ABSTRACT
Tight junctions form a barrier to the diffusion of apical and basolateral membrane proteins thus regulating membrane polarity. They also regulate the paracellular movement of ions and water across epithelial and endothelial cells so that functionally they constitute an important permselective barrier. Permselectivity at tight junctions is regulated by claudins, which confer anion or cation permeability, and tightness or leakiness, by forming several highly regulated pores within the apical tight junction complex. One interesting feature of claudins is that they are, more often than not, localized to the basolateral membrane, in intracellular cytoplasmic vesicles, or in the nucleus rather than to the apical tight junction complex. These intracellular pools of claudin molecules likely serve important functions in the epithelium. This review will address the widespread prevalence of claudins that are not associated with the apical tight junction complex and discuss the important and emerging non-traditional functions of these molecules in health and disease.
KEYWORDS:
Introduction
Tight junctions have a conserved 3-D structure of which our current knowledge has advanced considerably as described in numerous recent reviews.Citation1-4 Tight junctions were initially described from electron micrographs as the fusion of lateral membranes from the most apical part of adjacent cells with no intercellular space present over a variable distance.Citation5 By freeze-fracture replica electron microscopy, this interesting structure was further shown to consist of a band of anastomosing intramembrane strands that run horizontal to the cell surface and confer either leakiness or tightness to the epithelium.Citation6,7 In conventional electron micrographs, tight junctions were shown to contain numerous “kisses” at the plasma membrane of adjoining cells that interacted with the actin cytoskeleton.Citation8 These structures were later found to correlate with highly regulated transmembrane pores made by claudin molecules.Citation3 Through this system of paracellular pores that are regulated by a set of interacting transmembrane and accessory proteins linked to the actin cytoskeleton, tight junctions were thus determined to regulate paracellular ion transport across the epithelial sheet.Citation1,9 The nature of this complicated membrane structure also conferred a “fence” function that allowed for the sorting of apical and basolateral membrane proteins above and below the tight junction, respectively. The claudin family of tight junction proteins currently consists of 27 known members,Citation10 with good evidence to suggest that they form the tight junction strands visualized by freeze-fracture replica electron microscopy.Citation4 Considerable evidence exists to support the notion that cells express numerous claudin molecules simultaneously, which are chosen on a function-based paradigm to regulate paracellular ion and water permeability.
The general architecture of apical tight junctions is the same in epithelial cells from many organs including intestine, kidney, and lung.Citation11-15 Sertoli cells of the testes also have a classical tight junction structure, which in part helps to create the blood-testis barrier.Citation16 Although the epidermis consists of a stratified, rather than a simple epithelium, classical tight junctions form in one of the distinct cellular layers to regulate paracellular permeability and ion transport.Citation17 Endothelial cells, particularly those forming the blood-brain barrier, also have classical tight junction architecture.Citation18 Even some tissues without cells that have an apical and basolateral polarity form tight junctions to regulate ion permeability, including cardiac muscle cells in the heart and myelinated neurons.Citation19-22
Despite a considerable focus on the role of apical tight junctions in regulating paracellular ion transport and membrane polarity, mounting evidence suggests that tight junction claudin proteins are localized to sites outside of the tight junction complex. Tight junction transmembrane and accessory proteins, such as occludin and junction adhesion molecule (JAM-A) are not usually present in these “extra-tight junction” sites, suggesting that claudins serve important functions outside of their classical role in regulating ion permselectivity. Thus, the aim of this article is to review our present knowledge about the location and function of these “extra-tight junction” claudin proteins and to highlight their emerging role in regulating cellular signaling, cell adhesion, epithelial to mesenchymal transformation (EMT), cell migration/invasion/metastasis, and the transcription of genes that, at least in part, regulate cell survival and proliferation. Rather than categorizing the extra-tight junction claudins as “mislocalized” tight junction proteins, we suggest naming them differently. This might include tCLDN for tight-junction associated claudins, bCLDN for claudins localized to the basolateral membrane, and cnCLDN for claudins that shuttle between the cytoplasm and nucleus. Creating a new designation for these proteins may be necessary to fully consider their important functional role outside of the tight junction complex in health and in disease.
Basolateral membrane-associated claudins and cellular signaling
When freeze fracture replica electron microscopy of stomach epithelial cells was done in 1973 by Goodenough and Claude,Citation7 discontinuous tight junction-like structures (strands) were found along the basolateral membrane that were somewhat dismissed because they were thought to play no role in generating the transepithelial electrical resistance. Because claudin molecules constitute at least one component of tight junction strands,Citation4 these results suggested that claudin proteins are localized at the basolateral membrane in addition to the tight junction. Immunolocalization studies confirmed the basolateral localization of some claudin molecules in stomach () and similar results were obtained in many different tissues ().
Figure 1. Confocal micrographs of the stomach mucosa stained for tight junction associated claudin-4 and basolateral membrane-associated claudin-18. Note the absence of green fluorescence signal at apical tight junctions in the surface of tissues stained for claudin-4 but the strong signal at apical tight junctions of epithelial cells in the neck and base (arrows). In contrast, claudin-18 is highly concentrated at the basolateral membrane of all epithelial cells (arrowheads) in the stomach mucosa with particularly robust expression in surface epithelial cells. MM, muscularis mucosa.
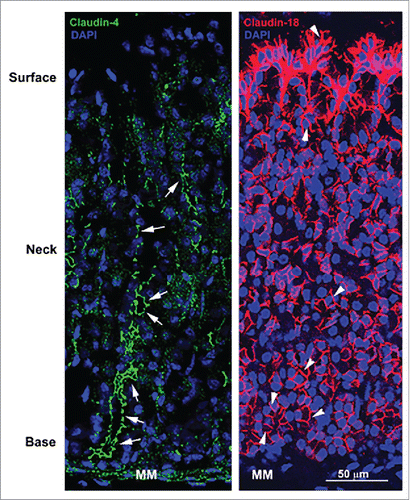
Table 1. Tissues and cultured cells in which tight junction proteins are found outside the apical tight junction complex.Footnote*
One of the first demonstrations that claudin molecules localize to sites other than the apical tight junction complex in tissues was by Rahner et al,Citation23 who showed that claudins can be expressed along the basolateral membrane of gastrointestinal (GI) epithelial cells and additionally that multiple claudins co-localize at these sites (). Studies in mouse and human GI tissues confirmed and extended this initial finding ().Citation24-26 In rat epididymis, the expression of basolateral claudin-1 was even more interesting because some tissue segments showed solely lateral localization below the tight junction, some segments showed very strong basal versus lateral localization, and one segment showed extremely strong apical localization specific to progenitor cells (basal cells) that sit along the basement membrane and never reach the epididymal surface ().Citation27 These results suggest that the lateral and basal membrane compartments have specific functional requirements that include claudin molecules. The important roles currently identified for basal claudins will be discussed in the next section.
One pertinent question to ask is “why is there a pool of basolateral membrane claudins in the first place”? Although little is currently known about the role of basolateral membrane claudins, it is possible that they represent a pool of claudin molecules that are available to recycle to the apical tight junction complex when needed (). It is well-established that the apical tight junction complex is highly dynamic and undergoes continual remodeling,Citation28 using a microtubule-based transport system to carry-out continuous endocytic recycling,Citation29,30 which is largely clathrin-mediated.Citation31 Apically-directed tight junction proteins are sorted in the trans-Golgi network (TGN), and targeted to the apical tight junction complex by specific trafficking proteins ().Citation32,33 In contrast, virtually nothing is known about how basolateral claudin molecules are sorted or targeted to the basolateral membrane and whether they follow conventional or canonical pathways.Citation34 Despite the vast literature on regulation of apically-targeted tight junction protein formation, trafficking, recycling, and turn-over, little evidence exists to support that a basolateral pool of claudins is needed for remodeling the apical tight junction (). Perhaps we do not have the appropriate tools currently available to evaluate membrane recycling from this pool.
