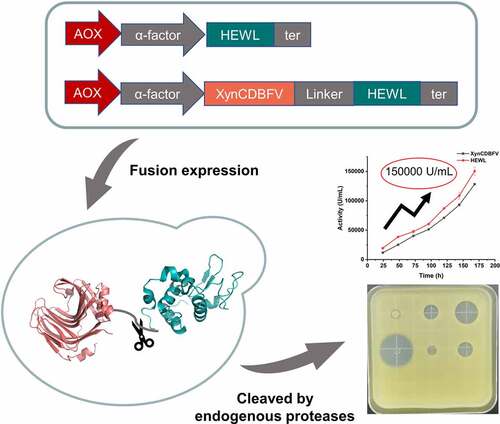
ABSTRACT
Due to its bacteriolytic activity, hen egg white lysozyme (HEWL) is widely used in the feed, food, and pharmaceutical industries. However, its application is hindered by low protein expression levels in microbial expression systems. In this work, a novel fusion protein expression strategy was proposed for increasing the expression level of HEWL. First, HEWL, fused with a highly expressed fusion protein partner xylanase XynCDBFV, is expressed in Pichia pastoris. Secondly, a linker including endogenous protease cleavage sites was introduced between two fusion proteins in order to separate them directly during the secretion process. Finally, the results show that the supernatant of XynCDBFV-HEWL has a higher HEWL expression level and activity compared with HEWL only. It should be noted that the expression of HEWL reaches to about 3.5 g/L, and the activity of HEWL against Micrococcus lysodeikticus reaches to 1.50 × 105 U/mL in a fed-batch fermentation, which is currently the highest level of recombinant expression of an egg white-derived lysozyme. Taken together, we acquired bioactive HEWL for large-scale recombinant production in Pichia pastoris using a novel fusion protein expression strategy, which could then be used for a variety of applications.
Graphical Abstract
1. Introduction
Multi-drug resistance caused by the abuse of antibiotics is a current global issue that should be solved urgently [Citation1,Citation2]. Lysozymes (EC 3.2.1.17) play an important role in the innate immune system to defend against microbial pathogen invasions, as they can lyse bacterial cell walls by destroying the β-1,4-glycosidic bond between the N-acetylmuramic acid and N-acetylglucosamine peptidoglycan, thereby exerting an antibacterial effect [Citation3]. Lysozyme is effective against gram-positive bacteria, and also peptides by protease digestion of lysozyme show broad-spectrum antibacterial effects in vitro [Citation4]. Previous studies show that lysozyme has a significant effect in dealing with lung infections caused by Pseudomonas aeruginosa, broiler necrotic enteritis caused by Clostridium perfringens as well as enterotoxigenic Escherichia coli infection in neonatal piglets [Citation5–8]. Moreover, lysozyme has also anti-viral and anti-tumor effects [Citation9,Citation10]. Hen egg white lysozyme (HEWL) stems from the egg white. As early as 1998, it was already considered safe as a food additive by the United States Food and Drug Administration (FDA), and has also been used for pharmaceutical application [Citation6,Citation11]. Despite its many advantages as an alternative antibiotic, large-scale production is still a challenge. Currently, HEWL is mainly extracted from eggs, which is limited by the lack of raw materials and high purification costs. Therefore, it is necessary to develop a novel strategy for the heterologous production of HEWL in microbial expression systems to cater to market requirements.
HEWL heterologous expression of HEWL has been attempted in E. coli., Aspergillus Niger and yeast expression systems, but low expression levels are still a problem, which limits its application [Citation12–14].
In recent years, fusion expression technology has been reported in several studies to improve the expression of recombinant proteins, Zhan et al. successfully expressed the derivative peptide of antimicrobial peptide LL37 in Pichia pastoris (P. pastoris) by fusion with a small ubiquitin-like modifier (SUMO), and the fusion protein in the supernatant of the fermentation broth reached 154.8 mg/L [Citation15]. In addition, fused with human serum albumin (HSA) can improve the secretion and stability of antimicrobial peptide apidaecin, the expression level of fusion protein HSA-apidaecin could achieve >700 mg/L [Citation16]. Although this technology can improve the expression of some proteins, it also has some problems. For example, fusion proteins usually need to be cleaved with specific proteases in vitro to obtain active target proteins, and it is relatively expensive and time-consuming [Citation15–17].
The aim of this study was to increase the production of recombinant HEWL by fusion expression technology and achieve in vivo cleavage in P. pastoris. We hypothesized that HEWL with the assistance of a highly expressed fusion protein partner (xylanase) may increase the expression amount. Furthermore, the fusion protein can be directly cleaved during the secretion process by introducing endogenous proteases Kex2 and Ste13 cleavage sites [Citation18]. The results of this study provide a new fusion strategy for the expression and application of recombinant HEWL.
2. Materials and methods
2.1. Strains, plasmids, and reagents
E. coli. TOP10 was purchased from Biomed (Beijing, P. R. China) and used as a host for vector construction and amplification. P. pastoris GS115 and the expression vector pPIC9K (Invitrogen, Carlsbad, CA) were used for protein expression. Restriction enzymes, Taq DNA polymerase, and recombinase used for plasmid construction were purchased from TAKARA (Beijing, P. R. China). DNA and protein markers were purchased from Tiangen (Beijing, P. R. China). Micrococcus lysodeikticus (M. lysodeikticus) used for the lysozyme activity assay was stored in our laboratory. Beechwood xylan was purchased from Sigma-Aldrich (St. Louis, MO). All chemicals used in this study were biological reagent grade and commercially available.
