ABSTRACT
The mammalian intestine harbors a highly complex and abundant ensemble of bacteria that flourish in a nutrient-rich environment while profoundly influencing many aspects of host biology. The intestine coevolved with its resident microbes in a manner where the mucosa developed a barrier function to segregate the resident microbes from the rest of the body, and yet paradoxically, allowing integration of microbial signals for the host benefit. In this review, we provided a comprehensive overview of why the gut microbiota is key to the efficient development and maintenance of the intestinal barrier. We also highlighted how a destabilized equilibrium between gut microbiota and the host may eventuate in a wide range of intestinal diseases characterized by the disrupted intestinal barrier. Finally, the review delineated how microenvironmental changes in the injured mucosa result in an enrichment of a pro-regenerating consortium of bacteria, which augments mucosal wound repair and restoration of barrier functions.
Introduction
The mammalian intestine harbors a taxonomically diverse microbial community, which acts as an accessory organ system with distinct functions fundamentally critical in the well-being of their host.Citation1–Citation5 This symbiotic and mutualistic host-microbiota relationship has evolved to the extent that the intestine has established a barrier function separating the colonized microbes from the systemic tissues, and yet, also providing a gateway for the cross-talk between the two. The gut barrier function is central to health and breakdown of the barrier is involved in a wide variety of clinical conditions.Citation5–Citation8 Although the intestinal barrier depends on the coordinated contributions from a complex network of cellular, immunological, biochemical or microbial factors, ultimately it is the monolayer of tightly junctioned columnar epithelial cells that confer the selective barrier function.Citation9–Citation11 Compromised epithelial barrier is a hallmark of IBD, irritable bowel syndrome, obesity and the metabolic syndrome and necrotizing enterocolitis.Citation7,Citation12–Citation14 However, mechanistically, it is not well understood how the intestinal resident microbiota influences the efficient maintenance and/or repair of the epithelial barrier. Damage to the barrier -the intestinal wound- is observed in IBD, enteric infections, as well as following surgical trauma, and environmental insults. Intestinal wound regeneration is a process that restores homeostasis through a complex process of cellular proliferation and migration.Citation15–Citation17 This repair process is orchestrated by a coordinated network of different cellular, immunological, biochemical, and also microbial influences.Citation18 The mechanisms of commensal gut resident microbiota in the restoration of epithelial barrier function and epithelial wound healing is only recently being delineated. Interestingly, several recent studies described the dynamic changes in the physiology and inflammatory responses of the wound microenvironmentCitation19-Citation22 and the consequent alterations in the bacterial population. Furthermore, it was determined that these events drive a spatiotemporal alteration of the microbial community structure in the mucosal wound microenvironments resulting in the enrichment of a mucosa-associated microbial consortium, which in turn, augments gut epithelial wound healing.Citation19 This review will particularly focus on the importance of gut microbiota and their products on (1) epithelial barrier functions, (2) resolution of mucosal inflammation, and (3) repair of intestinal wounds.
The composition of the commensal bacteria of the intestine during homeostasis
Mammals are not autonomous entities, rather can be considered holobiontsCitation23 that harbor microbial communities of astounding density and taxonomic diversity.Citation2,Citation24 Holobionts represent the host macroorganism which exists a close symbiosis with the associated microorganisms. The microbiota refers to all of the microorganisms associated with an animal or plant host. The microbiome, on the other hand, is defined as the sum of the genetic information of the microbiota. The components of the gut microbiota – bacteria, viruses, and eukaryotes – interact with one another and with the host immune system that ensure the intestinal homeostasis, and also, at times provoke the development of the disease. This review will focus on the fundamental role of the intestinal bacterial population during repair of the injured mucosa, and also in the restoration of homeostasis. Therefore, the term microbiota will refer to the commensal enteric bacteria in this review.
The past decade has observed a dramatic rise in meta’omics studies of IBD and related inflammatory diseases. As a result, we are beginning to develop an ecological or community-wide understanding of the critical role of the microbiome in intestinal diseases.Citation25,Citation26 Recently, these findings have begun to be translated into a functional mechanistic interpretation of the microbiome in the development as well as exacerbation of the inflammatory diseases.Citation7,Citation13,Citation25,Citation26 The gut microbiota is currently being surveyed using 16S rRNA gene amplicon sequencing or whole genome sequencing (WGS) techniques. 16S rRNA gene sequencing is popularly used for phylogenetic reconstruction and quantification of microbial diversity. In contrast, WGS explores the functions of the encoded genetic material as well as assesses bacterial diversity.Citation7,Citation24,Citation27,Citation28
The neonatal intestinal tract is rapidly colonized by an array of commensal enteric bacteria after birth eventually forming a complex and dynamic ecosystem. However, the temporal patterns of these changes remain unique to each infant.Citation29–Citation32 Furthermore, by around 2.5 years of age, the composition, diversity and functional capabilities of the infant microbiota resemble those of adult.Citation30–Citation32 Members of the microbiota may exist in a planktonic state, free-living in the luminal stream; they may be partially adherent to the intestinal mucosa or mucus layer; or they may be resident in the cecal and proximal-colonic crypt structures. The total number of commensal enteric bacteria vary greatly from ~ 10Citation11,Citation12 cells/gram of intestinal luminal content within the ascending colon, to ~ 10Citation7,Citation8 in the distal ileum, and ~ 10Citation2,Citation3 in proximal ileum and jejunum. Recent human studies identified more than 2100 species classified into 12 different phyla. However, high throughput sequencing analyses 16S rRNA genes have revealed that more than 90% of bacterial species found within the gut belong to 4 phyla including Proteobacteria, Actinobacteria, Firmicutes and Bacteroidetes.Citation24 The higher redundancy of the gut bacterial species is reflected in the richness and diversity of the microbial community and their genomes, which ensure diverse functionality and overall stability. Nonetheless, the relatively resilient composition of the adult gut microbiota is still subject to perturbation by different factors including diet and host genetics leading to the alteration in microbial community structure.Citation30,Citation31,Citation33
At homeostasis, the composition and number of the intestinal microbiota community are shaped by physiological (oxygen, reactive oxygen species, acids), biochemical (enzymes, metabolites), nutritional and immunological gradients (antimicrobials, sIgA) along the transverse and longitudinal axis of the GI tract.Citation21,Citation22,Citation34 For example, the pH in the mouth is close to neutral and the saliva contains enzymes inhibiting bacterial growth. On the other hand, the stomach is extremely acidic (pH 2). The pH gradually increases in the small intestine and the colon (pH 4–5 and 6.7 respectively.Citation35 Hence, within single individuals, the bacterial community of the small intestine is different from that of the colon. Similarly, the luminal and mucosal compositions of the microbiota are significantly divergent. Moreover, several environmental factors have been implicated in influencing the microbiota including geographical location, surgery, smoking, depression and living arrangements (urban or rural). In addition, antibiotics and other drugs alter the physiology and microbial community of the active human gut microbiome. Therefore, the relatively stable composition of the adult gut microbiota is still subject to perturbation by different factors.Citation30,Citation33
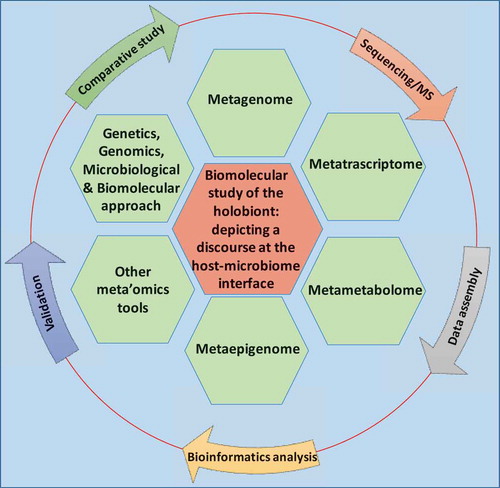
Complex microbial communities are an integral component of our bodies in health and disease. In fact, the taxonomically diverse human microbiome complements us with over 100 times more genes than those in our genome. Hence, microbes, their metagenome, and their biochemical activities are crucial factors determining the balance between health and disease state of the hosts.Citation4 Recently, an extensive catalog of the functional capacity of the human gut microbiome has been determined, where 9,879,896 genes were identified.Citation24,Citation36,Citation37 However, the microbiota’s nearly 10 million genes and their metabolic functions remained largely unrecognized.Citation7,Citation36,Citation38,Citation39 The early studies involving microbial community composition of a given ecosystem has moved well beyond this observational stage, progressing towards functional analysis of the genes and pathways carried by a microbial community, i.e., metagenome. High-throughput analytical approaches, such as metagenomics, metatranscriptomics, metametabolomics and other meta’omics analysis provide comprehensive technologies to explore functional and biomolecular activities of the microbiome and its host.Citation39,Citation28,(, Mass spectrometry-based and chromatography-based techniques have recently been applied to host–microbiome studies for targeted and untargeted metabolomic and metaproteomic strategies to systematically discover the chemical diversity of the commensal bacteria and the hosts.Citation28 This multidisciplinary approach in bioinformatics and molecular biology to discover genes, products and pathways of the holoorganism are important for dissecting disease mechanisms as well as pivotal in the development of the potential therapeutic, diagnostic, or prognostic applications ().
Function of gut microbiota during intestinal homeostasis
Intestinal barrier function is key to the maintenance of gut homeostasis. This is consistent with the fact that many of the IBD susceptibility genes are related to intestinal barrier function. Furthermore, investigations with germ-free mice have demonstrated that the resident bacteria profoundly influence epithelial barrier function, metabolism, proliferation, differentiation and survival.Citation1,Citation2,Citation40–Citation42 Although recent studies have implicated the gut microbiota in numerous aspects of the proper development and efficient functions of the intestine, this section will limit the discussion on the influence of commensal microbiota on (1) perception of gut microbiota (2) epithelial proliferation and turnover, (3) epithelial selective barrier, and (4) mucus barrier.
Intestinal perception of the microbiota
Intestinal homeostasis, and also, the development of many inflammatory diseases, are regulated in part by tolerance towards the commensal microbiota that influences the development and training of innate and adaptive immune systems,Citation43 processes modulated by the broad class of pattern recognition receptors (PRRs).Citation44 PRRs include Toll-like receptors (TLRs), NOD-like receptors (NLRs), C-type lectin receptors (CLRs) and also some G-protein coupled receptors (GPCRs).Citation45 TLRs are characterized structurally by the presence of leucine-rich repeat (LRR) motif in their extracellular ligand-binding domain, a transmembrane domain and a conserved Toll/IL-1R homology domain in the cytosolic region for signal transduction. PRRs recognize microbe-associated molecular patterns (MAMPs) which are conserved molecular motifs derived from commensals and pathogens. TLRs initiate immune responses via signaling adaptors, such as myeloid-differentiation primary response gene 88 (MyD88), which is required for several TLRs.Citation45 Additinonally, G-protein coupled receptors (GPCRs) are seven transmembrane (7 TM) receptors, and upon ligand binding, signal inside the cell through a complex network of signaling, adaptor and regulatory proteins, including heterotrimeric G proteins. Several GPCRs, including N-formyl peptide receptors (FPRs), have been demonstrated to mediate host-microbiota interactions by sensing microbial peptides, small molecules, and metabolites.
Microbial effects on epithelial proliferation and protection via epithelial prrs
During homeostasis, the epithelial surface is continually renewed by the proliferation and differentiation of pluripotent intestinal epithelial stem cells of the crypts. The differentiation results in the development of six specialized cell types with defined functions: enterocytes, enteroendocrine cells, Paneth cells, tuft cells, goblet cells and microfold (M) cells. The intestinal epithelium is equipped with different PRRs that recognizes microbial components, and ultimately, elicits cell signaling pathways involved in cellular proliferation. Rakoff-Nahoum et al. have established that commensal enteric bacteria are recognized by TLRs (TLR2 and TLR4) and that this interaction plays a crucial role in the maintenance of intestinal epithelial homeostasis as well as protection from mucosal injuries in a mouse model of chemically-induced colitis.Citation46 TLR/MyD88 and Nucleotide oligomerization domain (NOD)-like receptors (NLRs) are essential for the regulation of inflammation and production of antimicrobial proteins, such as RegIIIγ, cathelicidins, and defensins, which prevent encroachment of epithelial cells by gut microbiota and thereby maintain homeostasis.Citation47 For example, it has been previously shown that the development of gut inflammation in flagellin receptor TLR5-deficient mice was associated with failure to restrict pro-inflammatory proteobacteria, particularly in the post-weaning period.Citation48,Citation49 Direct activation of epithelial TLR signaling has been shown to alter intestinal homeostasis by affecting the proliferation and apoptosis of epithelial cells.Citation45,Citation50 How gut microbiota drives the intestinal epithelial cell self-renewal and turnover via the PRRs, and hence maintain, epithelial homeostasis, are matters of immense current interest.