Figure 2. Schematic diagram of potential functions for basolateral claudin molecules. (A) Basolateral claudins may function in the trafficking of endosomal vesicles (red circles) from the baolateral membrane to tight junctions for rapid remodeling. (B) Basolateral claudins may function as accessory pores to facilitate further discretion of ion transport at tight junctions (dotted arrow) and/or create signaling hubs via interaction with claudin/claudin complexes and the action cytoskeleton. EpCAM, epithelial cell adhesion molecule; TGN, trans-Golgi network.
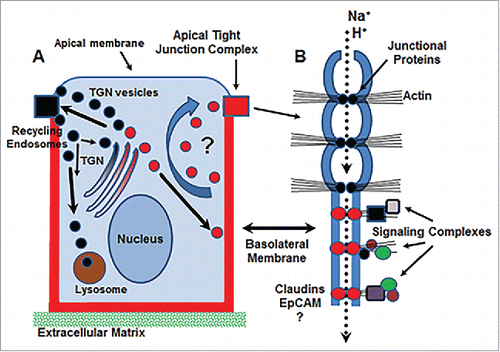
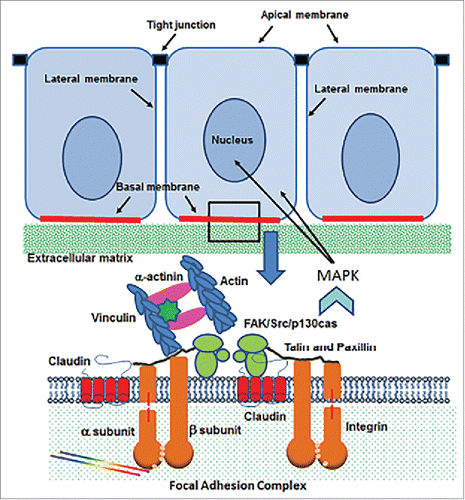
Figure 3. Schematic diagram of the cell/extracellular matrix interface in epithelial cells. Claudin molecules interact with integrin -α and -β subunits within the focal adhesion complex, which provides and anchor for cells to the extracellular matrix. The presence of claudins results in MAPK signaling through the focal adhesion kinase (FAK)/Src/p 130cas complex.
Three other possibilities, at the least, could explain the function of basolateral membrane-associated claudins. First, basolateral membrane claudins may function as an accessory structure to the tight junction, serving as an additional permeability barrier to increase the transit time of charged molecules (). This notion is supported by studies using CLDN18A2.1 (stomach isoform)-deficient mice, which express no stomach-specific claudin-18 at the basolateral membrane and show a faster rate of H+-diffusion,Citation35 suggesting that claudin 18A2.1 normally forms channels or pores along the basolateral membrane to impede paracellular cation movement. Second, it is possible that claudins are able to diffuse freely along the basolateral membrane and are trapped into tight junction fibrils as they oligomerize within the apical tight junction complex. Although this type of membrane trapping has been described for exchange between basolateral and junctional pools of E-cadherin,Citation36-38 it has not yet been studied for claudins within the apical tight junction complex. Lastly, basolateral claudins may function as a signaling hub, signaling complex, or cluster, which integrates, codifies, and transports information into the cell (). This possibility has the strongest supporting data. To form a signaling complex or cluster along the basolateral membrane, claudin molecules should interact with one another and be immobilized in specific domains rather than diffusing randomly in the plane of the membrane (). Such an interaction occurs in T84 and Caco-2 intestinal cells, which express both claudin-1 and -7 along the basolateral membrane ().Citation39 It was recently shown that these 2 claudins additionally interact with and bind to epithelial cell adhesion molecule (EpCAM), forming a functional unit along the basolateral cell membrane.Citation39 This interaction was required to stabilize claudins at the basolateral membrane and was also required to regulate their rate of endocytosis and subsequent lysosomal degradation.Citation39 The interaction with EpCAM occurred with basolateral claudins and not with apical tight junction-associated claudins,Citation39 which can be retained within the tight junction complex by phosphorylation and not trafficked to and degraded by lysosomes.Citation40 Overall, these data suggest that cells expressing EpCAM, or other yet undefined claudin-binding partners, organize claudin molecules into discrete functional units along the basolateral membrane. Furthermore, these structures may be the source of discontinuous strands along the basolateral membrane that were originally identified by freeze fracture replica electron microscopy.Citation7 Although this possibility is likely, the interpretations are not yet fully supported by data. With little known in this area of tight junction biology, a significant opportunity exists to determine novel claudin binding partners and to evaluate their functional role along the basolateral membrane.
Basal membrane claudins-cellular/extracellular matrix interactions
The basal membrane of epithelial cells is adjacent to an extracellular matrix (ECM) that, with the assistance of cellular ECM receptors (integrins), regulates cell adhesion, migration, differentiation, and the survival of epithelial cells ().Citation41 Integrins perform these functions by organizing a signaling complex consisting of focal adhesion kinase (FAK), Src, and p130cas that is held together by their association with accessory proteins like talin, paxillin, vinculin, α-actinin, and the actin cytoskeleton ().Citation41
At least 3 claudin molecules, claudins-1, -2, and -7, have been implicated in the regulation of cell/ECM interactions by integrating with integrin molecules in focal adhesions () and regulating the phosphorylation of FAK to ensure adhesion of epithelial cells to the ECM. This function is required for normal cellular homeostasis but can also be used by cancer cells to facilitate the adhesion of metastatic cells.
Recent studies demonstrated that claudin-7 and the integrin-β1 subunit (lung) or the integrin-α2 subunit (with claudin-1, intestine) form a macromolecular complex that restricts claudin-7 to focal adhesions along the basal membrane ().Citation42-44 Little is known about the configuration of claudins within the focal adhesion complex. The downregulation of claudin-7, by genetic silencing methods, disrupts adhesion to the extracellular matrix, dysregulates the phosphorylation of FAK, and results in the downregulation of integrins, cell cycle, cell survival, and cell proliferation markers.Citation42,44 To highlight the importance of this claudin-integrin interaction along the basal membrane, intestinal epithelial cells are unable to attach to the underlying mucosa in CLDN7-deficient mice in vivo, resulting in ulcerative lesions that promote inflammation and death shortly after birth.Citation42 Overall, these results suggest that claudin-7 interactions are essential to stabilize cell/matrix interactions and to regulate cell proliferation and survival by integrating, in some way, with effectors in the focal adhesion complex. Although it was originally thought that claudin-7 per se may transcriptionally regulate the expression of numerous effectors, particularly the expression of cell proliferation and survival genes, recent genomic profiling and pathway analysis demonstrated that claudin-7 indirectly regulates gene transcription via a cell signaling network with MAPK's as the central node ().Citation43 Other claudins also show strong expression patterns along the basal membrane (), suggesting that they are also involved in interactions at the focal adhesion complex. Further work would be required to test the role of these additional basally-localized claudins in various cell types.