2.2. Culture media
Luria-Bertani (LB) medium supplemented with 50 μg/mL kanamycin was used to select positive transformants. The media used to culture P. pastoris included minimal dextrose (MD), yeast extract peptone dextrose (YPD) alone or supplemented with 3.5 mg/L G418, buffered glycerol-complex medium (BMGY), buffered methanol-complex medium (BMMY); fermentation basal salts medium and PTM1 trace salt solution were prepared based on the Pichia Expression Kit (Invitrogen). Solid media were prepared by adding 2% (w/v) agar.
2.3. Codon optimization and expression vector construction
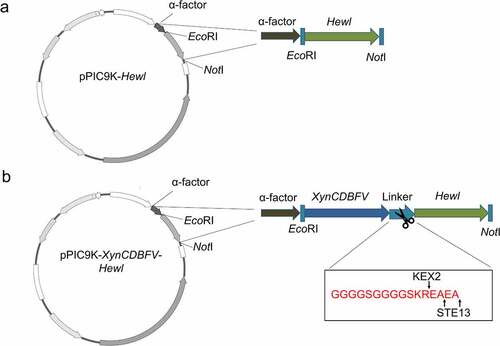
The vectors pPIC9K and pPIC9K-XynCDBFV were maintained in our laboratory. The Hewl gene (GenBank accession number: AAL69327.1) was optimized based on P. pastoris codon usage bias from the codon usage database (http://www. kazusa.or.jp/codon/). The optimized Hewl gene sequence was then synthesized by Biomed Co., Ltd. (P. R. China). The Hewl gene and XynCDBFV-Hewl fusion gene were amplified by specific primers, and then inserted into the EcoRI and NotI sites of pPIC9K (Table S1). The recombinant plasmid was transformed into E. coli Top 10. Positive transformants were further confirmed by PCR amplification with 5-AOX and 3-AOX primers. The sequence was further verified by Sanger sequencing and used for the next transformation.
2.4. P. pastoris transformation and screening
Competent P. pastoris GS115 cells were prepared according to the method described by Invitrogen. The plasmid of pPIC9K-Hewl or pPIC9K-XynCDBFV-Hewl was linearized by BglII (TAKARA, Japan), followed by transformation into P. pastoris GS115 by electroporation [Citation19]. Positive transformants were screened using MD plates without histidine, and high copy transformants were then screened by YPD plates containing 3.5 g/L geneticin G418 [Citation20].
2.5. Strain cultivation in shaking flasks
The shake flask fermentation was carried out according to the method described by He et al. [Citation21]. The recombinant P. pastoris GS115 strains were inoculated into 300 mL of BMGY media and cultivated for 36 h at 30°C and 220 rpm. When the OD600 value of the cells reached 2–6, they were harvested by centrifugation at 4500 rpm for 5 min at 25°C, and resuspended in 200 mL of BMMY media with 1% (v/v) methanol. Subsequently, 0.5% absolute methanol was added to the medium every 12 h. The cultivated supernatants were collected every 24 h for the antimicrobial activity assay and sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) analysis.
2.6. Assay of xylanase activity
The xylanase activity was assayed by determining the release of reducing sugars from beechwood xylan using the dinitrosalicylic acid (DNS) method [Citation22]: 900 μL of 1% beech xylan substrate was added to the test tube at pH 5.5 and placed in a 55-°C water bath for pre-heating for 5 min, 100 μL of appropriately diluted enzyme solution was added, and the mixture kept in 55°C warm water. After 10 min, 1.5 mL of DNS was added to stop the reaction, and the mixture was then boiled for 5 min. After cooling to room temperature, the absorbance was measured at 540 nm wavelength with a micro plate reader. One unit (U) of enzyme activity is defined as the amount of enzyme that released 1 μM of reducing sugars per minute under the given conditions.
2.7. Muramidase activity assay and kinetic analysis.
The bacteriolytic activity of the recombinant HEWL against M. lysodeikticus was determined according to the standard spectrophotometry method [Citation23]. 100 μL fermentation supernatant was mixed with 2.9 mL cell suspension with OD450 at 0.7 ± 0.1 in 100 mM phosphate buffer. A decrease in the OD value at 450 nm was recorded every 15 s for 3 min, and the difference between 1 and 3 min was used to calculate the antibacterial activity of HEWL. The amount of enzyme decreasing the OD at 450 nm by 0.001 per minute was defined as one U of enzyme activity [Citation24].
The maximal velocity (Vmax) and the Michaelis constant (Km) for the purified HEWL and XynCDBDV-HEWL were determined in a 0.1 M phosphate buffer at pH 6.2 containing 0.3–0.7 OD450/mL M. lysodeikticus cell suspension under standard conditions. All experimental data were replicated three times. Michaelis−Menten equation was used for the experimental data calculation. The catalytic efficiency (kcat/Km) was also calculated.