G-protein-coupled receptors (GPCRs) mediate myriad cellular functions including proliferation, differentiation, adhesion, and migration and play pivotal roles in development, homeostasis, inflammation, immunity, and oncogenesis. GPCRs are also important pharmaceutical targets, as about half of the drugs currently used are either agonists or antagonists for various GPCRs. Recent studies have shown that GPCRs can mediate host-microbiota interactions by sensing microbial products and small molecules. N-formyl peptide receptors (FPRs) can sense and bind an array of formylated and nonformylated peptides as well as nonpeptide ligands synthesized either endogenously by the host or exogenously by the microbiota.Citation51 Although FPRs are not considered as typical leucine rich repeat bearing PRRs such as TLRs and Nods, the FPRs are clearly, by definition, postulated to function as the pattern recognition receptors that recognize and respond to bacterial products. FPRs are expressed on the surface of neutrophils and macrophages, where they perceive bacterial products and stimulate phagocyte functions. While aberrant PRR activation, including TLRs and NOD2 is associated with the onset of inflammation,Citation52,Citation53 we have shown FPR can induce non-proinflammatory signaling in epithelial cells in vitro and in vivo.Citation54 Humans have three FPR paralogs.Citation55 FPR1 is the high-affinity receptor for the bacterially synthesized fMLF. FPR2 has been shown to respond to endogenous ligands including annexin A1, lipoxin A4, and serum amyloid A.Citation55,Citation56 Importantly, FPRs are also expressed in and localized to the apical and basolateral surface of the intestinal epithelial cells.Citation57 Although FPRs are well characterized in phagocytes, their roles in intestinal epithelial cells are not well understood. Recent studies demonstrated that commensal bacteria can activate FPR1 to stimulate the proliferation of intestinal epithelial cells in a redox-dependent manner.Citation58,Citation59 In a recent review article (please see refs.Citation53 and Citation59), Jones et al. comprehensively discussed how physiological level of reactive oxygen species (ROS) induced by the gut microbiota promotes epithelial proliferation and intestinal barrier functions.Citation59 The study demonstrated that human commensal lactobacilli induce ROS generation in intestinal epithelial cells via the catalytic action of Nox enzymes ultimately resulting in the efficient cellular proliferation.Citation60 Moreover, using a Drosophila model, mono-colonized Lactobacillus plantarum induced the dNox-dependent generation of cellular ROS, and ROS-stimulated epithelial cell proliferation in axenic larval flies.Citation60 Furthermore, L. rhamnosus GG was shown to induce ROS generation and cell proliferation in the murine intestine.Citation58,Citation61,Citation62 Importantly, it has been shown that specific probiotic bacteria activate FPR1 and epithelial NOX1 to stimulate physiological ROS that increase phosphorylation of ERKCitation58,Citation63 via oxidative inactivation of the redox-sensitive and ERK-specific phosphatase called DUSP3 (dual specific phosphatase 3), and consequently induce cell proliferation.Citation58 The molecular mechanism by which ROS control cell signaling pathways is by the oxidation of reactive cysteine residues within regulatory proteins.Citation64 A number of redox-sensitive regulatory proteins have been characterized and shown to be sensitive to microbiota-induced ROS generation, which includes the lipid phosphatase (PTEN), low-molecular weight (LMW)-PTP, and enzymes involved in ubiquitination processes.Citation61,Citation62,Citation65,Citation66 Together, these data from the cultured human enterocytes, Drosophila and murine models demonstrate that commensal bacteria and their products activate ROS signaling in an FPR-dependent manner and define a conserved mechanism by which cellular ROS enhance epithelial development and homeostasis. In addition, these discoveries establish the function for PRRs, including FPRs, in perceiving the commensal enteric microbial products and mediating an efficient epithelial turnover and barrier integrity.
Microbial effects on intestinal epithelial barrier
Intestinal epithelial cells establish and maintain a continuous barrier to avert potential encroachment of harmful luminal components; nonetheless, it remains selectively and dynamically permeable. The tasks of regulating paracellular transport and intercellular connections between the polarized columnar epithelial cells are achieved by a series of intercellular junctions, which are conferred by complex junctional structures, the tight junctions (TJs), adherens junctions, the desmosomes and the gap junctions.Citation10,Citation11,Citation14 A network of actin and myosin supports these types of junctions, which are composed of transmembrane proteins, scaffolding proteins, and junctional molecules.Citation67 The physiological structures, properties, and functions of the epithelial apical junctional complex have been reviewed extensively elsewhere (please see refs. Citation9,Citation11,Citation14, and Citation67). In response to different stimuli, these junctional complexes also dynamically change epithelial permeability by rapid assembly and disassembly.Citation68,Citation69 Increased epithelial permeability and dysfunctional epithelial integrity are widely acknowledged to play a critical role in the pathophysiology of various intestinal disorders, including the enteric infectious disease, inflammatory bowel disease (IBD), irritable bowel syndrome obesity Graft-versus-host disease (GVHD), celiac disease, the metabolic syndrome, necrotising enterocolitis and even in human immunodeficiency virus HIV/AIDS syndrome.Citation14,Citation70,Citation71 Transcriptional, structural and functional changes have been reported in the TJs of intestinal epithelial cells in patients with IBD, including claudin proteins.Citation68,Citation70
It is widely reported that commensal enteric bacteria can have profound effects on epithelial permeability and integrity, particularly, on TJ repair and maintenance ().Citation72–Citation76 For example, in 1996, Mao et al. demonstrated that the human L. plantarum and the rat-originating strain L. reuteri could reduce permeability dysfunction in a methotrexate-induced colitis model in the rat. Intriguingly, this study determined that the rats administered L. plantarum show less Enterobacteriaceae and Gram-negative anaerobes in the intestine. Ewaschuk et al. demonstrated that soluble factors produced by B. infantis elevated expression of occludin, which resulted in decreased paracellular permeability.Citation77 It has been shown that bifidobacteria isolated from infants and cultured on human milk oligosaccharides (HMO) increased expression of ZO-1, occludin in HT-29 cells. Furthermore, both B. infantis and B. bifidum grown on HMO prevented the significant intracellular redistribution of occludin.Citation78 In addition, a recent study demonstrated that specific B. animalis strain protected barrier functions by decreasing intestinal permeability in a mouse model characterized by low grade inflammation.Citation79 Importantly, this strain specifically normalized the level of several TJ proteins, in particular for claudin-4, and thereby, potently ameliorated compromised epithelial barrier in the mouse. A separate study demonstrated that L. rhamnosus and Faecalibacterium prausnitzii exhibit a protective effect in a murine model of induced-barrier hyper-permeability. An oral administration of these bacteria restored epithelial permeability defect by increasing the levels of tight junction proteins occludin and E-cadherin in murine colonic epithelial cells. The authors also postulated that these events were mediated by the microbiota-induced reduction of NF-κB activation leading to the amelioration of epithelial permeability dysfunction.Citation80 Using a neonatal mouse NEC model, Bergmann et al. demonstrated that bifidobacteria stabilize claudins at tight junctions and prevent epithelial barrier dysfunction. In mice, administration of B. infantis attenuated increases in intestinal permeability, preserved claudin 4 and occludin localization at TJs, and decreased NEC incidence in mice.Citation81 Taken together, this body of data collectively demonstrates that endogenous bacteria can have beneficial effects on the physiology of the intestinal barrier ().
Table 1. Microbial effects on intestinal epithelial barrier.
Microbial effects on mucus barrier
The intestinal epithelium surface is covered by a mucus layer which differs in composition, organization, and thickness at different locations along the intestinal tract, and represents the first site of mucosal contact with the gut microbiota. The mucus layer is a dynamic and complex structure that is mainly composed of the glycoprotein mucin-2 (MUC2). MUC2 is a large 5,197-amino acid protein core containing two mucin domains rich in proline, threonine, and serine repeats that become heavily O-glycosylated.Citation82 Due to the cross-links, MUC2 forms a large, net-like polymer that is secreted by the goblet cells. Mice deficient in Muc2 demonstrate a defective mucus layer and development of spontaneous colitis. Importantly, biopsy samples of ulcerative colitis patients also show alterations in MUC2 expression and depletion of the mucus layer.Citation83 The colonic mucus consists of two layers, a loose outer layer, and a dense epithelium-attached inner layer.Citation84 In the distal colon, the inner layer acts as a physical barrier restricting bacteria from accessing the epithelium, whereas the outer layer is the habitat and food source of a distinct population of commensal microbes. In contrast, the small intestine is coated with the single thin layer of mucus, which is loose and penetrable by a much less numerous and diverse microbiotaCitation85,Citation86, likely due to the secreted antimicrobial peptides from small intestinal Paneth cells.Citation47 In the colon, some commensal bacterial contact with the epithelial cells occursCitation87 For example, the close association of bacteria with the epithelial surface and within crypts have been demonstrated in the mouse proximal colon and cecum.Citation88,Citation89 The inner mucus layer of this region is less dense and more penetrable by bacteria.Citation86 Interestingly, the crypt-dwelling bacteria are distinct from the luminal community, and predominantly include Proteobacteria, Firmicutes, some Bacteroidetes. It is postulated that the deep channel of the crypts and transverse folds of the proximal colon provide a protected microhabitat for these bacteria.Citation89 Similarly, it has been recently shown that during regeneration of epithelial wounds, a consortium of anaerobic, mucinophilic bacteria transiently bloom in the close proximity of the murine intestinal epithelial cells.Citation19 In addition, the segmented filamentous bacteria (SFB) adhere tightly to epithelial cells of villi and Peyer’s patches in the ileum of mice, which confers pronounced immunomodulatory effects to the host.Citation90
Certain commensal microbes are enzymatically equipped to utilize mucin glycans as their energy source. However, an aberrant abundance of these bacteria has been reported to be associated with different inflammatory diseases. For example, Ruminococcus gnavus and Ruminococcus torques, two prominent mucin degraders, are increased in IBD.Citation91,Citation92 Interestingly, a number of studies also provided evidence that gut microbiota is essential for the proper function and integrity of the intestinal mucus layers. For example, the mucus layer of the germ-free mice is thinner in comparison with the conventionally raised animals, and monocolonization of the germ-free mice with a commensal bacteria resulted in the expansion of the volume of mucus layer.Citation93,Citation94 Furthermore, germ-free mice monocolonized with short chain fatty acids (SCFA)-producing B. thetaiotaomicron or Faecalibacterium prausnitzii increased goblet cell differentiation and elevated mucus production.Citation93 Thus, during intestinal homeostasis, the mucin-utilizing bacteria are required for the proper turnover of the mucin protein to ensure an efficient barrier. Paradoxically, the aberrant abundance of the mucin-degrading bacteria may result in the dysfunction in the mucus barrier.
Together, these studies emphasized the importance of intestinal commensal microbiota, which modulates intestinal barrier function by regulating epithelial cell self-renewal and turnover, production of mucus layer or importantly, by influencing tight junctions of the intestinal epithelial cells.