The localization of claudin-7 and its interaction with integrins in particular areas of a tissue (like crypt vs. villus in intestine) or segments of the same tissue (like proximal vs. distal vs. collecting ducts in the kidney) may be a key regulator of gene expression including the expression of tight junction-associated claudins, and thus of the permeability characteristics in different segments of the same tissue. One example to support this notion is in kidney, where proximal tubule epithelium, which is classified as a “leaky” epithelium due to robust claudin-2 expression, has low claudin-7 expression.Citation45 In the absence of β1 integrin, which was genetically deleted, the same cells changed phenotype to that of a “tight” epithelium as is found in the distal tubules and collecting ducts, which have minimal claudin-2 expression and high claudin-7 expression.Citation45 This conversion (claudin-2/claudin-7), driven by the absence of integrin β1, resulted in significant functional abnormalities in the kidney due to the lack of conventional proximal tubule function.Citation45 Although the relationship of claudin-7 expression and the expression of cell matrix effectors was not investigated,Citation45 it would be interesting to know whether or not high levels of claudin-7 expression stabilized the epithelium at focal contacts, activated MAPK or other signaling pathways, and transcriptionally regulated the expression of claudin-2 and other effectors that are constitutively expressed in distal tubule or collecting duct cells. These data however, suggest that cell signaling from focal adhesions on the basal membrane of epithelial cells, via an integrin-claudin pathway, could be a more generalized mechanism for conferring regional specificity to the mucosal barrier in a given tissue. More data, however, is needed to lend support to this interpretation.
Although integrin/claudin-7 interactions at the basal membrane are part of the story, claudin-7 is also known to interact directly with EpCAM along the basal membrane.Citation46 This association occurs in many tissues including colon and pancreas and in numerous cancers, for which the interaction is thought to mediate tumor progression.Citation47 Although there are several phosphorylation sites on claudin molecules that could regulate their activity, palmitolyation in particular regulates the ability of claudin-7 to interact with integrins, recruit EpCAM, and concomitantly associate with the actin cytoskeleton.Citation48 The histological features of intestinal epithelial cells in EpCAM-deficient mice look similar to CLDN7-deficient mice, most prominently with intestinal epithelial cells unable to attach to the underlying mucosa.Citation49 These results suggest that, at least for claudin-7, EpCAM interactions are essential for cell/ECM interactions that confer mucosal homeostasis.
In contrast to the important role claudin-7/integrin/EpCAM interactions play in normal mucosal function, the expression of claudin molecules in metastatic cells likely plays a detrimental role by facilitating cell/ECM interactions in cancer progression. One example is in breast cancer cells, where claudin-2 expression drives the cell surface expression of integrins α2β1 and α5β1, forming an integrin-claudin adhesion complex to facilitate the formation of liver metastases.Citation50 Perhaps the most novel use of claudins as adhesion receptors is in HIV infection, where the virus was shown to anchor claudin-7 to its envelop and use this protein to adhere-to and infect CD4(-) target cells.Citation51 Thus, while the expression of claudins at the basal membrane function to regulate beneficial cell/ECM interactions and cell signaling, this important function can also be used to promote cellular adhesion in disease processes that are not beneficial such as the adhesion of metastatic cells to promote cancer progression or to promote HIV infection.
Basolateral membrane claudins-regulation of epithelial-mesenchymal transformation, cell migration, invasion, and tumorigenesis
EMT is defined as the transformation of polarized epithelial cells, which are attached to one another in the epithelial sheet by tight and adherens junction complexes, into individual cells with a migratory (spindle-shaped) phenotype that is accompanied by the loss of membrane-associated E-cadherin, the expression of mesenchymal markers including nuclear β-catenin, the activation of stemness genes including CD44, and the activation of numerous cell signaling pathways.Citation52 EMT also occurs in wound healing and in tumorigenesis, allowing for the migration of epithelial cells. One interesting aspect of EMT is that although adherens junctions disappear, which should lead to the dissolution of tight junctions and the downregulation of tight junction protein expression, select claudin molecules are highly expressed. In nearly every article written about the role of claudins in EMT/wound healing and in tumorigenesis, the role of claudins in permeability at the apical tight junction complex is discussed when in fact the newly expressed claudin molecules are localized to the basolateral membrane, intracellular compartments, or the nucleus. Little is known about the role of claudins when they are expressed in these locations. While literature on the topic of claudins in EMT and cancer progression is rather extensive, the studies typically correlate changes in claudin expression to disease pathogenesis and/or patient outcome. The few mechanistic studies outlined below have shed light on the role of claudins in EMT and cell/ECM interactions, and cell migration/invasion in cancer pathogenesis.
Numerous studies have shown that claudins regulate the expression and activation of matrix metalloproteinases (MMT's),Citation53-56 which causally link claudins to ECM remodeling that is required for EMT, cell migration, and invasion in cancer pathogenesis. MMP's are secreted at the basolateral surface of epithelial cells and function to change the cell/ECM interaction to promote migration and invasion. Claudins have been shown to recruit membrane-type MMP's, enhancing the activation of pro-MMP, although the exact role of claudin molecules in this process is unknown.Citation57 In addition to matrix remodeling, MMP's have also been shown to activate important transcriptional regulators, like EGFR, which promotes cell signaling cascades that alter claudin expression.Citation58 Claudins may synergize to promote MMP activation, as was recently demonstrated by claudin-1/claudin-6 interactions in human AGS cells.Citation59 In that study, claudin-6 induced MMP-2 activation via claudin-1 expression, which accelerated the rate of cell migration and invasion.Citation59 Lastly, the invasiveness of cancer cells can be regulated by epigenetic means including DNA methylation, which modulates claudin expression by inhibiting (or stimulating) claudin gene promoter activity.Citation60
In in vivo studies, CLDN7-deficient mice showed robust MMP expression that accompanied the inability of cells to attach to the underlying mucosa,Citation42 suggesting a direct correlation between matrix remodeling and cell/ECM attachment. It is not clear whether all claudins have this capability and as such, further work is needed.