2.8. Zone of inhibition test
The Oxford cup method was used to assess antibacterial activity [Citation25]. M. lysodeikticus was inoculated into an LB medium and incubated overnight. The next day, the bacterium solution was 100 times diluted to then heat the LB medium with 2% agar. After cooling to 40–50°C, the medium was mixed well and spread into sterile petri dishes. Then, the Oxford cups were placed on the surface of the solid LB medium mixed with 1% M. lysodeikticus, and 200 μL fermentation supernatants or their dilutions of different transformants were added into the holes of Oxford cups, 40 ug/mL ampicillin was used as positive control. After 24 h incubation at 37°C, the inhibition zone in solid plates was observed and recorded.
2.9. Fed-batch fermentation
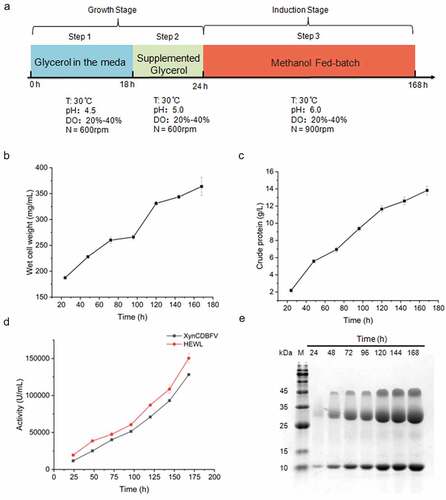
The high-density fermentation of P. pastoris was performed with a 15 L fermenter (EastBio Co. Ltd., Jiangsu, P. R. China). A single clone of the chosen transformant was inoculated into 50 mL YPD shake flasks, grown for 24 h at 30°C in a shaker at 220 rpm, and then transferred to a 600 mL YPD medium and cultured under the same conditions for 12 h. The cell culture was transferred into the fermenter once the OD600 reached 5–6. High-density fermentation was then carried out according to the standard procedures [Citation26]. In brief, the culture was used to inoculate a 15-L fermenter containing 7.5 L basal salt medium (0.93 g/L CaSO4, 18.20 g/L K2SO4, 14.90 g/L MgSO4 · 7H2O, 4.13 g/L KOH, 40.00 g/L glycerol, 26.70 mL/L H3PO4, and 4.35 mL/L. PTM1 trace salt solution, which consists of 6.00 g/L CuSO4 · 5H2O, 0.08 g/L NaI, 3.00 g/L MnSO4·H2O, 0.20 g/L Na2MoO4 · 2H2O, 0.02 g/L H3BO3, 0.50 g/L CoCl2, 20.00 g/L ZnCl2, 65.00 g/L FeSO4 · 7H2O, 0.20 g/L biotin, and 5 mL H2SO4 per liter), the temperature was controlled at 30°C, dissolved oxygen (DO) was controlled at 20–40% by adjusting the rotational speed, the whole process was adjusted with ammonia water to stabilize the pH between 4.5 and 6.0, and the final volume is about 10 L. During cultivation, the samples were collected every 24 h and analyzed for their wet cell weight, and the supernatant was used to detect the expression level of the fusion protein.
2.10. Purification of fusion protein
The clarified supernatant after fermentation was obtained by centrifugation, and the buffer was exchanged to 20 mM citrate phosphate buffer at pH 6.0 using hollow-fiber ultrafiltration. Size-exclusion chromatography was then performed using a Superdex-75 pg column (GE Healthcare) [Citation27]. The collected fractions were confirmed by SDS-PAGE (12% acrylamide).
2.11. Protein identification
The protein bands were extracted from the gel for protein identification using a Q Exactive HF system coupled to an Easy-nLC 1000 (Thermo Fisher Scientific, Waltham, MA, USA) via a nanoelectrospray ion source. To identify the protein bands, the MS/MS data were searched against an expected protein sequence using the in-house PEAKS Studio software (version 10.5, Bioinformatics Solutions Inc, Waterloo, ON, Canada) [Citation28].
3. Results
The aim of this study was to increase the expression of HEWL in P. pastoris. We hypothesized that HWEL fusion with highly expressed xylanase protein could increase the expression amount in P. pastoris. Experimental results indicate that fusion with xylanase can increase the expression of HEWL in P. pastoris by comparing the expression levels of HEWL expressed alone and fused with xylanase. The expression level of HEWL through high-density fermentation reached 1.50 ± 0.07 × 105 U/mL in the supernatant, which is the highest level reported so far.
3.1. Construction of expression plasmids of pPIC9K-Hewl and pPIC9K-XynCDBDV-Hewl in P. pastoris
To enhance the expression level of HEWL, the nucleotide sequence of the Hewl gene was optimized based on P. pastoris codon usage bias. After optimization, the GC content of the Hewl gene was reduced from 57.6% to 47.6%, and the codon adaptation index of the original Hewl was improved from 0.57 to 0.91, which was more conducive to the expression of recombinant protein in P. pastoris (Fig. S1). For XynCDBFV-Hewl, the gene length of XynCDBFV and Hewl was 675 bp and 387 bp, encoding 225 and 129 amino acids, with predicted molecular weights of 24.4 kDa and 14.3 kDa, respectively. In addition, a flexible linker GGGGSGGGGS and the Kex2 and Ste13 cleavage sites (KREAEA) were designed between XynCDBFV and HEWL to ensure correct folding and separation. The schematic details of the two plasmids are shown in ().