Microbial effects on intestinal inflammation
Intestinal homeostasis can be disturbed by the absence or over-representation of certain bacterial groups or altered abundance of microbial genes and products in the intestine. An unfavorable imbalance of microbial composition, altered diversity and defective functons of the intestinal microbiota is termed dysbiosis.Citation6–Citation8,Citation12,Citation13 Microbial dysbiosis is intricately associated with the host immune system and barrier functions. As a result, dysbiosis of the gut microbiota is associated with different diseases including IBD, liver diseases, gastrointestinal cancers, metabolic diseases, respiratory diseases, mental or psychological diseases, autoimmune diseases, and also infectious diseases.Citation7,Citation13
Dysbiosis of gut microbiota in IBD
This microbial dysbiosis in IBD is often attributed by the decrease of bacteria with anti-inflammatory capacities and the elevated abundance of bacteria with inflammatory potentials. Furthermore, dysbiosis can also be associated with major impairments in many crucial microbial metabolic functions with critical impact on the host.Citation6,Citation8,Citation13,Citation25,Citation95 For example, a lower abundance of Firmicutes and the increases in abundance of Proteobacteria and Bacteroidetes have been reported to be associated with IBD.Citation96 It has been reported that F. prausnitzii, Blautia faecis, Roseburia inulinivorans, and C. lavalense, all of which belong to Firmicutes, are depleted in patients with CD.Citation8,Citation97 Moreover, the number of F. prausnitzii is correlated with the risk of relapse of ileal CD after surgery. Furthermore, a significant decrease of Roseburia spp. was shown in the gut microbiota of healthy individuals with a high genetic risk for IBD. Interestingly, F. prausnitzii has been reported to have a pronounced anti-inflammatory effect by fermenting carbohydrate into butyrate, an abundant short chain fatty acid. Moreover, human peripheral blood mononuclear cells stimulated with F. prausnitzii induce the production of IL-10 and inhibit the generation of inflammatory cytokines, such as IL-12 and IFN-γ.Citation98 Similarly, a decreased abundance of Akkermansia has been reported in both UC and CD patients.Citation99 Akkermansia has been demonstrated to induce anti-inflammatory immune response and restore intestinal barrier defects.Citation100,Citation101 In contrast, a relative increase in pro-inflammatory Proteobacteria, mainly E. coli, was observed in CD patients, in particular, on mucosa-associated microbiota compared to fecal samples.Citation102 CD-associated E. coli with pro-inflammatory properties includes adhesion-invasive E. coli (AIEC), which was originally isolated from adult CD patients. In addition, increased abundance of mucosa-associated bacteria in IBD have been reported. Different mucolytic bacteria, including Ruminococcus gnavas and R. torques, may degrade the mucin layers in IBD patients, and thereby, may facilitate the colonization of the epithelial surface by an elevated abundance of proinflammatory bacteria.Citation92,Citation99,Citation103 Thus the microbial dysbiosis is often contributed by the aberrant balance between the bacteria with anti-inflammatory capacities and the bacteria with pro-inflammatory potentials.
Microbial effects on the pathogenesis of intestinal inflammation and colitis
Although the precise cause of IBD remains unknown, the most accepted hypothesis of IBD pathogenesis is that an aberrant immune response against the gut commensal is elicited by environmental factors in a genetically susceptible host.Citation12,Citation13,Citation104 The disruption of the intestinal homeostasis, and thereby the development of the inflammation is initiated in part by the failure of tolerance towards the commensal microbiota that influences the development and training of innate and adaptive immune systems.Citation43,Citation44,Citation105 Therefore, many of the known IBD susceptibility genes are associated with the recognition and processing of microbes, which is correlated with a role of the gut microbiota in the pathogenesis of IBD.
Recent studies using different animal models of inflammatory bowel diseases have provided evidence to establish a causal link between gut microbiota, aberrant immune response and disrupted barrier functions in a genetically susceptible host. For example, Kim et al. performed a mono-colonization study, in which a single strain of bacteria was inoculated into germ-free IL-10 knockout mice. The authors demonstrated that different commensal bacterial species selectively initiate CD4 + T cell-mediated intestinal inflammation with distinctly different kinetics and anatomic distribution in the same host. Specifically, E. coli induced cecal inflammation, Enterococcus faecalis induced distal colitis, and Pseudomonas fluorescens failed to cause colitis.Citation106 A separate study also reported that the mouse enteric commensal bacteria Helicobacter hepaticus exacerbated colitis in IL-10-deficient mice. In addition, a disproportionate outgrowth of a non-culturable Clostridium-related species of segmented filamentous bacteria promotes T cell-mediated pro-inflammatory immune responses.Citation107 These studies provided the evidence that the aberrant microbial composition can elicit intestinal immune responses even in an animal with the same genetic background. Interestingly, Garrett et al. demonstrated that mice deficient in both Tbx21/T-bet and Rag developed spontaneous colitis, which was ameliorated by the administration of antibiotics. Tbx21/T-bet is an essential transcription factor for Th1 differentiation, and Rag is central to the development of the acquired immune system. Importantly, wild-type mice co-housed with colitis-prone T-bet/Rag deficient mice also demonstrated similar colonic inflammaiton and colits. These findings suggested that a dysbiotic gut microbiota is communicable and can cause intestinal inflammation without genetic manipulation.Citation108 In addition, these results highlight that gut microbial dysbiosis may contribute to the intestinal inflammation.
Disequilibrium in the abundance microbial metabolites and small molecules are also involved in the regulation of inflammatory response. Microbially fermented short-chain fatty acids (SCFAs) can bind and activate GPCRs, which are expressed on the surface of epithelial cells and immune cells and have numerous regulatory functions on host physiology and immunity. These GPCRs include GPR41, GPR43 and GPR109A. One of the important functions of the SCFA-sensing GPCRs is the regulation of the effector and regulatory T cell development and expansion. Specifically, SCFAs influence peripheral T cell numbers and function, particularly regulatory T (Treg) cells, through epigenetic regulator histone deacetylases (HDAC) inhibition. SCFA-dependent inhibition of HDAC9 increased forkhead box P3 (FOXP3) expression, elevated Treg cell numbers, and finally, enhanced the immunosuppressive function of FOXP3+ Treg cells, which inhibits inflammatory response in a murine model of colitis.Citation109 In addition, activation of IEC-specific GPR43 and GPR109A by microbial SCFAs increases production of the cytokine IL-18, and further, prevents inflammation in a murine model of colitis via the activation of inflammasome assembly.Citation110–Citation112 Thus, a dysbiotic gut microbiota characterized by the depletion of SCFA producing bacteria may result in the defective immunosuppressive function of various cell types leading to the development of intestinal inflammation and colitis. Findings from animal models have revealed diverse roles of the gut microbiota in protective or pro-inflammatory functions. As discussed above, recent studies performed in animal models clearly provided data supporting an etiological role of gut microbiota in development of model colitis. However, a direct cause and effect relationship between dysbiosis and IBD has not been definitively established in humans.Citation104
Effect of gut microbiota on intestinal epithelial cells during repair of injured mucosa
Mucosal wound repair is coordinated by a spatiotemporal network of different cell types, which include the epithelium itself, inflammatory cells, endothelial cells, and mesenchymal cells. Intermolecular and intercellular communication among these cells creates a microenvironment conducive for epithelial regeneration and mucosal healing, which is characterized by efficient migration and proliferation of intestinal epithelial cells followed by tissue remodeling.Citation113 Recent studies provided evidence that commensal microbiota profoundly influences the repair of the injured mucosa by regulating the functions of the wound-associated cellular network and ultimately modulating migration and proliferation of epithelial cells adjacent to the wounded area.
Effect of microbiota on epithelial migration during wound healing
In larger wounds, the denuded mucosal surface is initially covered by the migrating epithelial cells adjacent to the wound. The migration of epithelial cells as a sheet is also referred to as ‘‘epithelial restitution’’, which requires dynamic and coordinated remodeling of cell-cell and cell-matrix adhesions. Within minutes to hours, epithelial restitution begins when epithelial cells undergo de-differentiation, reorganize the actin cytoskeleton and migrate to cover the damaged area.Citation114 The organization and remodeling of the actin cytoskeleton are controlled by the Rho family of small GTPases, which includes Rho, Rac, and Cdc42. These proteins regulate the formation of stress fibers, lamellipodia, and filopodia.Citation115 In addition, formyl peptide receptor-1 (FPR1) signaling activates Rac1 and Rho kinase to regulate intestinal epithelial cell migration and wound repair.Citation57,Citation116
Investigations in germ-free mice revealed an impaired rate of intestinal epithelial cell migration, a crucial cellular process of wound repair.Citation15,Citation117 This finding demonstrated that commensal bacteria are required for efficient migration and wound healing. Cell migration is regulated by a coordinated restructuring of the actin cytoskeleton at the advancing edge of the cell called focal adhesions (FA) of the extracellular matrix. FAs are large macromolecular assemblies that link the extracellular matrix (ECM) to the cytoplasmic cytoskeleton of an interacting cell. Cell migration, and thus wound repair, is critically dependent upon the dynamic assembly and disassembly of focal adhesions.Citation113 The dynamics of the FA assembly is controlled by an enzyme called focal adhesion kinase (FAK), which is a protein tyrosine kinase. FAK remains in its inactive state under the dephosphorylating influences of the redox-sensitive tyrosine phosphatases LMW-PTPase and SHP-2. Furthermore, interactions between growth factors and integrins at the basement membrane trigger Nox1-dependent cellular ROS production which inactivates PTPase. These events result in the elevated levels of phosphorylated FAK thereby initiating FA turnover and cell motility. We demonstrated that specific commensal bacteria induce the generation of ROS in intestinal epithelia, especially elevated at the leading edge of the migrating epithelial sheet, where there is constant remodeling of the actin and turnover of FA. Furthermore, our data demonstrated that the bacteria-mediated ROS generation induced rapid and transient oxidative inactivation of target cysteines in the redox-sensitive tyrosine phosphatases, LMW-PTP and SHP-2. Both of these are known regulators of FAK phosphorylation, and therefore, FAK activity.Citation62 Thus, these events resulted in increased phosphorylation of FAK. Consequently, phosphorylation of FAK substrate proteins, focal adhesion formation, and cell migration were all significantly enhanced in response to commensal bacteria in both in vitro and in vivo models of wound closure. These results supported that commensal bacteria regulate cell migration and restoration of intestinal barrier functions via induced generation of ROS in epithelial cells.
The NADPH oxidases (Nox) enzymes are widely conserved across the animal, and plant kingdoms. Cellular Nox enzymes often serves as the catalytic sources for ROS generation in response to bacterial stimuli. Activation of NOX2 is responsible for neutrophil and macrophage ROS generation. However, apart from the microbiocidal effect at a high concentration in mammalian cells, the physiological level of ROS may also serve as critical secondary messengers in multiple signal transduction pathways. In humans, paralogs of NOX2 are found in many tissues, two of which are NOX1 and DUOX2. Both of these genes are mostly expressed in the colonic tissue. Recently, Leoni at al. demonstrated that FPR ligand, annexin A1, and its cleavage product Ac2-26, mediate activation of ROS by activating the intestinal epithelial NOX1.Citation116 Furthermore, the study showed that epithelial cell migration was regulated by this signaling cascade through oxidative inactivation of the redox-sensitive regulatory phosphatases PTEN and PTP-PEST, with consequent activation of focal adhesion kinase (FAK) and paxillin.Citation116 In vivo studies using intestinal epithelial-specific Nox1−/-IEC mice demonstrated defects in intestinal mucosal wound repair, while systemic administration of ANXA1 promoted wound recovery in a NOX1-dependent fashion.Citation116 Subsequently, we reported that specific members of the gut microbiota stimulate FPR1 on intestinal epithelial cells and generate reactive oxygen species (ROS) via enterocyte NOX1.Citation58 Ultimately, NOX1-derived ROS caused a rapid increase in the level of phosphorylated FAK and enhanced epithelial migration and promoted wound closure in mouse models.Citation62,Citation63 Thus, our work demonstrated that the enteric microbiota activates FAK and thereby enhances epithelial restitution and promotes epithelial repair of mucosal wounds in a redox-dependent manner.Citation59,Citation62,Citation63,Citation116 Interestingly, germ-free mice show diminished expression of NOX1 and DUOX2 genes along the intestinal tract in comparison with the conventionally raised mice, suggesting the microbiota can stimulate expression of these enzymes.Citation118 Moreover, the promoter region for NOX-1 contains binding elements for AP-1, NFκB, CREB, STATs, and Interferon, indicating a role of cytokine-dependent regulation of gene expression. These studies support that microbiota has a crucial function in the regulation of the NOX-mediated and redox-dependent cellular signaling either by triggering receptor-mediated functional activation or by modulating expression of Nox genes.
