antioxidant, plays an important role in cellular redox maintenance in tumor cells [Citation43,Citation44]. The GSH level decreased in PAAD cells when treated with CDC25C inhibitors. Similarly, GRD 1 protein was also observed to be reduced. These results reinforce CDC25C’s protective role in PAAD cells by enhancing GSH levels. In light of all results, we speculate that CDC25C may promote energy metabolism by regulating mitochondrial function.
5. Conclusion
In summary, overexpression of CDC25C was correlated with poor prognosis in PAAD. Furthermore, our results demonstrate that CDC25C inhibition disrupted mitochondrial homeostasis with the accumulation of ROS and lipid peroxides, and suppressed glycolysis metabolism. These findings imply that CDC25C may represent a potential biological marker and promising therapeutic target in PAAD.
Authors’ contributions
Yanchun Li and Ying Wang conceived and designed the experiments. Chaoting Zhou, Luyang Wang, Wanye Hu performed the experiments. Lusheng Tang, Ping Zhang, Yan Gao analyzed the data. Yanchun Li, Chaoting Zhou, Luyang Wang wrote the paper. Jing Du and Ying Wang revised and finalized the manuscript. All authors read and approved the final manuscript.
Supplemental Material
Download JPEG Image (238.1 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/21655979.2022.2078940
Additional information
Funding
References
- Siegel RL, Miller KD, Fuchs HE, et al. Cancer statistics, 2021 (vol 71, pg 7, 2021)[J]. Ca-a Cancer J Clinicians. 2021;71(4):359.
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries[J]. Ca-a Cancer J Clinicians. 2018;68(6):394–424.
- Mai CW, Gan LL, Hii LW, et al. Molecular mechanisms and potential therapeutic reversal of pancreatic cancer-induced immune evasion[J]. Cancers (Basel). 2020;12(7):1872.
- Lee B, Lipton L, Cohen J, et al. Circulating tumor DNA as a potential marker of adjuvant chemotherapy benefit following surgery for localized pancreatic cancer[J]. Ann Oncol. 2019;30(9):1472–1478.
- Jin FF, Yang LQ, Wang WX, et al. A novel class of tsRNA signatures as biomarkers for diagnosis and prognosis of pancreatic cancer[J]. Mol Cancer. 2021;20(1):95.
- Khomiak A, Brunner M, Kordes M, et al. Recent discoveries of diagnostic, prognostic and predictive biomarkers for pancreatic cancer[J]. Cancers (Basel). 2020;12(11):3234.
- Hasan S, Jacob R, Manne U, et al. Advances in pancreatic cancer biomarkers[J]. Oncol Rev. 2019;13(1):410.
- Xun R, Lu H, Wang X. Identification of CDC25C as a potential biomarker in hepatocellular carcinoma using bioinformatics analysis[J]. Technol Cancer Res Treat. 2020;19(6):153303382096747.
- Aressy B, Ducommun B. Cell cycle control by the CDC25 phosphatases[J]. Anticancer Agents Med Chem. 2008;8(8):818–824.
- Fei F, Qu J, Liu K, et al. The subcellular location of cyclin B1 and CDC25 associated with the formation of polyploid giant cancer cells and their clinicopathological significance[J]. Lab Invest. 2019;99(4):483–498.
- Liu K, Zheng M, Lu R, et al. The role of CDC25C in cell cycle regulation and clinical cancer therapy: a systematic review[J]. Cancer Cell Int. 2020;20(1):213.
- B-z L, Chen Z-L, Shi -S-S, et al. Overexpression of Cdc25C predicts response to radiotherapy and survival in esophageal squamous cell carcinoma patients treated with radiotherapy followed by surgery[J]. Chin J Cancer. 2013;32(7):403–409.
- Topno R, Nazam N, Kumari P, et al. Integrative genome wide analysis of protein tyrosine phosphatases identifies CDC25C as prognostic and predictive marker for chemoresistance in breast cancer[J]. Cancer Biomarkers. 2021;32(4):491–504.
- Barrett T, Wilhite SE, Ledoux P, et al. NCBI GEO: archive for functional genomics data sets-update[J]. Nucleic Acids Res. 2013;41(D1):D991–D995.
- Edgar R, Domrachev M, Lash AE. Gene expression omnibus: NCBI gene expression and hybridization array data repository[J]. Nucleic Acids Res. 2002;30(1):207–210.
- Chen F, Chandrashekar DS, Varambally S, et al. Pan-cancer molecular subtypes revealed by mass-spectrometry-based proteomic characterization of more than 500 human cancers[J]. Nat Commun. 2019;10(1):5679.
- Chandrashekar DS, Bashel B, Balasubramanya SAH, et al. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses[J]. Neoplasia. 2017;19(8):649–658.
- Tang Z, Kang B, Li C, et al. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis[J]. Nucleic Acids Res. 2019;47(W1):W556–W560.
- Damian S, Gable AL, Nastou KC, et al. The STRING database in 2021: customizable protein–protein networks, and functional characterization of user-uploaded gene/measurement sets[J]. Nucleic Acids Res. 2020 (D1);48:D1.
- Szklarczyk D, Gable AL, Lyon D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets[J]. Nucleic Acids Res. 2019;47(D1):D607–D613.