3.2. HEWL expression level and activity
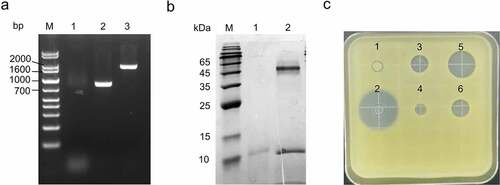
The expression plasmids encoding HEWL (pPIC9K-Hewl) and fusion protein XynCDBFV-HEWL (pPIC9K-XynCDBFV-Hewl) were linearized and individually transformed into P. pastoris GS115. Subsequently, the positive transformants were screened. Then, the clone with the highest expression level for HEWL was chosen by SDS-PAGE and zone of inhibition tests. The integration of the recombinant plasmid into the two chosen positive transformants was confirmed by PCR ()). To gain further insights into the difference in the expression level and activity between HEWL and XynCDBFV-HEWL, the selected two strains were verified in shaking flasks. After the induction of methanol for 3 days, one weak band was detected at the expected size for HEWL in the supernatant of pPIC9K-Hewl transformant, while two bands, XynCDBFV and HEWL were detected in the supernatant of pPIC9K-XynCDBFV- Hewl transformant, indicating that XynCDBFV and HEWL were expressed and separated as expected ()). Unsurprisingly, the apparent molecular mass of XynCDBFV was much larger than the predicted 24.4 kDa as shown in (), the expression level of HEWL in pPIC9K-XynCDBFV-Hewl transformant was much higher than in pPIC9K-Hewl transformant, indicating that the fusion of XynCDBFV improved the expression of HEWL in P. pastoris. The lytic activity against M. lysodeikticus of the culture supernatant in both transformants was also tested with the zone of inhibition method. The results showed that a much larger zone of inhibition was formed in pPIC9K-XynCDBFV-Hewl transformant compared within pPIC9K-Hewl transformant. Moreover, a ten times dilution of the supernatant in pPIC9K- XynCDBFV-Hewl transformant showed an almost similar size of the zone of inhibition as that in pPIC9K-Hewl transformant, while the supernatant in XynCDBFV transformant did not show antibacterial activity ()). Taken together, HEWL fused with XynCDBFV showed a higher expression level and activity than HEWL only.
Figure 2. Hen egg white lysozyme (HEWL) fused to xylanase XynCDBFV shows higher expression level and activity than HEWL only. (a) The integration of plasmids was identified by PCR amplification in genome of transformants. M: DNA marker; 1: GS115 control; 2: pPIC9K-Hewl transformant; 3: pPIC9K-XynCDBFV-Hewl transformant. (b) SDS-PAGE assay of the recombinant HEWL only and XynCDBFV-HEWL proteins in flask. M: protein marker; 1: HEWL only; 2: XynCDBFV-HEWL. (c) Antibacterial activity against M. lysodeikticus of the culture supernatant in different transformants. 1: XynCDBFV; 2: Positive control Ampicillin; 3: HEWL only; 4: ten times dilution of HEWL only; 5: XynCDBFV-HEWL; 6: ten times dilution of XynCDBFV-HEWL.
3.3. Heterologous production of XynCDBFV-HEWL in large-scale fermentations
The high-density fermentation in a 15-L fermenter was attempted to produce HEWL in large quantity. The fermentation process mainly consists of three stages: glycerol batch phase, glycerol fed-batch phase, and methanol feed phase ()). The glycerol fed-batch phase begins when DO increases rapidly. The feed medium contained 50% glycerol and 12 mL/L PTM1 solution. The glycerol feed rate was 18 mL/h/L for an additional 6 h until the culture wet weight reached 187.30 ± 1.53 g/L. During the fermentation process, the temperature was maintained at 30°C, DO was maintained at 20–40% by changing the rotation speed, and the pH was maintained at 5.0.
The yeast cells accumulated continuously during the methanol induction phase, and the wet cell weight reached 363.67 ± 17.39 g/L after 168 h of induction ()). Secreted crude proteins also began to accumulate after methanol induction, increasing from 2.18 ± 0.19 g/L at the beginning to 13.82 ± 0.51 g/L at the end of fermentation ()). The activity of lysozyme and xylanase in the fermentation broth increased over time, reaching 1.50 ± 0.07 × 105 U/mL for the final lysozyme activity and 1.28 ± 0.01 × 105 U/mL for the final xylanase activity ()). Using SDS-PAGE gel, the accumulation of XynCDBFV and HEWL was also observed (). Unexpectedly, a band of approximately 30 kDa was detected compared with the SDS-PAGE analysis results of the previous shaking flasks culture experiment (), which may have been caused by different degrees of glycoslation of XynCDBFV under different culture conditions. Based on the SDS-PAGE analysis and the concentration of crude proteins, 13.82 ± 0.51 g/L at the end of fermentation (), approximately 3.5 g/L HEWL was produced after 168 h of induction. These results indicate that the proteins XynCDBFV and HEWL could be produced separately in large quantity using our fusion strategy.