Effect of microbiota on epithelial proliferation during wound healing
Epithelial proliferation is an integral cellular process for the repair of larger wounds. The intestinal epithelium is comprised of simple columnar epithelial cells that are highly dynamic as they are continually renewed by stem cells residing in the crypt units. These cells are located in the crypts adjacent to the wounded area and undergo active proliferation to increase the number of proliferating progenitor cells within the crypts (). Ultimately, they emanate out of the crypts as a monolayer of non-proliferating wound-associated epithelial cells (), which migrate further and re-epithelialize the injured area, and thereby contribute to mucosal wound closure.Citation17,Citation63,Citation119 Many studies demonstrated that various growth factors including transforming growth factor (TGF)-α and TGF-β, epidermal growth factor (EGF), insulin-like growth factor (IGF), hepatocyte growth factor (HGF), keratinocyte growth factor (KGF), fibroblast growth factor (FGF), the cytokine interleukin (IL)-1β and IL-2, and trefoil peptides enhance proliferation of IECs. Citation120–Citation123 Remarkably, germ-free mice show a decreased level of TGF-β expression in comparison with the conventional mice. Besides, several clostridial, bifidobacteria and lactobacilli species were shown to modulate TGF-β, KGF, and EGF–mediated cellular functions in the colonic epithelial cells.
Figure 2. Host-microbiome interface of the mucosal wounds: dynamic changes in physiological and inflammatory responses of the wound microenvironment drive a spatiotemporal alteration of the microbial community structure resulting in the enrichment of a mucosa-associated microbial consortium, which in turn augments re-epithelialization of the mucosa, achieved by enhanced migration and subsequent proliferation of epithelial cells.
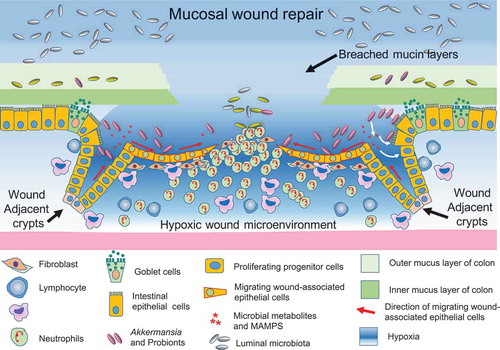
Activation of specific cell signaling pathways is involved in intestinal epithelial wound repair. For example, TGF-α promotes enterocyte proliferation through the activation of extracellular signal-regulated kinase ERK1/ERK2 mitogen-activated protein kinase (MAPK).Citation124 Similarly, FPRs have been shown to be important for intestinal epithelial cell proliferation and wound closure via the activation of ERK1/ERK2 MAPK. Although the role of FPRs in microbial perception and effector function in phagocytes goes back decades, only recently have they been shown to promote wound closure in colonic, stomach, lung and retinal epithelial cells. Recently, FPR-deficient mice have been reported to show an exacerbated colonic epithelial injury and delayed repair in a mouse model of chemically induced mucosal injury. In addition, during the resolution phase of inflammation, there is a temporal increase in pro-resolving mediators including resolvin D1 and lipoxin A4, which activate resolution via the FPR2 receptor.Citation63,Citation113,Citation116 Importantly, by using a mouse model of wound regeneration, we recently demonstrated that specific members of the gut microbiota promoted wound closure by stimulating enhanced proliferation of intestinal epithelial cells in the crypts which are located adjacent to the injured mucosa. Furthermore, our study demonstrated that the microbiota-induced proliferation of the crypt epithelial cells resulted from the rapid phosphorylation of ERK1/ERK2 MAPK, which requires activation of the FPR1 and enterocyte-specific NOX1-mediated redox axis.Citation58,Citation60,Citation61,Citation63,Citation125 These studies support that commensal microbiota are important for efficient proliferation and regeneration of intestinal wounds.
Effect of microbial mamps on cytoprotection and control of epithelial proliferation during wound healing
The innate immune system perceives the microbial invasion or epithelial injury via PRRs and confers immediate cellular responses during epithelial reneration. For example, in 2004, Rakoff-Nahoum et al. have demonstrated that commensal bacterial recognition by TLRs plays a crucial role in the protection of intestinal epithelia.Citation46,Citation126 This was the first evidence of microbial effect, particularly the released-MAMPs, in preserving gut homeostasis, promoting tissue repair and host survival upon recognition of bacterial products through PRRs. In addition, this discovery was surprising because up until then, TLRs were implicated in the stimulation of inflammation. Furthermore, Fukata et al. later demonstrated that TLR4 and MyD88- dependent protection from injury required cyclooxygenase 2 (Cox2)- mediated generation of prostaglandin E2 (PGE2).Citation127 However, the MyD88-Cox2 signal that promotes regeneration is largely provided by macrophages which migrate toward the site of injury to stimulate the proliferation of epithelial progenitors.Citation15,Citation128 The mechanism for improved epithelial repair may be through PGE2-dependent activation of epidermal growth factor receptor (EGFR). In addition, TLR2 signaling has been demonstrated to be required for the induction of Foxp3+ Tregs and IL-10 production by PSA produced by the Bacteroides fragilis. Epithelial TLR/MyD88 signaling is essential for the regulation of antimicrobial peptides and molecules, which are required to prevent microbial encroachments towards the intestinal mucosa.Citation47 Neal et al. recently demonstrated that intestinal stem cells (ISCs) also express TLR4 and microbial ligand-mediated direct activation of TLR4 on ISCs, especially Lgr5-positive ISCs, potently inhibits cellular proliferation in intestinal crypts.Citation50,Citation129 Furthermore, LPS-mediated direct activation of enterocyte-specific TLR4 resulted in the increased apoptosis of the ISCs in a mechanism that is dependent upon the activation of the p53-up-regulated modulator of apoptosis (PUMA) in the pathogenesis of NEC. Hence, further studies are needed to define the role of TLRs in intestinal stem cells. The cytosolic innate immune sensor Nod2 has been shown to confer a cytoprotective effect. The Intestinal epithelial Lgr5+ stem cells, which are located within the intestinal crypts, constitutively express the cytosolic innate immune sensor Nod2 at levels much higher than in Paneth cells. Moreover, muramyl dipeptide (MDP), a peptidoglycan motif common to all bacteria, stimulates stem cell survival of intestinal organoids. These events result in an efficient cytoprotection against oxidative stress-mediated cell death.Citation130 These findings are of special importance in the context of examining the crypt-microbiota interface during regeneration of injured mucosa. For example, Pedron et al. previously identified the presence of a “crypt-specific core microbiota” in both the ceacal and colonic crypts of mice.Citation88 The authors later postulated that the presence of the correct crypt-resident microbiota, particularly the released MDP, may have a protective effect on stem cells, making them more reactive to MDP itself and more resistant to death during injury.Citation130 Additionally, the presence of bacteria in the crypt had been shown in patients suffering from ulcerative colitis.Citation131 Furthermore, our group reported that during intestinal wound regeneration, the epithelial cells remain in direct contact with the specific “probionts”, or bacteria that promote pro-restitutive effects.Citation19 These studies, along with the findings of PRR expression in ISCs, are important in determining potential microbiota-ISC interactions, which might be mediated through direct contact or release of products and/or metabolites during regeneration of injured mucosa.Citation130 Presence of the correct crypt/wound-associated bacteria and their products may dictate the fate of the wound regeneration. Overall, these findings support that microbial activation of PRRs-dependent signaling affects proliferation and survival of intestinal epithelial cells and thereby plays a pivotal role in the repair of intestinal wounds.
Microbiota-mediated immunomodulation and mucosal wound healing
Gut microbiota is integral to the activation of immune response and repair of intestinal damage. For example, gut symbionts serve a critical role by augmenting Interleukin-22 (IL-22) dependent mucosal wound repair. Recently, it has been demonstrated that IL-22 promotes intestinal stem cell-mediated epithelial regeneration. The study demonstrated that the recombinant IL-22 directly targeted ISCs, augmenting the growth of both mouse and human intestinal organoids, increasing proliferation and promoting ISC expansion. It has been previously described that the interactions between gut commensals, a range of lymphoid cells, and other environmental signals ensure the fine-tuning of IL-22 levels, such that they maintain the barrier function and repair without provoking overt inflammation.Citation132 Remarkably, commensal microbiota potently induces retinoic acid production (a metabolite of vitamin A) by dendritic cells (DCs), which can, in turn, induce IL-22 production by innate lymphoid cells type 3 (ILC3s) and γδ T-cells.Citation133,Citation134 ILCs are derived from the common lymphoid progenitor. ILCs participate in the control of pathogens, tolerance of commensal bacteria and also help repair of the intestinal injuries. These cells express signature transcription factors, including T-bet (ILC1), GATA3 (ILC2), and RORγt (ILC3). ILCs also include T-bet+ natural killer cells and Id3+ regulatory ILC.Citation135 Importantly, enteric commensals have been proposed to have a direct influence on IL-22 mediated response by producing specific metabolites, which are ligands to the aryl hydrocarbon receptor (AHR). AHR functions as a ligand-dependent transcriptional factor, which senses environmental and endogenous compounds generated by commensal, dietary, or also cellular metabolism. Importantly, it has been shown that AHR is a key regulator of ILC3, which produces IL-22, a crucial mediator of intestinal wound repair.Citation136 Furthermore, many gut microbiota, including certain lactobacilli, produces ligands of AHR from bacterial metabolism and may activate AHR signaling pathways. Microbial AHR ligands include tryptophan, indole-3-aldehyde, and indole-3-acetic acid. Additionally, tissue-resident intraepithelial lymphocytes (IELs) express high levels of the AHR. A subgroup of IELs, termed γδ T cells, also express AHR. The intestinal epithelial surface is the home of γδ IEL, which can be activated by AHR ligands resulting in augmented mucosal healing following acute injury.Citation137 This tissue repair function of γδ IEL has been linked to the upregulated expression of keratinocyte growth factor (KGF), which stimulates proliferation of colonic epithelial progenitors.Citation138 Activation of the γδ IEL during DSS-induced colonic injuries of the mucosa mounts a coordinated transcriptional program involved in the regulation of cytoprotective and immunomodulatory factors. Furthermore, studies in germ-free mice indicated that commensal microbiota modulates critical components of this transcriptional program, thus delineating an interaction between gut bacteria and γδ IEL during repair of the injured mucosa.Citation138 Similarly, the microbial fermentation product SCFA induces functional colonic Treg pool via epigenetic up-regulation of the Foxp3 gene, which efficiently suppresses inflammation and enhances mucosal tissue repair.Citation109
Macrophages are important components of the resolution of inflammation and epithelial wound healing. Depending on the tissue microenvironment, macrophages polarizes to become classically activated M1 (pro-inflammatory) or alternatively activated M2 (pro-regenerative). Microbial metabolites or cell wall components facilitate polarization of macrophages towards M2 and also enhances migration and wound-healing properties in M2 macrophages leading to augmented wound regeneration in mouse models of colitis.Citation139 In addition, Trem2 (triggering receptor expressed on myeloid cells 2) is a cell surface receptor that is specifically induced in macrophages by IL-4/IL-13. Activation of Trem2 signaling augments an efficient wound healing of colonic mucosal injuries by inhibiting cytokines that stimulate M1 macrophage activation, and by promoting cytokines that induce M2 macrophage activation.Citation119 Interestingly, the indigenous ligands, as well as bacterially synthesized ligands for Trem2, have been identified in the anionic residues present on the surface of Gram-positive and negative bacteria, suggesting a potential role of the intestinal microbiota in regulating macrophage activation through the Trem-2 during mucosal wound healing. Interestingly, the macrophages also play important beneficial roles in wound healing by secretion of pro-resolution IL-10, activation of epithelial cAMP response element–binding protein (CREB) and by subsequent synthesis and secretion of the pro-repair WNT1-inducible signaling protein 1 (WISP-1) in a microbiota independent manner.Citation140 In addition, mice deficient in IL-36R exhibited defective recovery following DSS-induced damage and impaired closure of colonic mucosal biopsy wounds in the distal colon. Interestingly, germ-free mice failed to induce L-36γ in response to dextran sodium sulfate (DSS)-induced damage, suggesting that gut microbiota is involved in its induction.Citation141 These findings demonstrate a cross-talk between immune cells, intestinal microbiota, and their metabolites eventuating in an efficient mucosal epithelial repair.