Nuclear claudins as transcriptional regulators
The nuclear localization of tight junction proteins was first shown for ZO-1 and ZO-2 (),Citation61,62 and subsequently their role in gene transcription via interaction with ZONAB/DbpA or c-myc/E-box elements, respectively, was elucidated.Citation63,64 The presence of claudins in the nucleus was highlighted by Dhawan et al,Citation65 who showed that the expression of claudin-1 was localized to the nucleus in tissues from patients with human primary colon cancer, in colon cancer-derived liver metastases, and in metastasis to lymph nodes, whereas there was no nuclear claudin-1 expression in normal human colonic mucosa. Rather, claudin-1 was localized to the basolateral membrane in normal human colonic mucosa.Citation65 Colon cancer cell lines had differential claudin-1 expression, with SW480 and HCT116 cells expressing little to no claudin-1, respectively, and SW620 cells expressing high levels of claudin-1.Citation65 Immunostaining studies in the colonic cancer cell lines showed that the highly expressing line had strong nuclear localization of claudin-1, and cell fractionation studies consisting of nuclear, cytoplasmic, and membrane fractions validated that highly expressing cell lines contained claudin-1 in the nuclear fraction whereas those with no nuclear claudin-1 did not.Citation65 The nuclear localization of claudin-1 was also validated by immunostaining and by fractionation followed by Western blots using transformed nasopharyngeal carcinoma cells compared with non-transformed cells, which do not express nuclear claudin-1.Citation66 Numerous subsequent studies also showed that claudin-1 localized to the nucleus in melanoma cells, smooth muscle cells from asthmatic subjects, and thyroid carcinoma cells.Citation67-69 Additionally, claudin-2 localized to the nucleus in human lung adenocarcinoma cells,Citation70 claudin-3 localized to the nucleus in breast cancer cell lines,Citation71 and claudin-4 localized to the nucleus in endometrial cancer cells.Citation72 Using genetically modified colon cancer cells, robust expression of nuclear claudin-1 was associated with an increase in invasive capacity, high MMP-9 activity, and a decrease in non-contact mediated cell death (anoikis) rather than affecting cellular proliferation, which is the target of ZO-1 and -2 localized to the nucleus.Citation63,65,73
The nuclear localization of claudin-1 in colon cancer cells occurred without changes in the molecular weight, suggesting that no cleavage or modification was needed to drive this protein into the nucleus.Citation65 In contrast, the prominent nuclear localization of claudin-2 in human lung adenocarcinoma and in cultured lung adenocarcinoma cells was upregulated by dephosphorylation.Citation70 Forskolin, via increases in cAMP, was proposed to activate protein phosphatase activity that in turn dephosphorylates claudin-2 to facilitate its nuclear localization.Citation70 Ultimately, claudin-2 retained ZONAB and cyclin D1 in the nucleus to enhance cell proliferation.Citation70 In melanoma cells, claudin-1 showed both cytoplasmic and weak nuclear localization in isolated cell fractions and by immunostaining.Citation67 When claudin-1 was tagged with a strong nuclear localization signal (NLS), the cytoplasmic fraction disappeared in exchange for complete nuclear localization.Citation67 This effect was activated by protein kinase A (PKA) and blocked by phosphorylation site mutations, suggesting that claudin-1 also utilizes phosphorylation to regulate its nuclear localization. Different cell types also localize claudin-1 to the nucleus but rather by protein kinase C (PKC) activation.Citation69
ZO proteins were shown to have 2 NLS's, one in the PDZ domain and the other in the GK domain, with robust nuclear localization under sparse plating conditions but little localization in confluent monolayers that are contact inhibited.Citation62,74 Although early work demonstrated that claudins -1 and -2 can gain access to the nucleus, the investigators were unable to show that either claudin had a putative NLS sequence.Citation65,70 Rather, it was suggested that without an NLS sequence, claudins may utilize their PDZ domain or other mechanism for transport into the nucleus.Citation65
While claudin-2 does not have a strong NLS sequence, the cNLS mapper program (see for the http:// site) identified putative NLS sequences in many of the 27 known claudin molecules from mouse that could facilitate transport to the nucleus (). The highest cNLS score was for claudin-12, followed by claudins 10, 23, 15, and 16 (). For these claudins, and the remainder of claudin molecules with cNLS scores of 3–5, the putative NLS sequence can be localization to the cytoplasm and/or to the nucleus (). Although it is not known how claudins enter the nucleus, claudin-12 is highly localized to the nucleus in bladder epithelium from mouse and rat, and in human Caco-2 intestinal cells treated with vitamin D.Citation75,76 Similarily, claudin-15 localized to the nucleus in Caco-2 cells.Citation76 Furthermore, claudin-23 is highly expressed in the nucleus of pancreatic cancer cells when compared with its tight junction localization.Citation77
Table 2. Putative nuclear localization sequence (NLS) for mouse claudins determined from cNLS Mapper at http://nls-mapper.iab.keio.ac.jp/cgi-bin/NLS_Mapper_form.cgi
The nuclear localization of claudins suggests that these proteins function to regulate gene transcription but is there any evidence to support this notion? Molecular studies by Dhawan et al,Citation65 using cultured colon cancer cell lines, lend considerable insight. For this, the nuclear localization of claudin-1 in genetically manipulated colon cancer cell lines, from highly expressing to virtual knockouts, resulted in 1) changes in luciferase activity of the E-cadherin promoter that were consistent with the level of nuclear claudin-1 expression (inhibition of nuclear claudin-1 expression resulted in the activation of E-cadherin reporter activity) and 2) concomitant changes in β-catenin/Tcf/Lef and Wnt/β-catenin/Tcf/Lef signaling that affected the expression of downstream target genes such as c-Myc.Citation65 Overall, these studies demonstrate a direct transcriptional role for nuclear claudin-1 in E-cadherin expression that affected indirectly the β-catenin/Tcf/Lef signaling pathway in cultured colon cancer cells.Citation65
Although it seems puzzling that a membrane-spanning protein as large as claudin molecules (20–27 kDa) would transport and enter the nucleus, there is emerging data to support that similarly sized tetraspanin molecules do so and also regulate gene transcription. It was recently shown that TM4SF3 (26 kDa), also known as tetraspanin-8, enters the nucleus of prostate cancer cells.Citation78 The efficiency of nuclear accumulation is dependent on a direct interaction with the androgen receptor, which provides stability for TM4SF3.Citation78 Once in the nucleus, TM4SF3 contributes to androgen-dependent gene expression, particularly to the expression of genes regulating cellular proliferation.Citation78 CD9 (25 kDa), also known as tetraspanin-29, was shown to enter the nucleus in breast cancer cells.Citation79 CD9 associated with IgFS8 in the cytoplasm and IgFS8 and CEP97 in the nucleus, suggesting that CD9 is part of a larger complex that, in some way, translocates from the cell membrane to the nucleus to regulate gene transcription.Citation79 Recent studies demonstrated that CD9 transits to and enters the nucleus using a novel extracellular vesicle-derived system that delivers cargo to the nucleus via Rab7-positive late endosomal vesicle fusion with the nuclear envelope.Citation80 E-cadherin, a type-1 membrane spanning protein, also has a nuclear localization pattern. To gain access to the nucleus, the large E-cadherin molecule is cleaved by metaloproteinases and by γ-secretase into E-cadherin C-terminal fragments, which along with a chaperone protein (p120) translocates to the nucleus and regulates gene transcription.Citation81,82 Results from Dhawan et alCitation65 argue against claudins entering the nucleus as fragments because the band for claudin-1 in Western blots was the same size in membrane-derived, cytoplasmic, and nuclear fractions of colonic cancer cells.
Overall, many claudins have a putative NLS, or multiple NLS's, which map to the intermembrane domain alone or to the extracellular loop 2/intermembrane domain area of the claudin structureCitation83 that would facilitate transport into the nucleus. Although constitutive and disease-related shuttling of claudins into the nucleus occurs, little is known about conditions that facilitate this event. Although the existing data support that nuclear claudin molecules interact with transcriptional regulators to impact gene expression and the regulation of cell attachment, cell proliferation/cell death, and cell proliferation effectors, further data are needed to identify putative binding partners in the cytoplasm and in the nucleus.
Conclusions
There is a wealth of supporting data in the literature to conclude that claudin molecules are purposely localized to sites outside of the apical tight junction complex and as such, serve important roles in both mucosal homeostasis and in disease pathogenesis. In particular, most if not all of the claudin family members localize to numerous other sites, being renegades by not fitting into a single functional paradigm at the tight junction.
To move this field ahead, it is possible that new tools may be needed to properly determine whether or not the basolateral pool of claudins is a reservoir of extra material that can be recruited to the tight junction when needed. If it is, we also need to understand what regulates this function. Furthermore, it is essential to understand whether or not basolateral claudins are actually expressed within the tight junction complex, or is the proximity so close that our current instrumentation cannot resolve them spatially. If solely along the basolateral membrane, do basolateral claudins form a secondary permeability barrier that acts in series to the tight junction barrier to lend additional transit time for the molecules crossing into the basolateral compartment? Do basolateral claudins, via their PDZ domain, nucleate a yet undefined macromolecular complex that organizes signaling or other functions to the basolateral membrane and do they also use this feature of their structure to gain access to the nucleus? What is the role of cytoplasmic and nuclear claudins and how are these claudins regulated? Do nuclear claudins directly regulate transcription by forming DNA binding complexes within the nucleus or indirectly regulate transcription, via signaling pathways from the cytoplasm? Similarly, do the nuclear claudins work in concert to promote or inhibit the action of classic transcription factors? These questions and numerous others pose an exciting new era in barrier biology that will take us well into the future.