3.4. Purification and identification of proteins in the culture supernatant
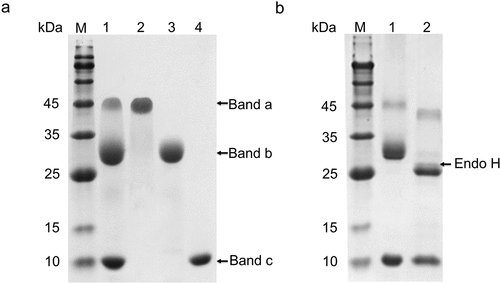
The SDS-PAGE analysis result showed three main bands in the fermentation culture supernatant (), with a molecular weight of approximately 45 kDa (band a), 30 kDa (band b), and 10 kDa (band c). In contrast, there were only two main bands of approximately 45 kDa and 10 kDa in the supernatant of the flask culture ()). To analyze the protein character, the proteins in the fermentation supernatant were purified by a Superdex-75 pg column. The samples of the three elution peaks were analyzed by SDS-PAGE, and the data showed that the three proteins had been separated ()).
Figure 4. Purification and identification of proteins in the culture supernatant. (a) SDS-PAGE assay of the fermentation supernatant and purified proteins. Lane M: molecular mass marker; Lane 1 shows the protein in the fermentation supernatant; Lane 2: band a; Lane 3: band b; Lane 4: band c; (b) De-glycosylation of the protein in the fermentation supernatant using Endo H. Lane M: molecular mass marker; Lane 1 shows the protein in the fermentation supernatant; Lane 2: the protein in the fermentation supernatant treated with Endo H.
The kinetics parameters results were shown in (). XynCDBFV-HEWL and HEWL showed a specific activity of 27,884 ± 192 and 24,807 ± 175 U/mg, respectively, similar Vmax (49,749 ± 8353 vs. 49,100 ± 9564 mg/mL·min) and kcat (11,939 ± 2004 vs. 11,784 ± 2295 min−1) values, but different Km values (0.70 ± 0.02 vs. 0.99 ± 0.06 mg/mL).
Table 1. Kinetics parameters of HEWL and XynCDBFV-HEWL*.
To determine what is represented by protein bands a, b, and c, the activity of the three proteins was measured. Both band a and b showed only xylanase activity, while band c showed lysozyme activity only ()). Furthermore, the three bands extracted from the SDS-PAGE gel were analyzed by LC-MS/MS. The data demonstrate that bands a and b were XynCDBFV, and band c was HEWL (Fig. S2). The values of both bands a and b were larger than the expected molecular weight of XynCDBFV, 24.4 kDa, indicating that they may have been glycosylated. To verify the hypothesis, the culture supernatant was incubated with Endo H to remove the N-glycosylation, and then analyzed by SDS-PAGE. The data showed that the values of both bands a and b decreased, indicating that both bands had been N-glycosylated ()). However, there were still three bands in the supernatant after removing N-glycosylation, suggesting that there may have been other forms of glycosylation, such as O-glycosylation ()). These results indicate that both XynCDBFV and HEWL were released and separated efficiently in the supernatant, and that XynCDBFV has been glycosylated to different degrees.
4. Discussion
HEWL is a potential alternative to antibiotics: however, it is difficult to express in microbial expression systems in large quantity, which greatly limits its application [Citation29]. To date, heterologous expression of HEWL has been attempted in microbial expression systems, but low expression levels are still a problem and an issue to be solved. For example, the secretory expression production of HEWL under the control of promoter GAP in P. pastoris reached 54 mg/L with a bacteriolytic activity of 1,680 U/mL in a 50-L fermenter [Citation13]. In Aspergillus Niger, under the control of the Aspergillus awamori glucoamylase promoter, the expression level of HEWL reached 12 mg/L [Citation12]. E. coli lacks the post-translational modification system of eukaryotes, and it is difficult to form the correct folded conformation, which causes the formation of inclusion bodies [Citation30]. Studies have reported that the expression level of soluble lysozyme in E. coli is only 10 ng/mL [Citation14].
The methylotrophic yeast P. pastoris has been successfully employed for heterologous protein expression in fundamental research, biotechnology, and the biopharmaceutical industry because of its high product yields and convenient purification process [Citation31]. With the alcohol oxidase 1 (AOX1) promoter and the α-factor signal sequence, P. pastoris was able to produce high levels of recombinant proteins, such as insulin, chitinase, or vaccines [Citation32–34].
Xylanase has a wide range of sources in nature, mainly from bacteria, fungi, actinomycetes, protozoa, and marine algae [Citation35]. To date, many recombinant xylanases have been stably and efficiently expressed in P. pastoris [Citation36]. XynCDBFV, a thermally stable and alkalophilic xylanase, has been identified in the ruminal fungus Neocallimastix patriciarum, is considered one of the most efficient xylanases, and has been recombinantly expressed with high expression levels in P. pastoris [Citation37,Citation38]. Recombinant production of the fusion protein XynCDBFV-somatostatin has been achieved in P. pastoris, and somatostatin immunogenicity and xylanase activity have been well retained [Citation39].