The microenvironment of wound mucosa undergoing active repair
Intestinal microenvironment and physiological parameters of the injured mucosa are remarkably different from intact mucosa. Therefore, it is plausible that the microbial community structure-associated with the injured mucosa are distinctly divergent from the intact mucosa or the luminal bacteria. Indeed, recent studies provided evidence that this alteration in microbial community is driven by the altered microenvironment of the intestinal tract during inflammation and active mucosal regeneration. The biochemical and biophysical components of the wound microenvironment are contributed by the damaged and regenerating epithelial cells, resident and transmigrating immune cells and mesenchymal cells as well as the wound-associated bacteria.Citation113,Citation142 Therefore, sites of intestinal mucosal inflammation and subsequent restitution are characterized by multifaceted alterations in the tissue microenvironment, including the appearance of pro-resolving mediators, antimicrobials, bacterial small molecules and metabolites, enzymes, cytokines, mucins and changes in reactive oxygen species and oxygen tension.Citation21,Citation22,Citation143 The functional role of antimicrobials, IgA and ROS in the regulation of bacterial population have been studied for decades and recently discussed (For reviews, please see refs.144–146).Citation144–Citation146
During inflammation of the gut, mucosal microenvironment is changed due to depletion of the oxygen and generation of the reactive oxygen species. Transmigrating neutrophils accumulate in the injured intestinal mucosa and play a key role in altering the physiological parameters of the wound microenvironment. Also, the expression levels of the mucins, which predominantly supply the glycan for the foraging microbiota, are altered in the mucosal wounds.Citation147 Recently, we have demonstrated that FPR and neutrophilic NOX2 are required for the rapid depletion of microenvironmental oxygen and compensatory responses, resulting in a dramatic enrichment of an anaerobic bacterial consortium.Citation19 Similarly, Campbell et al. demonstrated that the NOX2-mediated oxidative burst rapidly depletes the oxygen in the inflamed epithelia within recruited neutrophils during trinitrobenzenesulphonic acid (TNBS)-induced colitis. Concurrently, the study showed that intestinal epithelial cells increased synthesis of a mucin protein, MUC3.Citation20 Recently, we performed high-throughput sequencing (HTS) sequence analysis of the Bacterial 16S rRNA genes (V4 region) to delineate the bacterial population colonized in mucosal wounds undergoing active repair in a mouse model of colonic wound regeneration.Citation19 As expected the initial composition of phyla present in the wound bed most closely resembled that of the intact mucosa, though interestingly, the community structure markedly changed over the several day healing period, and the microbiota composition of the resealing wounds differed distinctly from that of intact mucosa and the colonic luminal contents. Our study identified seven anaerobic and microaerophilic commensal bacterial genera that increased in abundance, specifically in the early regenerative mucosa. Most notably, the relative abundance of anaerobic mucinophilic Akkermansia (phyla: Verrucomicrobia) increased dramatically and additional bacteria that are represented in the wound-associated consortium includes the anaerobic taxa Coprococcus, Mucispirillum, Odoribacter, Prevotella, Oscillospira and aerotolerant lactobacilli. These findings indicate that, during repair of gut mucosal injury, temporally dynamic local environmental conditions in the wound favor the growth of specific taxa, and define a wound-associated microbiota consortium that preferentially thrives for several days and resolves to the original state as the wound repairs.Citation19 Moreover, the dominant member of this wound-mucosa-associated microbiota, Akkermansia muciniphila, an anaerobic, mucinophilic gut symbiontCitation148, stimulated proliferation and migration of enterocytes adjacent to the colonic wounds.Citation19 These findings thus demonstrate how wound microenvironments induce the rapid emergence of prorestitutive symbionts, designated probiont,’ which contribute to an enhanced repair of mucosal injuries.Citation19 Other workers have shown administration of the mucin utilizing A. muciniphila, to mice prevents the development of high-fat diet-induced obesity and ameliorates metabolic endotoxemia-induced inflammation through the restoration of the gut barrier.Citation148–Citation150 Some of these effects are the result of increased mucin secretion and intestinal tight junction proteins, which emphasize the dynamic role played by mucin utilizers in their interaction with the host. Also, the protective function of A. muciniphila could be recapitulated using A. muciniphila purified membrane protein or the pasteurized bacterium.Citation150
Overall, these results suggest that the microbiota can adapt to local environmental changes to form definable and transient consortia. Importantly, members of this consortium can stimulate pro-restitutive signaling and increased migration and proliferation leading to enhanced epithelial wound healing. Mucosal wound environment provides a classic model of ecological succession, where the hypoxic environment, secondary to the influx of transmigrating neutrophils, of the wound bed encourages the growth of A. muciniphila and several other types of anaerobic bacteria. Consequently, two to four days post-biopsy injuries in the colon, anaerobic Akkermansia and related consortium enrich to a peak of about 8% of the mucosa-associated bacteria, which in turn enhance epithelial regeneration, and then diminish in abundance along with the disappearance of neutrophils and alterations in the hypoxic condition. Thus, this study delineates a ecological feedback loop and cross-talk induced by the injured microenvironment and gut microbiota.Citation19 Taken together, these findings support that commensal enteric bacteria play a crucial role in epithelial wound healing and restoration of barrier functions. These studies also highlight that discovery, development, and designing of the future therapies and interventions targeted for the inflammatory gastrointestinal diseases should be aligned with the mechanism of microenvironmental alterations and enrichment of beneficial microbiota during mucosal wound healing.
Concluding remarks
Numerous studies determined that the mammalian intestine houses taxonomically diverse and numerically vast prokaryotic microorganisms. However, mere identification of the microbial species and observation of their effect does not ensure deciphering the molecular mechanisms of the host-microbial interactions. The mammalian microbiota generates a diverse array of metabolites -metabolites are small molecules synthesized by the cells- in the intestine. Molecular functions of the microbial metabolites generated in the microenvironment to modulate the host cellular events are fundamentally crucial to determining the mechanism of diseases and development of potential therapeutics. Recently, there has been growing interest in the identification and functional characterization of microbial metabolites in the intestine. Future studies will delineate mechanistic functions of microbial products and small molecules in the pathogenesis of barrier disruption. Moreover, future research studies will be forging a multidisciplinary approach in computational, quantitative analytical chemistry and experimental biology to systematically discover genes, pathways, and molecules essential for dissecting a the novel etiology of diseases as well as will be pivotal in the development of the potential therapeutic application, diagnosis of a disease, or prognosis of an intervention.
Disclosure of potential conflicts of interest
No potential conflict of interest were disclosed.
Additional information
Funding
References
- Hooper LV, Wong MH, Thelin A, Hansson L, Falk PG, Gordon JI. Molecular analysis of commensal host-microbial relationships in the intestine. Science. 2001;291(5505):881–884. doi:10.1126/science.291.5505.881.
- Neish AS. Microbes in gastrointestinal health and disease. Gastroenterology. 2009;136(1):65–80. S0016-5085(08)01978-1 [pii]. doi:10.1053/j.gastro.2008.10.080.
- Backhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science. 2005;307(5717):1915–1920. doi:10.1126/science.1104816.
- Lloyd-Price J, Abu-Ali G, Huttenhower C. The healthy human microbiome. Genome Med. 2016;8(1):51. PMID:PMC4848870. doi:10.1186/s13073-016-0307-y.
- Xavier RJ, Podolsky DK. Unravelling the pathogenesis of inflammatory bowel disease. Nature. 2007;448(7152):427–434. doi:10.1038/nature06005.nature06005[pii].
- Imhann F, Vich Vila A, Bonder MJ, Fu J, Gevers D, Visschedijk MC, Spekhorst LM, Alberts R, Franke L, van Dullemen HM, et al. Interplay of host genetics and gut microbiota underlying the onset and clinical presentation of inflammatory bowel disease. Gut. 2018;67(1):108–119. PMID:PMC5699972. doi:10.1136/gutjnl-2016-312135.
- Gilbert JA, Quinn RA, Debelius J, Xu ZZ, Morton J, Garg N, Jansson JK, Dorrestein PC, Knight R. Microbiome-wide association studies link dynamic microbial consortia to disease. Nature. 2016;535(7610):94–103. doi:10.1038/nature18850.
- Sartor RB, Wu GD. Roles for intestinal bacteria, viruses, and fungi in pathogenesis of inflammatory bowel diseases and therapeutic approaches. Gastroenterology. 2017;152(2):327–339e324. PMID:PMC5511756. doi:10.1053/j.gastro.2016.10.012.
- Garcia-Hernandez V, Quiros M, Nusrat A. Intestinal epithelial claudins: expression and regulation in homeostasis and inflammation. Ann N Y Acad Sci. 2017;1397(1):66–79. PMID:PMC5545801. doi:10.1111/nyas.13360.
- Laukoetter MG, Bruewer M, Nusrat A. Regulation of the intestinal epithelial barrier by the apical junctional complex. Curr Opin Gastroen. 2006;22(2):85–89. doi:10.1097/01.mog.0000203864.48255.4f.
- Buckley A, Turner JR. Cell biology of tight junction barrier regulation and mucosal disease. Cold Spring Harb Perspect Biol. 2018;10(1). doi:10.1101/cshperspect.a029314.
- Morgan XC, Tickle TL, Sokol H, Gevers D, Devaney KL, Ward DV, Reyes JA, Shah SA, LeLeiko N, Snapper SB, et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012;13(9):R79. PMID:PMC3506950. doi:10.1186/gb-2012-13-9-r79.
- Hall AB, Tolonen AC, Xavier RJ. Human genetic variation and the gut microbiome in disease. Nat Rev Genet. 2017;18(11):690–699. doi:10.1038/nrg.2017.63.
- Farkas AE, Nusrat A. Pharmacological targeting of the inflamed intestinal barrier. Curr Pharm Design. 2016;22(35):5400–5414. doi:10.2174/1381612822666160726123857.
- Pull SL, Doherty JM, Mills JC, Gordon JI, Stappenbeck TS. Activated macrophages are an adaptive element of the colonic epithelial progenitor niche necessary for regenerative responses to injury. Proc Natl Acad Sci. 2005;102(1):99–104. doi:10.1073/pnas.0405979102.
- Lotz MM, Nusrat A, Madara JL, Ezzell R, Wewer UM, Mercurio AM. Intestinal epithelial restitution. Involvement of specific laminin isoforms and integrin laminin receptors in wound closure of a transformed model epithelium. Am J Pathol. 1997;150(2):747–760. PMID:1858283.
- Miyoshi H, Ajima R, Luo CT, Yamaguchi TP, Stappenbeck TS. Wnt5a potentiates tgf-beta signaling to promote colonic crypt regeneration after tissue injury. Science. 2012;338(6103):108–113. science.1223821[pii]. doi:10.1126/science.1223821.
- Peterson LW, Artis D. Intestinal epithelial cells: regulators of barrier function and immune homeostasis. Nat Rev Immunol. 2014;14(3):141–153. doi:10.1038/nri3608.
- Alam A, Leoni G, Quiros M, Wu H, Desai C, Nishio H, Jones RM, Nusrat A, Neish AS. The microenvironment of injured murine gut elicits a local pro-restitutive microbiota. Nat Microbiol. 2016;1:15021. doi:10.1038/nmicrobiol.2015.21.
- Campbell EL, Bruyninckx WJ, Kelly CJ, Glover LE, McNamee EN, Bowers BE, Bayless AJ, Scully M, Saeedi BJ, Golden-Mason L, et al. Transmigrating neutrophils shape the mucosal microenvironment through localized oxygen depletion to influence resolution of inflammation. Immunity. 2014;40(1):66–77. S1074-7613(13)00562-1 [pii]. PMID:3951457. doi:10.1016/j.immuni.2013.11.020.
- Barrett KE, Wu GD. Influence of the microbiota on host physiology - moving beyond the gut. J Physiol. 2017;595(2):433–435. PMID:PMC5233661. doi:10.1113/JP273451.
- Friedman ES, Bittinger K, Esipova TV, Hou L, Chau L, Jiang J, Mesaros C, Lund PJ, Liang X, FitzGerald GA, et al. Microbes vs. Chemistry in the origin of the anaerobic gut lumen. Proc Natl Acad Sci U S A. 2018;115(16):4170–4175. PMID:PMC5910840. doi:10.1073/pnas.1718635115.