Abbreviations
| CLDN | = | claudin |
| ECM | = | extracellular matrix |
| EpCAM | = | epithelial cell adhesion molecule |
| FAK | = | focal adhesion kinase |
| GI | = | gastrointestinal |
| JAM | = | junctional adhesion molecule |
| MAPK | = | MAP kinase |
| MMP | = | matrix metalloproteinase |
| NLS | = | nuclear localization signal (or sequence) |
| PKA | = | protein kinase A |
| PCK | = | protein kinase C |
| TGN | = | trans-Golgi network |
| ZO | = | zonula occludins. |
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by the Harvard Catalyst | The Harvard Clinical and Translational Science Center, National Center for Research Resources and the National Center for Advancing Translational Sciences, National Institutes of Health (NIH) Award 1UL1 TR001102–01 and financial contributions from Harvard University and its affiliated academic health care centers, Department of Surgery Bridge funds, Special Fund for Gastric Cancer Research, and NIH grant P30 DK034854.
References
- Van Itallie CM, Anderson JM. Architecture of tight junctions and principles of molecular composition. Semin Cell Dev Biol 2014; 36:157-65; PMID:25171873; https://doi.org/10.1016/j.semcdb.2014.08.011
- Anderson JM, Van Itallie CM. Physiology and function of the tight junction. Cold Spring Harb Perspect Biol 2009; 1:a002584; PMID:20066090; https://doi.org/10.1101/cshperspect.a002584
- Suzuki H, Tani K, Tamura A, Tsukita S, Fujiyoshi Y. Model for the architecture of claudin-based paracellular ion channels through tight junctions. J Mol Biol 2015; 427:291-7; PMID:25451028; https://doi.org/10.1016/j.jmb.2014.10.020
- Tsukita S, Furuse M, Itoh M. Multifunctional strands in tight junctions. Nat Rev Mol Cell Biol 2001; 2:285-93; PMID:11283726; https://doi.org/10.1038/35067088
- Farquhar MG, Palade GE. Junctional complexes in various epithelia. J Cell Biol 1963; 17:375-412; PMID:13944428
- Chalcroft JP, Bullivant S. An interpretation of liver cell membrane and junction structure based on observation of freeze-fracture replicas of both sides of the fracture. J Cell Biol 1970; 47:49-60; PMID:4935338
- Claude P, Goodenough DA. Fracture faces of zonulae occludentes from “tight” and “leaky” epithelia. J Cell Biol 1973; 58:390-400; PMID:4199658
- Madara JL. Intestinal absorptive cell tight junctions are linked to the cytoskeleton. Am J Physiol 1987; 253:C854-61; PMID:3605327
- Van Itallie CM, Anderson JM. Claudin interactions in and out of the tight junction. Tissue Barriers 2013; 1:e25247; PMID:24665401; https://doi.org/10.4161/tisb.25247
- Mineta K, Yamamoto Y, Yamazaki Y, Tanaka H, Tada Y, Saito K, Tamura A, Igarashi M, Endo T, Takeuchi K, et al. Predicted expansion of the claudin multigene family. FEBS Lett 2011; 585:606-612; PMID:21276448; https://doi.org/10.1016/j.febslet.2011.01.028
- Lu Z, Ding L, Lu Q, Chen YH. Claudins in intestines: Distribution and functional significance in health and disease. Tissue Barriers 2013; 1:e24978; PMID:24478939; https://doi.org10.4161/tisb.24978
- Luettig J, Rosenthal R, Barmeyer C, Schulzke JD. Claudin-2 as a mediator of leaky gut barrier during intestinal inflammation. Tissue Barriers 2015; 3:e977176; PMID:25838982; https://doi.org/10.4161/21688370.2014.977176
- Hou J. The kidney tight junction (review). Int J Mol Med 2017; 34:1451-7; PMID:25319473; https://doi.org/10.3892/ijmm.2014.1955
- Yu ASL. Claudins and the kidney. J Am Soc Nephrol 2015; 26:11-9; PMID:24948743; https://doi.org/10.1681/ASN.2014030284
- Frank JA. Claudins and alveolar epithelial barrier function in the lung. Ann N Y Acad Sci 2012; 1257:175-83; PMID:22671604; https://doi.org/10.1111/j.1749-6632.2012.06533.x
- Mruk DD, Cheng CY. The mammalian blood-testis barrier: its biology and regulation. Endocr Rev 2015; 36:564-91; PMID:26357922; https://doi.org//10.1210/er.2014-1101
- Brandner JM, Zorn-Kruppa M, Yoshida T, Moll I, Beck LA, De Benedetto A. Epidermal tight junctions in health and disease. Tissue Barriers 2015; 3:e974448; PMID:25838981; https://doi.org/10.4161/21688370.2014.974451
- Stamatovic SM, Johnson AM, Keep RF, Andjelkovic AV. Junctional proteins of the blood-brain barrier: New insights into function and dysfunction. Tissue Barriers 2016; 4:e1154641; PMID:27141427; https://doi.org/10.1080/21688370.2016.1154641
- Lisewski U, Shi Y, Wrackmeyer U, Fischer R, Chen C, Schirdewan A, Jüttner R, Rathjen F, Poller W, Radke MH, et al. The tight junction protein CAR regulates cardiac conduction and cell-cell communication. J Exp Med 2008; 205:2369-79; PMID:18794341; https://doi.org/10.1084/jem.20080897
- Itoh M, Nagafuchi A, Yonemura S, Kitani-Yasuda T, Tsukita S, Tsukita S. The 220-kD protein colocalizing with cadherins in non-epithelial cells is identical to ZO-1, a tight junction-associated protein in epithelial cells: cDNA cloning and immunoelectron microscopy. J Cell Biol 1993; 121:491-502; PMID:8486731
- Denninger AR, Breglio A, Maheras KJ, DeLuc G, Cristiglio V, Deme B, Gow A, Kirschner DA. Claudin-11 tight junctions in myelin are a barrier to diffusion and lack strong adhesive properties. Biophys J 2015; 109:1387-97; PMID:26445439; https://doi.org/10.1016/j.bpj.2015.08.012
- Devaux J, Fykkolodziej B, Gow A. Claudin proteins and neuronal function. Curr Top Membr 2010; 65:229-53; PMID:25013353; https://doi.org/10.1016/S1063-5823(10)65010-7
- Rahner C, Mitic LL, Anderson JM. Heterogeneity in expression and subcellular localization of claudins 2, 3, 4, and 5 in the rat liver, pancreas and gut. Gastroenterology 2001; 120:411-23; PMID:11159882
- Fujita H, Chiba H, Yokozaki H, Sakai N, Sugimoto K, Wada T, Kojima T, Yamashita T, Sawada N. Differential expression and subcellular localization of claudin-7, -8, 12-, -13, and -15 along the mouse intestine. J Histochem Cytochem 2006; 54:933-44; PMID:16651389; https://doi.org/10.1369/jhc.6A6944.2006
- Tamagawa H, Takahashi I, Furuse M, Yoshitake-Kitano Y, Tsukita S, Ito T, Matsuda H, Kiyono H. Characteristics of claudin expression in follicle-associated epithelium of Peyer's patches: preferential localization of claudin-4 at the apex oof the dome region. Lab Invest 2003; 83:1045-53; PMID:12861044