In this study, bioactive HEWL was produced in high quantity by fusing it to xylanase XynCDBFV compared with HEWL only. HEWL with the assistance of a fusion partner (xylanase) showed a markedly higher protein expression and activity, reaching 3.5 g/L and 1.50 × 105 U/mL, respectively, which is the highest expression level of HEWL reported so far, indicating that xylanase greatly improves HEWL expression.
Protein fusion technology plays an important role in the efficient production and purification of recombinant proteins [Citation40]. A large number of fusion tags, such as SUMO, maltose-binding protein, glutathione S-transferase, have been used to solve the problems related to the expression, stability, solubility, folding, and purification of their fusion partners [Citation41–43]. To improve the low yield expression of HEWL, the fusion protein strategy was conducted to enhance the expression level of HEWL in P. pastoris. Considering that the recombinant expression of xylanase XynCDBFV reached a very high level in P. pastoris, even up to approximately 50 g/L in our lab, xylanase XynCDBFV was consequently chosen as a fuse partner for assisting a high HEWL expression. As shown by the SDS-PAGE and muramidase activity assay results, xylanase XynCDBFV greatly promoted the expression level of HEWL in P. pastoris. In previous reports, fusion proteins usually improved the expression of recombinant proteins by helping the target protein to fold correctly and avoid being degraded by proteases [Citation44]. However, the mechanism of how xylanase XynCDBFV enhances the expression level of HEWL is unclear, which is an interesting topic warranting future investigation.
Usually, specific enzyme cleavage sites, such as variants of tobacco etch virus protease or enterokinase, are introduced between two fusion proteins to separate them in vitro [Citation45]. However, these specific enzymes increase the production costs and make downstream processing procedures more cumbersome [Citation41–45]. Both Kex2 and Ste13 are endogenously expressed in yeast, and it has been reported that the antimicrobial peptide CGA-N12 can be successfully expressed in P. pastoris by designing an endogenous protease cleavage site between the fusion proteins without in vitro cleavage treatment [Citation46]. The cleavage sites (KR and EAEA) of Kex2 and Ste13 were designed between HEWL and XynCDBFV following a flexible linker in the current experiment. It is supposed that the fusion protein is directly cleaved by the endogenous proteases after translation, which allows them to work separately. As indicated by the results, the fusion protein XynCDBFV-HEWL was cleaved efficiently by Kex2 and Ste13. However, a noteworthy phenomenon ()) indicated that the level of HEWL was about three times lower than that of XynCDBFV in the supernatant, indicating that most of HEWL did not secret to the supernatant successfully or was degraded. This may be related to the mechanism of unfolded protein response [Citation47,Citation48]; With the increase in protein secretion, more misfolded proteins are degraded by proteasome [Citation49]. Further research is needed to increase the level of HEWL in the supernatant.
Xylanase was used as a fusion protein to increase the HEWL expression. Moreover, xylanase is an effective feed additive in livestock feeds, which can improve animal growth performance and feed utilization [Citation50,Citation51]. It is also well known for its pleiotropic effects, including reduction of chyme viscosity, facilitation of nutrient release, and generation of probiotic xylo-oligosaccharides to modulate the gut flora [Citation52,Citation53]. Furthermore, lysozyme is also an effective feed additive in livestock feeds [Citation54], which can improve the gastrointestinal health of pigs, improve the metabolic status, and change the bacterial ecology of the gastrointestinal tract of pigs [Citation55]. In this study, both lysozyme and xylanase were obtained in the fermentation supernatant, which can directly be used in livestock feeds to solve growth promotion and disease prevention in the breeding industry.
Conclusion
In conclusion, through fusion with xylanase, codon optimization, introducing endogenous protease cleavage sites between fusion proteins, screening of high-copy transformants, and high-density fermentation, the final expression level of HEWL reached 1.50 × 105 U/mL in P. pastoris GS115, which is currently the highest level. The fusion expression strategy has the potential to improve other difficult-to-express or lowly expressed protein productivity.
Author contributions
Conceived and designed the experiments: Yuan Wang, Huoqing Huang. Performed the experiments: Lin Cui, Huoqing Huang. Analyzed the data: Xiaolu Wang, Xing Qin, Tao Tu. Contributed reagents/materials/analysis tools: Jie Zhang, XiaoyunSu, Huimin Yu, Huiying Luo, Bin Yao. Wrote the paper: Lin Cui, Honglian Zhang, Yuan Wang.
Supplemental Material
Download Zip (2.5 MB)Disclosure statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/21655979.2022.2084496
Additional information
Funding
References
- Ianiro G, Tilg H, Gasbarrini A. Antibiotics as deep modulators of gut microbiota: between good and evil. Gut. 2016;65(11):1906–1915.
- Ramirez J, Guarner F, Fernandez LB, et al. Antibiotics as major disruptors of gut microbiota. Front Cell Infect Microbiol. 2020;10:572912.
- Ragland SA, Criss AK. From bacterial killing to immune modulation: recent insights into the functions of lysozyme. PLoS Pathog. 2017 ;13:e1006512.
- Derde M, Lechevalier V, Guerin-Dubiard C, et al. Hen egg white lysozyme permeabilizes Escherichia coli outer and inner membranes. J Agric Food Chem. 2013;61(41):9922–9929.