- Rosenberg E, Zilber-Rosenberg I. Microbes drive evolution of animals and plants: the hologenome concept. mBio. 2016;7(2). doi:10.1128/mBio.01395-15.
- Lloyd-Price J, Mahurkar A, Rahnavard G, Crabtree J, Orvis J, Hall AB, Brady A, Creasy HH, McCracken C, Giglio MG, et al. Strains, functions and dynamics in the expanded human microbiome project. Nature. 2017;550(7674):61–66. PMID:PMC5831082. doi:10.1038/nature23889.
- Dantas G, Sommer MOA, Degnan PH, Goodman AL. Experimental approaches for defining functional roles of microbes in the human gut. Annu Rev Microbiol. 2013;67:459–475. PMID: 24024637. doi:10.1146/annurev-micro-092412-155642.
- Knights D, Silverberg MS, Weersma RK, Gevers D, Dijkstra G, Huang H, Tyler AD, van Sommeren S, Imhann F, Stempak JM, et al. Complex host genetics influence the microbiome in inflammatory bowel disease. Genome Med. 2014;6(12):107. PMID:PMC4292994. doi:10.1186/s13073-014-0107-1.
- Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M, et al. Ultra-high-throughput microbial community analysis on the illumina hiseq and miseq platforms. ISME J. 2012;6(8):1621–1624. ismej20128[pii]. PMID:3400413. doi:10.1038/ismej.2012.8.
- Knight R, Vrbanac A, Taylor BC, Aksenov A, Callewaert C, Debelius J, Gonzalez A, Kosciolek T, McCall LI, McDonald D, et al. Best practices for analysing microbiomes. Nat Rev Microbiol. 2018;16(7):410–422. doi:10.1038/s41579-018-0029-9.
- Kundu P, Blacher E, Elinav E, Pettersson S. Our gut microbiome: the evolving inner self. Cell. 2017;171(7):1481–1493. PMID: 29245010. doi:10.1016/j.cell.2017.11.024.
- Mehta RS, Abu-Ali GS, Drew DA, Lloyd-Price J, Subramanian A, Lochhead P, Joshi AD, Ivey KL, Khalili H, Brown GT, et al. Stability of the human faecal microbiome in a cohort of adult men. Nat Microbiol. 2018;3(3):347–355. PMID:PMC6016839. doi:10.1038/s41564-017-0096-0.
- Cong X, Xu W, Janton S, Henderson WA, Matson A, McGrath JM, Maas K, Graf J. Gut microbiome developmental patterns in early life of preterm infants: impacts of feeding and gender. PLoS One. 2016;11(4):e0152751. PMID:PMC4844123. doi:10.1371/journal.pone.0152751.
- Yassour M, Jason E, Hogstrom LJ, Arthur TD, Tripathi S, Siljander H, Selvenius J, Oikarinen S, Hyoty H, Virtanen SM, et al. Strain-level analysis of mother-to-child bacterial transmission during the first few months of life. Cell Host Microbe. 2018;24(1):146–154 e144. doi:10.1016/j.chom.2018.06.007.
- Kolde R, Franzosa EA, Rahnavard G, Hall AB, Vlamakis H, Stevens C, Daly MJ, Xavier RJ, Huttenhower C. Host genetic variation and its microbiome interactions within the human microbiome project. Genome Med. 2018;10(1):6. PMID:PMC5789541. doi:10.1186/s13073-018-0515-8.
- Tropini C, Earle KA, Huang KC, Sonnenburg JL. The gut microbiome: connecting spatial organization to function. Cell Host & Microbe. 2017;21(4):433–442. doi:10.1016/j.chom.2017.03.010.
- Fallingborg J. Intraluminal ph of the human gastrointestinal tract. Dan Med Bull. 1999;46(3):183–196.
- Li J, Jia H, Cai X, Zhong H, Feng Q, Sunagawa S, Arumugam M, Kultima JR, Prifti E, Nielsen T, et al. An integrated catalog of reference genes in the human gut microbiome. Nat Biotechnol. 2014;32(8):834–841. doi:10.1038/nbt.2942.
- Xiao L, Feng Q, Liang S, Sonne SB, Xia Z, Qiu X, Li X, Long H, Zhang J, Zhang D, et al. A catalog of the mouse gut metagenome. Nat Biotechnol. 2015;33(10):1103–1108. doi:10.1038/nbt.3353.
- Gonzalez A, King A, Robeson MS 2nd, Song S, Shade A, Metcalf JL, Knight R. Characterizing microbial communities through space and time. Curr Opin Biotechnol. 2012;23(3):431–436. PMID:PMC3311466. doi:10.1016/j.copbio.2011.11.017.
- Segata N, Boernigen D, Tickle TL, Morgan XC, Garrett WS, Huttenhower C. Computational meta’omics for microbial community studies. Mol Syst Biol. 2013;9:666. PMID:4039370. doi:10.1038/msb.2013.22.
- Ismail AS, Hooper LV. Epithelial cells and their neighbors. Iv. Bacterial contributions to intestinal epithelial barrier integrity. Am J Physiol-Gastr L. 2005;289(5):G779–G784. doi:10.1152/ajpgi.00203.2005.
- Madsen K, Cornish A, Soper P, McKaigney C, Jijon H, Yachimec C, Doyle J, Jewell L, De Simone C. Probiotic bacteria enhance murine and human intestinal epithelial barrier function. Gastroenterology. 2001;121(3):580–591. doi:10.1053/gast.2001.27224.
- Smith K, McCoy KD, Macpherson AJ. Use of axenic animals in studying the adaptation of mammals to their commensal intestinal microbiota. Semin Immunol. 2007;19(2):59–69. doi:10.1016/j.smim.2006.10.002.
- Sansonetti PJ, Medzhitov R. Learning tolerance while fighting ignorance. Cell. 2009;138(3):416–420. doi:10.1016/j.cell.2009.07.024.
- Chung HC, Kasper DL. Microbiota-stimulated immune mechanisms to maintain gut homeostasis. Curr Opin Immunol. 2010;22(4):455–460. doi:10.1016/j.coi.2010.06.008.
- Harris G, KuoLee R, Chen WX. Role of toll-like receptors in health and diseases of gastrointestinal tract. World J Gastroentero. 2006;12(14):2149–2160. doi:10.3748/wjg.v12.i14.2149.
- Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118(2):229–241. doi:10.1016/j.cell.2004.07.002S0092867404006610[pii].
- Vaishnava S, Yamamoto M, Severson KM, Ruhn KA, Yu X, Koren O, Ley R, Wakeland EK, Hooper LV. The antibacterial lectin regiiigamma promotes the spatial segregation of microbiota and host in the intestine. Science. 2011;334(6053):255–258. PMID:3321924. doi:10.1126/science.1209791.
- Carvalho FA, Koren O, Goodrich JK, Johansson ME, Nalbantoglu I, Aitken JD, Su Y, Chassaing B, Walters WA, Gonzalez A, et al. Transient inability to manage proteobacteria promotes chronic gut inflammation in tlr5-deficient mice. Cell Host Microbe. 2012;12(2):139–152. PMID:PMC4310462. doi:10.1016/j.chom.2012.07.004.
- Vijay-Kumar M, Aitken JD, Carvalho FA, Cullender TC, Mwangi S, Srinivasan S, Sitaraman SV, Knight R, Ley RE, Gewirtz AT. Metabolic syndrome and altered gut microbiota in mice lacking toll-like receptor 5. Science. 2010;328(5975):228–231. PMID:PMC4714868. doi:10.1126/science.1179721.
- Hou QH, Ye LL, Huang LL, Yu QH. The research progress on intestinal stem cells and its relationship with intestinal microbiota. Front Immunol. 2017;8. ARTN 599. doi:10.3389/fimmu.2017.00599.
- Migeotte I, Communi D, Parmentier M. Formyl peptide receptors: A promiscuous subfamily of g protein-coupled receptors controlling immune responses. Cytokine Growth Factor Rev. 2006;17(6):501–519. doi:10.1016/j.cytogfr.2006.09.009.
- Macdonald TT, Monteleone G. Immunity, inflammation, and allergy in the gut. Science. 2005;307(5717):1920–1925. 307/5717/1920[pii]. doi:10.1126/science.1106442.
- Jones RM, Neish AS. Recognition of bacterial pathogens and mucosal immunity. Cell Microbiol. 2011;13(5):670–676. doi:10.1111/j.1462-5822.2011.01579.x.
- Wentworth CC, Jones RM, Kwon YM, Nusrat A, Neish AS. Commensal-epithelial signaling mediated via formyl peptide receptors. Am J Pathol. 2010;177(6):2782–2790. S0002-9440(10)62907-0 [pii]. PMID:2993286. doi:10.2353/ajpath.2010.100529.
- Ye RD, Boulay F, Wang JM, Dahlgren C, Gerard C, Parmentier M, Serhan CN, Murphy PM. International union of basic and clinical pharmacology. Lxxiii. Nomenclature for the formyl peptide receptor (fpr) family. Pharmacol Rev. 2009;61(2):119–161. pr.109.001578 [pii]. PMID:2745437. doi:10.1124/pr.109.001578.
- Perretti M, Dalli J. Exploiting the annexin a1 pathway for the development of novel anti-inflammatory therapeutics. Br J Pharmacol. 2009;158(4):936–946. BPH483 [pii]. PMID:2785517. doi:10.1111/j.1476-5381.2009.00483.x.
- Babbin BA, Jesaitis AJ, Ivanov AI, Kelly D, Laukoetter M, Nava P, Parkos CA, Nusrat A. Formyl peptide receptor-1 activation enhances intestinal epithelial cell restitution through phosphatidylinositol 3-kinase-dependent activation of rac1 and cdc42. J Immunology. 2007;179(12):8112–8121. doi:10.4049/jimmunol.179.12.8112.
- Wentworth CC, Alam A, Jones RM, Nusrat A, Neish AS. Enteric commensal bacteria induce extracellular signal-regulated kinase pathway signaling via formyl peptide receptor-dependent redox modulation of dual specific phosphatase 3. J Biol Chem. 2011;286(44):38448–38455. M111.268938 [pii]. PMID:3207424. doi:10.1074/jbc.M111.268938.
- Jones RM, Mercante JW, Neish AS. Reactive oxygen production induced by the gut microbiota: pharmacotherapeutic implications. Curr Med Chem. 2012;19(10):1519–1529.
- Jones RM, Luo L, Ardita CS, Richardson AN, Kwon YM, Mercante JW, Alam A, Gates CL, Wu H, Swanson PA, et al. Symbiotic lactobacilli stimulate gut epithelial proliferation via nox-mediated generation of reactive oxygen species. The EMBO Journal. 2013;32(23):3017–3028. emboj2013224 [pii]. PMID:3844951. doi:10.1038/emboj.2013.224.
- Kumar A, Wu H, Collier-Hyams LS, Kwon YM, Hanson JM, Neish AS. The bacterial fermentation product butyrate influences epithelial signaling via reactive oxygen species-mediated changes in cullin-1 neddylation. J Immunology. 2009;182(1):538–546. doi:10.4049/jimmunol.182.1.538.
- Swanson PA 2nd, Kumar A, Samarin S, Vijay-Kumar M, Kundu K, Murthy N, Hansen J, Nusrat A, Neish AS. Enteric commensal bacteria potentiate epithelial restitution via reactive oxygen species-mediated inactivation of focal adhesion kinase phosphatases. Proc Natl Acad Sci U S A. 2011;108(21):8803–8808. 1010042108 [pii]. PMID:3102402. doi:10.1073/pnas.1010042108.
- Alam A, Leoni G, Wentworth CC, Kwal JM, Wu H, Ardita CS, Swanson PA, Lambeth JD, Jones RM, Nusrat A, et al. Redox signaling regulates commensal-mediated mucosal homeostasis and restitution and requires formyl peptide receptor 1. Mucosal Immunol. 2014;7(3):645–655. mi201384 [pii]. PMID:3999246. doi:10.1038/mi.2013.84.
- Ray PD, Huang BW, Tsuji Y. Reactive oxygen species (ros) homeostasis and redox regulation in cellular signaling. Cell Signal. 2012;24(5):981–990. doi:10.1016/j.cellsig.2012.01.008.