- Weber CR, Nalle SC, Tretiakova M, Rubin DT, Turner JR. Claudin-1 and claudin-2 expression is elevated in inflammatory bowel disease and may contribute to early neoplastic transformation. Lab Invest 2008; 88:1110-20; PMID:18711353; https://doi.org/10.1038/labinvest.2008.78
- Gregory M, Dufresne J, Hermo L, Cyr DG. Claudin-1 is not restricted to tight junctions in the rat epididymis. Endocrinology 2001; 142:854-63; PMID:11159859; https://doi.org/10.1210/endo.142.2.7975
- Shen L, Weber CR, Turner JR. The tight junction protein complex undergoes rapid and continuous molecular remodeling at steady state. J Cell Biol 2008; 181:683-95; PMID:18474622; https://doi.org/10.1083/jcb.200711165
- Chalmers AD, Whitley P. Continuous endocytic recycling of tight junction proteins: how and why? Essays Biochem 2012; 53:41-54; PMID:22928507; https://doi.org/10.1042/bse0530041
- Ivanov AI, Nusrat A, Parkos CA. Endocytosis of the apical junctional complex: mechanisms and possible roles in regulation of epithelial barriers. BioEssays 2005; 27:356-65; PMID:15770686; https://doi.org/10.1002/bies.20203
- Ivanov AI, Nusrat A, Parkos CA. Endocytosis of epithelial apical junctional proteins by a clathrin-mediated pathway into a unique storage compartment. Mol Biol Cell 2004; 15:176-88; PMID:14528017; https://doi.org/10.1091/mbc.E03-05-0319
- Lu R, Stewart L, Wilson JM. The scaffolding protein GOPC regulates tight junction structure. Cell Tissue Res 2015; 360:321-32; PMID:25616555; https://doi.org/10.1007/s00441-014-2088-1
- Lu R, Johnson DL, Stewart L, Waite K, Elliot D, Wilson JM. Rab14 regulation of claudin-2 trafficking modulates epithelial permeability and lumen morphogenesis. Mol Biol Cell 2014; 25:1744-54; PMID:24694596; https://doi.org/10.1091/mbc.E13-12-0724
- Apodaca G, Gallo LI, Bryant DM. Role of membrane traffic in the generation of epithelial cell asymmetry. Nat Cell Biol 2012; 14:1235-43; PMID:23196841; https://doi.org/10.1038/ncb2635
- Hayashi D, Tamura A, Tanaka H, Yamazaki Y, Watanabe S, Suzuki K, Suzuki K, Sentani K, Yasui W, Rakugi W, et al. Deficiency of claudin-18 causes paracellular H+ leakage, up-regulation of interleukin-1-beta, and atrophic gastritis in mice. Gastroenterology 2012; 142:292-304; PMID:22079592; https://doi.org/10.1053/j.gastro.2011.10.040
- Iino R, Koyama I, Kusumi A. Single molecule imaging of green fluorescent proteins in living cells: E-cadherin forms oligomers on the free cell surface. Biophys J 2001; 80:2667-77; PMID:11371443; https://doi.org/10.1016/S0006-3495(01)76236-4
- Hansen MDH, Ehrlich JS, Nelson WJ. Molecular mechanism for orienting membrane and actin dynamics to nascent cell-cell contacts in epithelial cells. J Biol Chem 2002; 277:45371-6; [PMID:12244058; https://doi.org/10.1074/jbc.M207747200
- Perez TD, Tamada M, Sheetz MP, Nelson WJ. Immediate-early signaling induced by E-cadherin engagement and adhesion. J Biol Chem 2008; 283:5014-22; PMID:18089563; https://doi.org/10.1074/jbc.M705209200
- Wu C-J, Mannan P, Lu M, Udey MC. Epithelial cell adhesion molecule (EpCAM) regulates claudin dynamics and tight junctions. J Biol Chem 2013; 288:12253-68; PMID:23486470; https://doi.org/10.1074/jbc.M113.457499
- Van Itallie CM, Tietgens AJ, LoGrande K, Aponte A, Gucek M, Anderson JM. Phosphorylation of claudin-2 on serine 208 promotes membrane retention and reduces trafficking to lysosomes. J Cell Sci 2012; 125:4902-12; PMID:22825868; https://doi.org/10.1242/jcs.111237
- Iwamoto DV, Calderwood DA. Regulation of integrin-mediated adhesions. Curr Opin Cell Biol 2015; 36:41-7; PMID:26189062; https://doi.org/10.1016/j.ceb.2015.06.009
- Ding L, Lu Z, Foreman O, Tatum R, Lu Q, Renegar R, Cao J, Chen Y-H. Inflammation and disruption of the mucosal architecture in claudin-7-deficient mice. Gastroenterology 2012; 142:305-15; PMID:22044670; https://doi.org/10.1053/j.gastro.2011.10.025
- Ding L, Wang L, Sui L, Zhao H, Xu X, Li T, Wang X, Li W, Zhao P, Kong L. Claudin-7 indirectly regulaters the integrin/FAK signaling pathway in human colon cancer tissue. J Hum Genet 2016; 61:711-20; PMID:27121327; https://doi.org/10.1038/jhg.2016.35
- Lu Z, Kim DH, Fan J, Lu Q, Verbanac K, Ding L, Renegar R, Chen Y-H. A non-tight junction function of claudin-7-interaction with integrin signaling in suppressing lung cancer cell proliferation and detachment. Mol Cancer 2015; 14:120; PMID:26081244; https://doi.org/10.1186/s12943-015-0387-0
- Elias BC, Mathew S, Srichai MB, Palamuttam R, Bulus N, Mernaugh G, Singh AB, Sanders CR, Harris RC, Pozzi A, et al. The integrin β1 subunit regulates paracellular permeability of kidney proximal tubule cells. J Biol Chem 2014; 289:8532-44; PMID:24509849; https://doi.org/10.1074/jbc.M113.526509
- Ladwein M, Pape U-F, Schmidt D-S, Schnölzer M, Fiedler S, Langbein L, Franke WW, Moldenhauer G, Zöller M. The cell-cell adhesion molecule EpCAM interacts directly with the tight junction protein claudin-7. Expt Cell Res 2005; 309:345-57; PMID:16054130; https://doi.org/10.1016/j.yexcr.2005.06.013
- Nübel T, Preobraschenski J, Tuncay H, Weiss T, Kuhn S, Ladwein M, Langbein L, Zöller M. Claudin-7 regulates EpCAM-mediated functions in tumor progression. Mol Cancer Res 2009; 7:285-99; PMID:19276185; https://doi.org/10.1158/1541-7786.MCR-08-0200
- Heiler S, Mu W, Zöller M, Thuma F. The importance of claudin-7 palmitoylation on membrane subdomain localization and metastasis-promoting activities. Cell 2015; 13:29; PMID:26054340; https://doi.org/10.1186/s12964-015-0105-y
- Kozan PA, McGeough MD, Pena CA, Mueller JL, Barrett KE, Marchelletta RR, Sivagnanam M. Mutation of EpCAM leads to intestinal barrier and ion transport dysfunction. J Mol Med 2015; 93:535-45; PMID:25482158; https://doi.org/10.1007/s00109-014-1239-x
- Tabaries S, Dong Z, Annis MG, Omeroglu A, Pepin F, Ouellet V, Russo C, Hassanain M, Metrakos P, Diaz Z, et al. Claudin-2 is selectively enriched in and promotes the formation of breast cancer liver metastases through engagement of integrin complexes. Oncogene 2011; 30:1318-28; PMID:21076473; https://doi.org/10.1038/onc.2010.518