- Griswold KE, Bement JL, Teneback CC, et al. Bioengineered lysozyme in combination therapies for Pseudomonas aeruginosa lung infections. Bioengineered. 2014;5(2):143–147.
- Sava G. Pharmacological aspects and therapeutic applications of lysozymes. EXS. 1996;75:433–449.
- Zhang G, Darius S, Smith SR, et al. In vitro inhibitory effect of hen egg white lysozyme on Clostridium perfringens type A associated with broiler necrotic enteritis and its alpha-toxin production. Lett Appl Microbiol. 2006;42(2):138–143.
- Huang G, Li X, Lu D, et al. Lysozyme improves gut performance and protects against enterotoxigenic Escherichia coli infection in neonatal piglets. Vet Res. 2018 20;49(1):20.
- Khan MI, Dowarha D, Katte R, et al. Lysozyme as the anti-proliferative agent to block the interaction between S100A6 and the RAGE V domain. PloS One. 2019;14(5):e0216427.
- Mahanta S, Paul S, Srivastava A, et al. Stable self-assembled nanostructured hen egg white lysozyme exhibits strong anti-proliferative activity against breast cancer cells. Colloids Surf B Biointerfaces. 2015;130:237–245.
- Wilken LR, Nikolov ZL. Process evaluations and economic analyses of recombinant human lysozyme and hen egg-white lysozyme purifications. Biotechnol Prog. 2011;27(3):733–743.
- Archer DB, Jeenes DJ, MacKenzie DA, et al. Hen egg white lysozyme expressed in and secreted from, aspergillus niger is correctly processed and folded. Nat Biotechnol. 1990;8(8):741–745.
- Jun L. Codon optimization, constitutive expression and antimicrobial characterization of hen egg white lysozyme (HEWL) in Pichia pastoris. Afr J Biotechnol. 2012;11(56):11887–11893.
- Fischer B, Perry B, Phillips G , et al. Physiological consequence of expression of soluble and active hen egg white lysozyme in Escherichia coli. Escherichia Appl Microbiol Biotechnol. 1993;39:537–540.
- Zhan N, Zhang L, Yang H, et al. Design and heterologous expression of a novel dimeric LL37 variant in Pichia pastoris. Microb Cell Fac. 2021;20(1):143
- Cao J, de la Fuente-Nunez C, Ou RW, et al. Yeast-based synthetic biology platform for antimicrobial peptide production. ACS Synth Biol. 2018;7(3):896–902.
- Wibowo D, Zhao CX. Recent achievements and perspectives for large-scale recombinant production of antimicrobial peptides. Appl Microbiol Biotechnol. 2019;103(2):659–671.
- Bourbonnais Y, Bolin D, Shields D. Secretion of somatostatin by Saccharomyces cerevisiae. Correct proteolytic processing of pro-alpha-factor- somatostatin hybrids requires the products of the KEX2 and STE13 genes. J Biol Chem. 1988;263(30):15342–15347.
- Xia W, Bai Y, Cui Y, et al. Functional diversity of family 3 β-glucosidases from thermophilic cellulolytic fungus Humicola insolens Y1. Sci Rep. 2016 Jun 8; 6(1):27062.
- Scorer CA, Clare JJ, McCombie WR, et al. Rapid selection using G418 of high copy number transformants of Pichia pastoris for high-level foreign gene expression. Nat Biotechnol. 1994;12(2):181–184.
- He H, Wu S, Mei M, et al. A combinational strategy for effective heterologous production of functional human lysozyme in Pichia pastoris. Front Bioeng Biotechnol. 2020;10(8):118.
- Miller GL. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem. 1959;31(3):426–428.
- Le Snierowski G, Kijowski J. Methods for testing enzymatic activity and quantification of lysozyme from chicken egg protein. Przem Spoz. 1995;12:476–479.
- Huang P, Sun Q, Shi W, et al. Efficient production of human goose-type lysozyme 2 in the methylotrophic yeast Pichia pastoris. J Biotechnol. 2018;275:44–52.
- Guo HN, Tong YC, Wang HL, et al. Novel hybrid peptide cathelicidin 2 (1-13)- Thymopentin (TP5) and its derived peptides with effective antibacterial, antibiofilm, and anti-adhesion activities. Int J Mol Sci. 2021;22(21):11681.
- Liu WC, Inwood S, Gong T, et al. Fed-batch high-cell-density fermentation strategies for Pichia pastoris growth and production. Crit Rev Biotechnol. 2019;39(2):258–271.
- Oroz J, Chang BJ, Wysoczanski P, et al. Structure and pro-toxic mechanism of the human Hsp90/PPIase/Tau complex. Nat Commun. 2018 31;9(1):4532.
- Huo X, Wu B, Feng M, et al. Proteomic analysis reveals the molecular underpinnings of mandibular gland development and lipid metabolism in two lines of honeybees (Apis mellifera ligustica. J Proteome Res. 2016;15(9):3342–3357.
- Ferraboschi P, Ciceri S, Grisenti P. Applications of lysozyme, an innate immune defense factor, as an alternative antibiotic. Antibiotics. 2021;10(12):1534.
- Chen R. Bacterial expression systems for recombinant protein production: e. coli and beyond. Biotechnol Adv. 2012;30(5):1102–1107.