- Kumar A, Wu H, Collier-Hyams LS, Hansen JM, Li T, Yamoah K, Pan ZQ, Jones DP, Neish AS. Commensal bacteria modulate cullin-dependent signaling via generation of reactive oxygen species. The EMBO Journal. 2007;26(21):4457–4466. doi:10.1038/sj.emboj.7601867.
- Tonks NK. Redox redux: revisiting ptps and the control of cell signaling. Cell. 2005;121(5):667–670. S0092-8674(05)00495-2 [pii]. doi:10.1016/j.cell.2005.05.016.
- Quiros M, Nusrat A. Rhogtpases, actomyosin signaling and regulation of the epithelial apical junctional complex. Semin Cell Dev Biol. 2014;36:194–203. doi:10.1016/j.semcdb.2014.09.003.
- Capaldo CT, Nusrat A. Claudin switching: physiological plasticity of the tight junction. Semin Cell Dev Biol. 2015;42:22–29. doi:10.1016/j.semcdb.2015.04.003.
- Shen L, Weber CR, Raleigh DR, Yu D, Tumer JR. Tight, junction pore and leak pathways: A dynamic duo. Annu Rev Physiol. 2011;73:283–309. doi:10.1146/annurev-physiol-012110-142150.
- Laukoetter MG, Nava P, Nusrat A. Role of the intestinal barrier in inflammatory bowel disease. World J Gastroenterol. 2008;14(3):401–407. PMID:2679128.
- Khounlotham M, Kim W, Peatman E, Nava P, Medina-Contreras O, Addis C, Koch S, Fournier B, Nusrat A, Denning TL, et al. Compromised intestinal epithelial barrier induces adaptive immune compensation that protects from colitis. Immunity. 2012;37(3):563–573. PMID:PMC3564580. doi:10.1016/j.immuni.2012.06.017.
- Zyrek AA, Cichon C, Helms S, Enders C, Sonnenborn U, Schmidt MA. Molecular mechanisms underlying the probiotic effects of escherichia coli nissle 1917 involve zo-2 and pkc zeta redistribution resulting in tight junction and epithelial barrier repair. Cell Microbiol. 2007;9(3):804–816. doi:10.1111/j.1462-5822.2006.00836.x.
- Khailova L, Dvorak K, Arganbright KM, Halpern MD, Kinouchi T, Yajima M, Dvorak B. Bifidobacterium bifidum improves intestinal integrity in a rat model of necrotizing enterocolitis. Am J Physiol-Gastr L. 2009;297(5):G940–G949. doi:10.1152/ajpgi.00141.2009.
- Seth A, Yan F, Polk DB, Rao RK. Probiotics ameliorate the hydrogen peroxide-induced epithelial barrier disruption by a pkc- and map kinase-dependent mechanism. Am J Physiol-Gastr L. 2008;294(4):G1060–G1069. doi:10.1152/ajpgi.00202.2007.
- Resta-Lenert SC, Barrett KE. Probiotics and commensals reverse tnf-alpha- and ifn-gamma-induced dysfunction in human intestinal epithelial cells. Gastroenterology. 2003;124(4):A477–A477. doi:10.1016/S0016-5085(03)82414-9.
- Ahrne S, Hagslatt MLJ. Effect of lactobacilli on paracellular permeability in the gut. Nutrients. 2011;3(1):104–117. doi:10.3390/nu3010104.
- Ewaschuk JB, Diaz H, Meddings L, Diederichs B, Dmytrash A, Backer J, Langen MLV, Madsen KL. Secreted bioactive factors from bifidobacterium infantis enhance epithelial cell barrier function. Am J Physiol-Gastr L. 2008;295(5):G1025–G1034. doi:10.1152/ajpgi.90227.2008.
- Chichlowski M, De Lartigue G, German JB, Raybould HE, Mills DA. Bifidobacteria isolated from infants and cultured on human milk oligosaccharides affect intestinal epithelial function. J Pediatr Gastr Nutr. 2012;55(3):321–327. doi:10.1097/MPG.0b013e31824fb899.
- Martin R, Laval L, Chain F, Miquel S, Natividad J, Cherbuy C, Sokol H, Verdu EF, Vlieg JVH, Bermudez-Humaran LG, et al. Bifidobacterium animalis ssp lactis cncm-i2494 restores gut barrier permeability in chronically low-grade inflamed mice. Front Microbiol. 2016;7. ARTN 608. doi:10.3389/fmicb.2016.00608.
- Laval L, Martin R, Natividad JN, Chain F, Miquel S, Desclee de Maredsous C, Capronnier S, Sokol H, Verdu EF, van Hylckama Vlieg JE, et al. Lactobacillus rhamnosus cncm i-3690 and the commensal bacterium faecalibacterium prausnitzii a2-165 exhibit similar protective effects to induced barrier hyper-permeability in mice. Gut Microbes. 2015;6(1):1–9. PMID:PMC4615674. doi:10.4161/19490976.2014.990784.
- Bergmann KR, Liu SXL, Tian RL, Kushnir A, Turner JR, Li HL, Chou PM, Weber CR, De Plaen IG. Bifidobacteria stabilize claudins at tight junctions and prevent intestinal barrier dysfunction in mouse necrotizing enterocolitis. Am J Pathol. 2013;182(5):1595–1606. doi:10.1016/j.ajpath.2013.01.013.
- Johansson MEV, Phillipson M, Petersson J, Velcich A, Holm L, Hansson GC. The inner of the two muc2 mucin-dependent mucus layers in colon is devoid of bacteria. P Natl Acad Sci USA. 2008;105(39):15064–15069. doi:10.1073/pnas.0803124105.
- Wenzel UA, Magnusson MK, Rydstrom A, Jonstrand C, Hengst J, Johansson MEV, Velcich A, Ohman L, Strid H, Sjovall H, et al. Spontaneous colitis in muc2-deficient mice reflects clinical and cellular features of active ulcerative colitis. PLoS One. 2014;9(6). ARTN e100217. doi:10.1371/journal.pone.0100217.
- Johansson MEV, Larsson JMH, Hansson GC. The two mucus layers of colon are organized by the muc2 mucin, whereas the outer layer is a legislator of host-microbial interactions. P Natl Acad Sci USA. 2011;108:4659–4665. doi:10.1073/pnas.1006451107.
- Ermund A, Gustafsson JK, Hansson GC, Keita AV. Mucus properties and goblet cell quantification in mouse, rat and human ileal peyer’s patches. PLoS One. 2013;8(12). ARTN e83688. doi:10.1371/journal.pone.0083688.
- Ermund A, Schutte A, Johansson ME, Gustafsson JK, Hansson GC. Studies of mucus in mouse stomach, small intestine, and colon. I. Gastrointestinal mucus layers have different properties depending on location as well as over the peyer’s patches. Am J Physiol Gastrointest Liver Physiol. 2013;305(5):G341–347. PMID:PMC3761247. doi:10.1152/ajpgi.00046.2013.
- Johansson MEV, Gustafsson JK, Holmen-Larsson J, Jabbar KS, Xia LJ, Xu H, Ghishan FK, Carvalho FA, Gewirtz AT, Sjovall H, et al. Bacteria penetrate the normally impenetrable inner colon mucus layer in both murine colitis models and patients with ulcerative colitis. Gut. 2014;63(2):281–291. doi:10.1136/gutjnl-2012-303207.
- Pedron T, Mulet C, Dauga C, Frangeul L, Chervaux C, Grompone G, Sansonetti PJ. A crypt-specific core microbiota resides in the mouse colon. mBio. 2012;3(3). ARTN e00116-12. doi:10.1128/mBio.00116-12.
- Lee SM, Donaldson GP, Mikulski Z, Boyajian S, Ley K, Mazmanian SK. Bacterial colonization factors control specificity and stability of the gut microbiota. Nature. 2013;501(7467):426. doi:10.1038/nature12447.
- Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, et al. Induction of intestinal th17 cells by segmented filamentous bacteria. Cell. 2009;139(3):485–498. PMID:2796826. doi:10.1016/j.cell.2009.09.033.
- Crost EH, Tailford LE, Monestier M, Swarbreck D, Henrissat B, Crossman LC, Juge N. The mucin-degradation strategy of ruminococcus gnavus: the importance of intramolecular trans-sialidases. Gut Microbes. 2016;7(4):302–312. PMID:PMC4988440. doi:10.1080/19490976.2016.1186334.
- Hall AB, Yassour M, Sauk J, Garner A, Jiang X, Arthur T, Lagoudas GK, Vatanen T, Fornelos N, Wilson R, et al. A novel ruminococcus gnavus clade enriched in inflammatory bowel disease patients. Genome Med. 2017;9(1):103. PMID:PMC5704459. doi:10.1186/s13073-017-0490-5.
- Willemsen LEM, Koetsier MA, van Deventer SJH, van Tol EAF. Short chain fatty acids stimulate epithelial mucin 2 expression through differential effects on prostaglandin e-1 and e-2 production by intestinal myofibroblasts. Gut. 2003;52(10):1442–1447. doi:10.1136/gut.52.10.1442.
- Johansson ME, Jakobsson HE, Holmen-Larsson J, Schutte A, Ermund A, Rodriguez-Pineiro AM, Arike L, Wising C, Svensson F, Backhed F, et al. Normalization of host intestinal mucus layers requires long-term microbial colonization. Cell Host Microbe. 2015;18(5):582–592. PMID:PMC4648652. doi:10.1016/j.chom.2015.10.007.
- Gevers D, Kugathasan S, Denson LA, Vazquez-Baeza Y, Van Treuren W, Ren B, Schwager E, Knights D, Song SJ, Yassour M, et al. The treatment-naive microbiome in new-onset crohn’s disease. Cell Host Microbe. 2014;15(3):382–392. S1931-3128(14)00063-8 [pii]. PMID:4059512. doi:10.1016/j.chom.2014.02.005.
- Manichanh C, Rigottier-Gois L, Bonnaud E, Gloux K, Pelletier E, Frangeul L, Nalin R, Jarrin C, Chardon P, Marteau P, et al. Reduced diversity of faecal microbiota in crohn’s disease revealed by a metagenomic approach. Gut. 2006;55(2):205–211. PMID:PMC1856500. doi:10.1136/gut.2005.073817.
- Fujimoto T, Imaeda H, Takahashi K, Kasumi E, Bamba S, Fujiyama Y, Andoh A. Decreased abundance of faecalibacterium prausnitzii in the gut microbiota of crohn’s disease. J Gastroen Hepatol. 2013;28(4):613–619. doi:10.1111/jgh.12073.
- Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermudez-Humaran LG, Gratadoux JJ, Blugeon S, Bridonneau C, Furet JP, Corthier G, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of crohn disease patients. P Natl Acad Sci USA. 2008;105(43):16731–16736. doi:10.1073/pnas.0804812105.
- Png CW, Linden SK, Gilshenan KS, Zoetendal EG, McSweeney CS, Sly LI, McGuckin MA, Florin TH. Mucolytic bacteria with increased prevalence in ibd mucosa augment in vitro utilization of mucin by other bacteria. Am J Gastroenterol. 2010;105(11):2420–2428. doi:10.1038/ajg.2010.281.
- Wlodarska M, Luo C, Kolde R, d’Hennezel E, Annand JW, Heim CE, Krastel P, Schmitt EK, Omar AS, Creasey EA, et al. Indoleacrylic acid produced by commensal peptostreptococcus species suppresses inflammation. Cell Host Microbe. 2017;22(1):25–37e26. PMID:PMC5672633. doi:10.1016/j.chom.2017.06.007.
- Kang CS, Ban M, Choi EJ, Moon HG, Jeon JS, Kim DK, Park SK, Jeon SG, Roh TY, Myung SJ, et al. Extracellular vesicles derived from gut microbiota, especially akkermansia muciniphila, protect the progression of dextran sulfate sodium-induced colitis. PLoS One. 2013;8(10):e76520. PMID:3811976. doi:10.1371/journal.pone.0076520.
- Nishino K, Nishida A, Inoue R, Kawada Y, Ohno M, Sakai S, Inatomi O, Bamba S, Sugimoto M, Kawahara M, et al. Analysis of endoscopic brush samples identified mucosa-associated dysbiosis in inflammatory bowel disease. J Gastroenterol. 2018;53(1):95–106. doi:10.1007/s00535-017-1384-4.