- Zheng J, Xie Y, Campbell R, Song J, Samira M, Razi M, Chiu R, Berenson J, Yang OO, Chen ISY, et al. Involvement of claudin-7 in HIV infection of CD4(-) cells. Retrovirology 2005; 2:79; PMID:16368003
- Gruzu S, Silveanu C, Fetyko A, Butiurca V, Kovacs Z, Jung I. Systematic review of the old and new concepts in the epithelial-mesenchymal transition of colorectal cancer. World J Gastroenterol 2016; 22:6764-75; PMID:27570416; https://doi.org/10.3748/wjg.v22.i30.6764
- Agarwal R, D'Souza T, Morin PJ. Claudin-3 and claudin-4 expression in ovarian epithelial cells enhances invasion and is associated with increased matrix metalloproteinase-2 activity. Cancer Res 2005; 65:7378-85; PMID:16103090; https://doi.org/10.1158/0008-5472.CAN-05-1036
- Yoon CH, Kim MJ, Park MJ, Park IC, Hwang SG, An S, Choi YH, Yoon G, Lee SJ. Claudin-1 acts through c-Abl-protein kinase cδ (PKCδ) signaling and has a causal role in the acquisition of invasive capacity in human liver cells. J Biol Chem 2010; 285:226-33; PMID:19897486; https://doi.org/10.1074/jbc.M109.054189
- Pope JL, Bhat AA, Sharma A, Ahmad R, Krishnan M, Washington MK, Beauchamp RD, Singh AB, Dhawan P. Claudin-1 regulates intestinal epithelial homeostasis through the modulation of Notch signaling. Gut 2014; 63:622-34; PMID:24997475; https://doi.org/10.1186/1476-4598-13-167
- Leotlela PD, Wade MS, Duray PH, Rhode MJ, Brown HF, Rosenthal DT, Dissanayake SK, Earley R, Indig FE, Nickoloff BJ, et al. Claudin-1 overexpression in melanoma is regulated by PKC and contributes to melanoma cell motility. Oncogene 2007; 26:3846-56; PMID:17160014; https://doi.org/10.1038/sj.onc.1210155
- Miyamori H, Takino T, Kobayashi Y, Tokai H, Itoh Y, Seiki M, Sato H. Claudin promotes activation of pro-matrix metalloproteinase-2 mediated by membrane-type matrix metalloproteinases. J Biol Chem 2001; 276:28204-11; PMID:11382769; https://doi.org/10.1074/jbc.M103083200
- Ikari A, Sato T, Watanabe R, Yamazaki Y, Sugatani J. Increase in claudin-2 expression by an EGFR/MEK/ERK/c-Fos pathway in lung adenocarcinoma A549 cells. Biochim Biophys Acta 2012; 1823:1118; PMID:22546605; https://doi.org/10.1016/j.bbamcr.2012.04.005
- Torres-Martinez AC, Gallardo-Vera JF, Lara-Holguin AN, Montano LF, Rendon-Huerta EP. Claudin-6 enhances cell invasiveness through claudin-1 in AGS human adenocarcinoma gastric cancer cells. Exp Cell Res 2017; 350:226-35; PMID:27914788; https://doi.org/10.1016/j.yexcr.2016.11.025
- Liu Y, Jin X, Li Y, Ruan Y, Lu Y, Yang M, Lin D, Song P, Guo Y, Zhao S, et al. DNA methylation of claudin-6 promotes breast cancer cell migration and invasion by recruiting MeCP2 and deacetylating H3Ac and H4Ac. J Exp Clin Cancer Res 2016; 35:120; PMID:27461117; https://doi.org/10.1186/s13046-016-0396-x
- Gottardi CJ, Arpin M, Fanning AS, Louvard D. The junction-associated protein, zonula occludens-1, localizes to the nucleus before the maturation and during the remodeling of cell-cell contacts. Proc Natl Acad Sci USA 1996; 93:10779-84; PMID:8855257
- Islas S, Vega J, Ponce L, González-Garcia M. Nuclear localization of the tight junction protein ZO-2 in epithelial cells. Exp Cell Res 2002; 274:138-48; PMID:11855865; https://doi.org/10.1006/excr.2001.5457
- Sourisseau T, Georgiadis A, Tsapara A, Ali RR, Pestell R, Matter K, Balda MS. Regulation of PCNA and cyclin D1 expression and epithelial morphogenesis by the ZO-1-regulated transcription factor ZONAB/DbpA. Mol Cell Biol 2006; 26:2387-98; PMID:16508013; https://doi.org/10.1128/MCB.26.6.2387-2398.2006
- Spadaro D, Tapia R, Pulimeno P, Citi S. The control of gene expression and cell proliferation by the epithelial apical junctional complex. Essays Biochem 2012; 53:83-93; PMID:22928510; https://doi.org/10.1042/bse0530083
- Dhawan P, Singh AB, Deane NG, No YR, Shiou S-R, Schmidt C, Neff J, Washington MK, Beauchamp RD. Claudin-1 regulates cellular transformation and metastatic behavior in colon cancer. J Clin Invest 2005; 115:1765-76; https://doi.org/10.1172/JCI24543
- Lee J-W, Hsiao W-T, Chen H-Y, Hsu L-P, Chen P-R, Lin M-D, Chiu S-J, Shih W-L, Hsu Y-C. Upregulated claudin-1 expression confers resistance to cell death of nasopharyngeal carcinoma cells. Int J Cancer 2010; 126:1353-66; PMID:19739116; https://doi.org/10.1002/ijc.24857
- French AD, Fiori JL, Camilli TC, Leotlela PD, O'Connell MP, Frank BP, Subaran S, Indig FE, Traub DD, Weeraratna AT. PKC and PKA phosphorylation affect the subcellular localization of claudin-1 in melanoma cells. Int J Med Sci 2009; 6:93-101; PMID:19305641
- Fujita H, Chalubinski M, Rhyner C, Indermitte P, Ing D, Meyer N, Ferstl R, Treis A, Gomez E, Akkaya A, et al. Claudin-1 expression in airway smooth muscle cells exacerbates airway remodeling in asthmatic subjects. J Allergy Clin Immunol 2011; 127:1612-21; PMID:21624620; https://doi.org/10.1016/j.jaci.2011.03.039
- Zwanziger D, Badziong J, Ting S, Mooeller LC, Schmid KW, Siebolts U, Wickenhauser C, Dralle H, Fuehrer D. The impact of CLAUDIN-1 on follicular thyroid carcinoma aggrressiveness. Endocr Relat Cancer 2015; 22:819-30; PMID:26219679; https://doi.org/10.1530/ERC-14-0502
- Ikari A, Watanabe R, Sato T, Taga S, Simobaba S, Yamaguchi M, Yamazaki Y, Endo S, Matsunaga T, Sugatani J. Nuclear distribution of claudin-2 increases cell proliferation in human lung adenocarcinoma cells. Biochim Biophys Acta 2014; 1843:2079-88; PMID:24907662; https://doi.org/10.1016/j.bbamcr.2014.05.017
- Todd MC, Petty HM, King JM, Piana Marshall BN, Sheller RA, Cuevas ME. Overexpression and delocalization of claudin-3 protein in MCF-7 and MDA-MB-415 breast cancer cell lines. Oncol Lett 2015; 10:156-62; PMID:26170992; https://doi.org/10.3892/ol.2015.3160
- Cuevas ME, Gaska JM, Gist AC, King JM, Sheller RA, Todd MC. Estrogen-dependent expression and subcellular localization of the tight junction protein claudin-4 in HEC-1A endometrial cancer cells. Int J Oncol 2015; 47:650-6; PMID:26043767; https://doi.org/10.3892/ijo.2015.3030