- Zhou Y, Zhou B, Pache L, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets[J]. Nat Commun. 2019 Apr 3;10(1):1523.
- Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles[J]. Nat Methods. 2015;12(5):453–+.
- Li T, Fan J, Wang B, et al. TIMER: a web server for comprehensive analysis of tumor-infiltrating immune cells[J]. Cancer Res. 2017;77(21):E108–E110.
- Yang C, Li Y, Hu W, et al. TEOA promotes autophagic cell death via ROS-mediated inhibition of mTOR/p70S6k signaling pathway in pancreatic cancer cells[J]. Front Cell Dev Biol. 2021 Oct 6;9:734818.
- Chatterjee N, Sanphui P, Kemeny S, et al. Role and regulation of Cdc25A phosphatase in neuron death induced by NGF deprivation or beta-amyloid[J]. Cell Death Discov. 2016;2:16083.
- Li Y, Wang X, Huang Z, et al. CISD3 inhibition drives cystine-deprivation induced ferroptosis[J]. Cell Death Dis. 2021;12(9):839.
- Du J, Wang T, Li Y, et al. DHA inhibits proliferation and induces ferroptosis of leukemia cells through autophagy dependent degradation of ferritin[J]. Free Radic Biol Med. 2019;131:356–369.
- Shao F, Li Y, Hu W, et al. Downregulation of CISD2 has prognostic value in non-small cell lung cancer and inhibits the tumorigenesis by inducing mitochondrial dysfunction[J]. Front Oncol. 2021;10:595524.
- Du J, Zhou Y, Li Y, et al. Identification of Frataxin as a regulator of ferroptosis[J]. Redox Biol. 2020;32:101483.
- Ren X, Li Y, Zhou Y, et al. Overcoming the compensatory elevation of NRF2 renders hepatocellular carcinoma cells more vulnerable to disulfiram/copper-induced ferroptosis[J]. Redox Biol. 2021;46:102122.
- Li Y, Xia J, Shao F, et al. Sorafenib induces mitochondrial dysfunction and exhibits synergistic effect with cysteine depletion by promoting HCC cells ferroptosis[J]. Biochem Biophys Res Commun. 2021;534:877–884.
- Franck C, Mueller C, Rosania R, et al. Advanced pancreatic ductal adenocarcinoma: moving forward[J]. Cancers (Basel). 2020;12(7):1955.
- C CY, E PJ, C PB, et al. Cell cycle-dependent Cdc25C phosphatase determines cell survival by regulating apoptosis signal-regulating kinase 1[J]. Cell Death Differ. 2015;22(10):1605–1617.
- Giono LE, Resnick-Silverman L, Carvajal LA, et al. Mdm2 promotes Cdc25C protein degradation and delays cell cycle progression through the G2/M phase[J]. Oncogene. 2017;36(49):6762–6773.
- Liu K, Zheng M, Zhao Q, et al. Different p53 genotypes regulating different phosphorylation sites and subcellular location of CDC25C associated with the formation of polyploid giant cancer cells[J]. J Exp Clin Cancer Res. 2020;39(1):83.
- Huang D, Guo W, Gao J, et al. Clinacanthus nutans (Burm. f.) Lindau ethanol extract inhibits hepatoma in mice through upregulation of the immune response[J]. Molecules. 2015;20(9):17405–17428.
- Jiang ZH, Liu ZX, Li MY, et al. Increased glycolysis correlates with elevated immune activity in tumor immune microenvironment[J]. Ebiomedicine. 2019;42:431–442.
- Kotlan B, Horvath S, Eles K, et al. Tumor-associated disialylated glycosphingolipid antigen-revealing antibodies found in melanoma patient’s immunoglobulin repertoire suggest a two-direction regulation mechanism between immune b cells and the tumor[J]. Front Immunol. 2019;10:650.
- Du J, Wang X, Li Y, et al. DHA exhibits synergistic therapeutic efficacy with cisplatin to induce ferroptosis in pancreatic ductal adenocarcinoma via modulation of iron metabolism[J]. Cell Death Dis. 2021;12(7):705.
- Ying H, Kimmelman AC, Lyssiotis CA, et al. Oncogenic kras maintains pancreatic tumors through regulation of anabolic glucose metabolism[J]. Cell. 2012;149(3):656–670.
- Wang P, Deng J, Dong J, et al. TDP-43 induces mitochondrial damage and activates the mitochondrial unfolded protein response[J]. PLoS Genet. 2019;15(5):e1007947.
- Huang S, Zhao J, Song J, et al. Interferon alpha-inducible protein 27 (IFI27) is a prognostic marker for pancreatic cancer based on comprehensive bioinformatics analysis. Bioengineered. 2021;12(1):8515–8528.
- Xia Y, Wang G, Jiang M, et al. A novel biological activity of the STAT3 inhibitor stattic in inhibiting glutathione reductase and suppressing the tumorigenicity of human cervical cancer cells via a ROS-Dependent pathway[J]. Onco Targets Ther. 2021;14:4047–4060.
- Freitas M, Baldeiras I, Proenca T, et al. Oxidative stress adaptation in aggressive prostate cancer may be counteracted by the reduction of glutathione reductase[J]. Febs Open Bio. 2012;2:119–128.