- Karbalaei M, Rezaee SA, Farsiani H. Pichia pastoris: a highly successful expression system for optimal synthesis of heterologous proteins. J Cell Physiol. 2020 Sep;235(9):5867–5881.
- Hazra P, Sreenivas S, Venkatesan K, et al. A novel peptide design aids in the expression and its simplified process of manufacturing of insulin glargine in Pichia pastoris. Appl Microbiol Biotechnol. 2021;105(8):3061–3074.
- Song W, Zhang N, Yang M, et al. Multiple strategies to improve the yield of chitinase a from Bacillus licheniformis in Pichia pastoris to obtain plant growth enhancer and GlcNAc. Microb Cell Fact. 2020;19(1):181.
- Wang M, Jiang S, Wang Y. Recent advances in the production of recombinant subunit vaccines in Pichia pastoris. Bioengineered. 2016;7(3):155–165.
- Ishida T, Parks JM, Smith JC. Insight into the catalytic mechanism of GH11 xylanase: computational analysis of substrate distortion based on a neutron structure. J Am Chem Soc. 2020;142(42):17966–17980.
- Chadha BS, Kaur B, Basotra N, et al. Thermostable xylanases from thermophilic fungi and bacteria: current perspective. Bioresour Technol. 2019;277:195–203.
- Yu J, Liu X, Guan L, et al. High-level expression and enzymatic properties of a novel thermostable xylanase with high arabinoxylan degradation ability from Chaetomium sp. suitable for beer mashing. Int J Biol Macromol. 2021;168:223–232.
- Han N, Miao H, Ding J, et al. Improving the thermostability of a fungal GH11 xylanase via site-directed mutagenesis guided by sequence and structural analysis. Biotechnol Biofuels. 2017;10(1):133.
- Huang K, Chu Y, Qin X, et al. Recombinant production of two xylanase-somatostatin fusion proteins retaining somatostatin immunogenicity and xylanase activity in Pichia pastoris. Appl Microbiol Biotechnol. 2021;105(10):4167–4175.
- Ki MR, Pack SP. Fusion tags to enhance heterologous protein expression. Appl Microbiol Biotechnol. 2020;104(6):2411–2425.
- Park AR, Kim SW, Kim SY, et al. Expression of antimicrobial peptide (AMP), cecropin B, in a fused form to SUMO tag with or without three-glycine linker in Escherichia coli and evaluation of bacteriolytic activity of the purified AMP. Probiotics Antimicro. 2021;13(6):1780–1789
- Xu SY, Fomenkov A, Chen TH, et al. Expression of human ACE2 N-terminal domain, part of the receptor for SARS-CoV-2, in fusion with maltose-binding protein, E. coli ribonuclease I and human RNase A. Front Microbiol. 2021;12:660149.
- Zhou LX, Liu ZY, Xu GY, et al. Expression of melittin in fusion with GST in Escherichia coli and its purification as a pure peptide with good bacteriostatic efficacy. Acs Omega. 2020;5(16):9251–9258.
- Esposito D, Chatterjee DK. Enhancement of soluble protein expression through the use of fusion tags. Curr Opin Biotechnol. 2006;17(4):353–358.
- Sequeira AF, Turchetto J, Saez NJ, et al. Gene design, fusion technology and TEV cleavage conditions influence the purification of oxidized disulphide-rich venom peptides in Escherichia coli. Microb Cell Fact. 2017;16(1):4.
- Li X, Fan Y, Lin Q, et al. Expression of chromogranin A-derived antifungal peptide CGA-N12 in Pichia pastoris. Bioengineered. 2020;11(1):318–327.
- Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011;334(6059):1081–1086.
- Huang J, Wang Q, Bu W, et al. Different construction strategies affected on the physiology of Pichia pastoris strains highly expressed lipase by transcriptional analysis of key genes. Bioengineered. 2019;10(1):150–161.
- Smith MH, Ploegh HL, Weissman JS. Road to ruin: targeting proteins for degradation in the endoplasmic reticulum. Science. 2011;334(6059):1086–1090.
- Li X, Dilokpimol A, Kabel MA, et al. Fungal xylanolytic enzymes: diversity and applications. Bioresour Technol. 2022;344:126290.
- De Maesschalck C, Eeckhaut V, Maertens L, et al. Effects of xylo-oligosaccharides on broiler chicken performance and microbiota. Appl Environ Microbiol. 2015;81(17):5880–5888.
- Ding XM, Li DD, Bai SP, et al. Effect of dietary xylooligosaccharides on intestinal characteristics, gut microbiota, cecal short-chain fatty acids, and plasma immune parameters of laying hens. Poult Sci. 2018;97(3):874–881.
- Su X, Yao B. Exploiting enzymes as a powerful tool to modulate the gut microbiota. Trends Microbiol. 2022;30(4):314–317.
- El-Deep MH, Amber KA, Eid YZ, et al. The influence of dietary chicken egg lysozyme on the growth performance, blood health, and resistance against Escherichia coli in the growing rabbits’ cecum. Front Vet Sci. 2020;7:579576.
- Oliver WT, Wells JE. Lysozyme as an alternative to growth promoting antibiotics in swine production. J Anim Sci Biotechnol. 2015;6(1):35.