- Huerta M, Munoz R, Tapia R, Soto-Reyes E, Ramirez L, Recillas-Targa F, González-Garcia M, Lopez-Bayghen E. Cyclin D1 is transcriptionally down-regulated by ZO-2 via an E box and the transcription factor c-myc. Mol Biol Cell 2017; 18:4826-36; PMID:17881732; https://doi.org/10.1091/mbc.E07-02-0109
- González-Garcia M, Islas S, Contreras RG, Garcia-Villegas MR, Betanzos A, Vega J, Diaz-Quinonez A, Martin-Orozco N, Ortiz-Navarrete V, Creijido M, et al. Molecular characterization of the tight junction protein ZO-1 in MDCK cells. Exp Cell Res 1999; 248:97-109; PMID:10094817
- Acharya P, Beckel J, Ruiz WG, Wang E, Rojas R, Birder L, Apodaca G. Distribution of the tight junction proteins ZO-1, occludin, claudin-4, -8, and -12 in bladder epithelium. Am J Physiol Renal Physiol 2004; 287:F305-18; PMID:15068973; https://doi.org/10.1152/ajprenal.00341.2003
- Fujita H, Sugimoto K, Inatomi S, Maeda T, Osani M, Uchiyama Y, Yamamoto H, Wada T, Kojima T, Yokozaki H, et al. Tight junction proteins claudin-2 and -12 are critical for vitamin D-dependent Ca2+ absorption between enterocytes. Mol Biol Cell 2008; 19:1912-21; PMID:18287530; https://doi.org/10.1091/mbc.E07-09-0973
- Wang W, Tan X, Zhou L, Gao F, Dai X. Involvement of the expression and redistribution of claudin-23 in pancreatic cancer cell dissociation. Mol Med Rep 2010; 3:845-50; PMID:21472324; https://doi.org/10.3892/mmr.2010.334
- Bhansali M, Zhou J, Shemshedini L. TM4SF3 and AR: A nuclear complex that stabilizes both proteins. Mol Endocrinol 2016; 30:13-25; PMID:26649804; https://doi.org/10.1210/me.2015-1075
- Rappa G, Green TM, Lorico A. The nuclear pool of tetraspanin CD9 contributes to mitotic processes in human breast carcinoma. Mol Cancer Res 2014; 12:1840-50; PMID:25103498; https://doi.org/10.1158/1541-7786.MCR-14-0242
- Rappa G, Santos MF, Green TM, Karbanova J, Hassler J, Bai Y, Barsky SH, Corbeil D, Lorico A. Nuclear transport of cancer extracellular vesicle-derived biomaterials through nuclear envelope invagination-associated late endosomes. Oncotarget 2017; 8:14443-61; PMID:28129640; https://doi.org/10.18632/oncotarget.14804
- Ferber EC, Kajita M, Wadlow A, Tobiansky L, Niessen C, Ariga H, Daniel J, Fujita Y. A role for the cleaved cytoplasmic domain of E-cadherin in the nucleus. J Biol Chem 2008; 283:12691-700
- Du W, Liu S, Fan G, Zhao X, Sun Y, Wang T, Zhao R, Wang G, Zhao C, Zhu Y, et al. From cell membrane to the nucleus: an emerging role of E-cadherin in gene transcriptional regulation. J Cell Mol Med 2017; 18:1712-9
- Cukierman L, Meertens L, Bertaux C, Kajumo F, Dragic T. Residues in a highly conserved claudin-1 motif are required for hepatitis C virus entry and mediate the formation of cell-cell contacts. J Virol 2009; 83:5477-84; PMID:19297469; https://doi.org/10.1128/JVI.02262-08
- Furuse M, Hata M, Furuse K, Yoshida Y, Haratake A, Sugitani Y, Noda T, Kubo A, Tsukita S. Claudin-based tight junctions are critical for the mammalian epidermal barrier: a lesson from claudin-1-deficient mice. J Cell Biol 2002; 156:1099-111; PMID:11889141; https://doi.org/10.1083/jcb.200110122
- Meng J, Holdcraft RW, Shima JE, Griswold MD, Braun RE. Androgens regulate the permeability of the blood-testis barrier. Proc Natl Acad Sci USA 2005; 102:16696-700; PMID:16275920; https://doi.org/10.1073/pnas.0506084102
- Resnick MB, Gavilanez M, Newton E, Konkin T, Bhattacharya B, Britt DE, Sabo E, Moss SE. Claudin expression in gastric adenocarcinomas: a tissue microarray study with prognostic correlation. Hum Pathol 2005; 36:886-92; PMID:16112005; https://doi.org/10.1016/j.humpath.2005.05.019
- Chang TL, Ito K, Ko TK, Liu Q, Salto-Tellez M, Yeoh KG, Fukamachi H, Ito Y. Claudin-1 has tumor suppressive activity and is a direct target of RUNX3 in gastric epithelial cells. Gastroenterology 2010; 138:255-65; PMID:19706291; https://doi.org/10.1053/j.gastro.2009.08.044
- Psáder R, Jakab C, Mathe S, Balka G, Papa K, Sterczer A. Expression of claudins in the normal canine gastric mucosa. Acta Vet Hung 2014; 62:13-21; PMID:24334088; https://doi.org/10.1556/AVet.2013.053
- Sanada Y, Oue N, Mitani Y, Yoshida K, Nakayama H, Yasui W. Down-regulation of the claudin-18 gene, identified through serial analysis of gene expression data analysis, in gastric cancer with an intestinal phenotype. J Pathol 2006; 208:633-42; PMID:16435283; https://doi.org/10.1002/path.1922
- Matsuda Y, Semba S, Ueda J, Fuku T, Hasuo T, Chiba H, Sawada N, Kuroda Y, Yokozaki H. Gastric and intestinal claudin expression at the invasive front of gastric carcinoma. Cancer Sci 2007; 98:1014-9; PMID:17459057; https://doi.org/10.1111/j.1349-7006.2007.00490.x
- Darido C, Buchert M, Pannequin J, Bastide P, Zalzali H, Mantamadiotis T, Burgaux J-F, Garambois V, Jay P, Blache P, et al. Defective claudin-7 regulation by Tcf-4 and SOX9 disrupts the polarity and increases the tumorgenicity of colorectal cancer cells. Cancer Res 2008; 68:4258-68; PMID:18519685; https://doi.org/10.1158/0008-5472.CAN-07-5805
- Li WY, Huey CL, Yu ASL. Expression of claudin-7 and -8 along the mouse nephron. Am J Physiol Lung Cell Mol Physiol 2004; 286:F1063-71; PMID:14722018; https://doi.org/10.1152/ajprenal.00384.2003
- Alexandre MD, Lu Q, Chen Y-H. Overexpresssion of claudin-7 decreases the paracellular Cl- conductance and increases the paracellular Na+ conductance in LLC-PK1 cells. J Cell Sci 2005; 118:2683-93; PMID:15928046; https://doi.org/10.1242/jcs.02406
- Gonzalez-Mariscal L, Namorado MDC, Martin D, Sierra G, Reyes JL. The tight junction proteins claudin-7 and -8 display a different subcellular localization at Henle's loops and collecting ducts of rabbit kidney. Nephrol Dial Transplant 2006; 21:2391-8; PMID:16766545; https://doi.org/10.1093/ndt/gfl255
- Coyne CB, Gambling TM, Boucher RC, Carson JL, Johnson LG. Role of claudin interactions in airway tight junctional permeability. Am J Physiol Lung Cell Mol Physiol 2003; 285:L1166-78; PMID:12909588; https://doi.org/10.1152/ajplung.00182.2003
- Marzesco A-M, Dunia I, Pandjaitan R, Recouvreur M, Dauzonne D, Benedetti EL, Louvard D, Zahraoui A. The small GTPase Rab13 regulates assembly of functional tight junctions in epithelial cells. Mol Biol Cell 2002; 13:1819-31; PMID:12058051; https://doi.org/ 02-02-0029