- Schultsz C, Van Den Berg FM, Ten Kate FW, Tytgat GN, Dankert J. The intestinal mucus layer from patients with inflammatory bowel disease harbors high numbers of bacteria compared with controls. Gastroenterology. 1999;117(5):1089–1097.
- Ni J, Wu GD, Albenberg L, Tomov VT. Gut microbiota and ibd: causation or correlation? Nat Rev Gastroenterol Hepatol. 2017;14(10):573–584. PMID:PMC5880536. doi:10.1038/nrgastro.2017.88.
- Lee YK, Mazmanian SK. Has the microbiota played a critical role in the evolution of the adaptive immune system? Science. 2010;330(6012):1768–1773. doi:10.1126/science.1195568.
- Kim SC, Tonkonogy SL, Albright CA, Tsang J, Balish EJ, Braun J, Huycke MM, Sartor RB. Variable phenotypes of enterocolitis in interleukin 10-deficient mice monoassociated with two different commensal bacteria. Gastroenterology. 2005;128(4):891–906. doi:10.1053/j.gastro.2005.02.009.
- Gaboriau-Routhiau V, Rakotobe S, Lecuyer E, Mulder I, Lan A, Bridonneau C, Rochet V, Pisi A, De Paepe M, Brandi G, et al. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper t cell responses. Immunity. 2009;31(4):677–689. doi:10.1016/j.immuni.2009.08.020.
- Garrett WS, Lord GM, Punit S, Lugo-Villarino G, Mazmanian SK, Ito S, Glickman JN, Glimcher LH. Communicable ulcerative colitis induced by t-bet deficiency in the innate immune system. Cell. 2007;131(1):33–45. doi:10.1016/j.cell.2007.08.017.
- Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly YM, Glickman JN, Garrett WS. The microbial metabolites, short-chain fatty acids, regulate colonic treg cell homeostasis. Science. 2013;341(6145):569–573. PMID:PMC3807819. doi:10.1126/science.1241165.
- Elinav E, Strowig T, Kau AL, Henao-Mejia J, Thaiss CA, Booth CJ, Peaper DR, Bertin J, Eisenbarth SC, Gordon JI, et al. Nlrp6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. 2011;145(5):745–757. doi:10.1016/j.cell.2011.04.022.
- Macia L, Tan J, Vieira AT, Leach K, Stanley D, Luong S, Maruya M, McKenzie CI, Hijikata A, Wong C, et al. Metabolite-sensing receptors gpr43 and gpr109a facilitate dietary fibre-induced gut homeostasis through regulation of the inflammasome. Nat Commun. 2015;6. ARTN 6734. doi:10.1038/ncomms7734.
- McKenzie CI, Mackay CR, Macia L. Gpr43-a prototypic metabolite sensor linking metabolic and inflammatory diseases. Trends Endocrin Met. 2015;26(10):511–512. doi:10.1016/j.tem.2015.07.009.
- Leoni G, Neumann PA, Sumagin R, Denning TL, Nusrat A. Wound repair: role of immune-epithelial interactions. Mucosal Immunol. 2015;8(5):959–968. doi:10.1038/mi.2015.63.
- Hopkins AM, Pineda AA, Winfree LM, Brown GT, Laukoetter MG, Nusrat A. Organized migration of epithelial cells requires control of adhesion and protrusion through rho kinase effectors. Am J Physiol-Gastr L. 2007;292(3):G806–G817. doi:10.1152/ajpgi.00333.2006.
- Hall A, Nobes CD. Rho gtpases: molecular switches that control the organization and dynamics of the actin cytoskeleton. Philos T R Soc B. 2000;355(1399):965–970. doi:10.1098/rstb.2000.0632.
- Leoni G, Alam A, Neumann PA, Lambeth JD, Cheng G, McCoy J, Hilgarth RS, Kundu K, Murthy N, Kusters D, et al. Annexin a1, formyl peptide receptor, and nox1 orchestrate epithelial repair. J Clin Invest. 2013;123(1):443–454. 65831 [pii]. PMID:3533303. doi:10.1172/JCI65831.
- Abrams GD, Bauer H, Sprinz H. Influence of the normal flora on mucosal morphology and cellular renewal in the ileum. A comparison of germ-free and conventional mice. Lab Invest. 1963;12:355–364.
- Larsson E, Tremaroli V, Lee YS, Koren O, Nookaew I, Fricker A, Nielsen J, Ley RE, Backhed F. Analysis of gut microbial regulation of host gene expression along the length of the gut and regulation of gut microbial ecology through myd88. Gut. 2012;61(8):1124–1131. PMID:PMC3388726. doi:10.1136/gutjnl-2011-301104.
- Seno H, Miyoshi H, Brown SL, Geske MJ, Colonna M, Stappenbeck TS. Efficient colonic mucosal wound repair requires trem2 signaling. Proc Natl Acad Sci U S A. 2009;106(1):256–261. 0803343106 [pii]. PMID:2629230. doi:10.1073/pnas.0803343106.
- Sturm A, Dignass AU. Epithelial restitution and wound healing in inflammatory bowel disease. World J Gastroentero. 2008;14(3):348–353. doi:10.3748/wjg.14.348.
- Dignass AU, Podolsky DK. Transforming growth-factor-beta plays a central role in the modulation of cytokine mediated intestinal epithelial-cell restitution in an invitro wounding model. Gastroenterology. 1993;104(4):A693–A693.
- Gilbert S, Lin J, Han XN. Constitutively active long myosin light chain kinase in intestinal epithelial cells impairs the activation of stat5 and delays intestinal mucosal wound healing. Gastroenterology. 2011;140(5):S633–S633. doi:10.1016/S0016-5085(11)62619-X.
- Iizuka M, Suzuki M, Hisamatsu T, Hashi H, Podolsky DK. Ligands of toll like receptors modulate wound healing in rat intestinal epithelial cells. Gastroenterology. 2002;122(4):A277–A277.
- Goke M, Kanai M, Lynch-Devaney K, Podolsky DK. Rapid mitogen-activated protein kinase activation by transforming growth factor alpha in wounded rat intestinal epithelial cells. Gastroenterology. 1998;114(4):697–705. doi:10.1016/S0016-5085(98)70583-9.
- Lin PW, Myers LE, Ray L, Song SC, Nasr TR, Berardinelli AJ, Kundu K, Murthy N, Hansen JM, Neish AS. Lactobacillus rhamnosus blocks inflammatory signaling in vivo via reactive oxygen species generation. Free Radic Biol Med. 2009. S0891-5849(09)00451-1 [pii]. doi:10.1016/j.freeradbiomed.2009.07.033.
- Rakoff-Nahoum S, Hao LM, Medzhitov R. Role of toll-like receptors in spontaneous commensal-dependent colitis. Immunity. 2006;25(2):319–329. doi:10.1016/j.immuni.2006.06.010.
- Fukata M, Chen A, Klepper A, Krishnareddy S, Vamadevan AS, Thomas LS, Xu R, Inoue H, Arditi M, Dannenberg AJ, et al. Cox-2 is regulated by toll-like receptor-4 (tlr4) signaling: role in proliferation and apoptosis in the intestine. Gastroenterology. 2006;131(3):862–877. PMID:PMC2169292. doi:10.1053/j.gastro.2006.06.017.
- Brown SL, Riehl TE, Walker MR, Geske MJ, Doherty JM, Stenson WF, Stappenbeck TS. Myd88-dependent positioning of ptgs2-expressing stromal cells maintains colonic epithelial proliferation during injury. J Clin Invest. 2007;117(1):258–269. PMID:1716207. doi:10.1172/JCI29159.
- Neal MD, Sodhi CP, Jia H, Dyer M, Egan CE, Yazji I, Good M, Afrazi A, Marino R, Slagle D, et al. Toll-like receptor 4 is expressed on intestinal stem cells and regulates their proliferation and apoptosis via the p53 up-regulated modulator of apoptosis. J Biol Chem. 2012;287(44):37296–37308. PMID:PMC3481327. doi:10.1074/jbc.M112.375881.
- Nigro G, Rossi R, Commere PH, Jay P, Sansonetti PJ. The cytosolic bacterial peptidoglycan sensor nod2 affords stem cell protection and links microbes to gut epithelial regeneration. Cell Host & Microbe. 2014;15(6):792–798. doi:10.1016/j.chom.2014.05.003.
- Sokol H, Vasquez N, Hoyeau-Idrissi N, Seksik P, Beaugerie L, Lavergne-Slove A, Pochart P, Marteau P. Crypt abscess-associated microbiota in inflammatory bowel disease and acute self-limited colitis. World J Gastroenterol. 2010;16(5):583–587. PMID:PMC2816270.
- Pickert G, Neufert C, Leppkes M, Zheng Y, Wittkopf N, Warntjen M, Lehr HA, Hirth S, Weigmann B, Wirtz S, et al. Stat3 links il-22 signaling in intestinal epithelial cells to mucosal wound healing. J Exp Med. 2009;206(7):1465–1472. jem.20082683 [pii]. PMID:2715097. doi:10.1084/jem.20082683.
- Konieczna P, Ferstl R, Ziegler M, Frei R, Nehrbass D, Lauener RP, Akdis CA, O’Mahony L. Immunomodulation by bifidobacterium infantis 35624 in the murine lamina propria requires retinoic acid-dependent and independent mechanisms. PLoS One. 2013;8(5):e62617. PMID:PMC3660574. doi:10.1371/journal.pone.0062617.
- Mielke LA, Jones SA, Raverdeau M, Higgs R, Stefanska A, Groom JR, Misiak A, Dungan LS, Sutton CE, Streubel G, et al. Retinoic acid expression associates with enhanced il-22 production by gammadelta t cells and innate lymphoid cells and attenuation of intestinal inflammation. J Exp Med. 2013;210(6):1117–1124. PMID:PMC3674702. doi:10.1084/jem.20121588.
- Kabat AM, Srinivasan N, Maloy KJ. Modulation of immune development and function by intestinal microbiota. Trends Immunol. 2014;35(11):507–517. doi:10.1016/j.it.2014.07.010.
- Geremia A, Arancibia-Carcamo CV. Innate lymphoid cells in intestinal inflammation. Front Immunol. 2017;8:1296. PMID:PMC5645495. doi:10.3389/fimmu.2017.01296.
- Li Y, Innocentin S, Withers DR, Roberts NA, Gallagher AR, Grigorieva EF, Wilhelm C, Veldhoen M. Exogenous stimuli maintain intraepithelial lymphocytes via aryl hydrocarbon receptor activation. Cell. 2011;147(3):629–640. doi:10.1016/j.cell.2011.09.025.
- Chen Y, Chou K, Fuchs E, Havran WL, Boismenu R. Protection of the intestinal mucosa by intraepithelial gamma delta t cells. Proc Natl Acad Sci U S A.. 2002;99(22):14338–14343. PMID:PMC137885. doi:10.1073/pnas.212290499.
- Vannella KM, Wynn TA. Mechanisms of organ injury and repair by macrophages. Annu Rev Physiol. 2017;79:593–617. doi:10.1146/annurev-physiol-022516-034356.
- Quiros M, Nishio H, Neumann PA, Siuda D, Brazil JC, Azcutia V, Hilgarth R, O’Leary MN, Garcia-Hernandez V, Leoni G, et al. Macrophage-derived il-10 mediates mucosal repair by epithelial wisp-1 signaling. J Clin Invest. 2017;127(9):3510–3520. PMID:PMC5669557. doi:10.1172/JCI90229.
- Medina-Contreras O, Harusato A, Nishio H, Flannigan KL, Ngo V, Leoni G, Neumann PA, Geem D, Lili LN, Ramadas RA, et al. Cutting edge: il-36 receptor promotes resolution of intestinal damage. J Immunology. 2016;196(1):34–38. PMID:PMC4684965. doi:10.4049/jimmunol.1501312.
- Brazil JC, Louis NA, Parkos CA. The role of polymorphonuclear leukocyte trafficking in the perpetuation of inflammation during inflammatory bowel disease. Inflamm Bowel Dis. 2013;19(7):1556–1565. PMID:PMC4110963. doi:10.1097/MIB.0b013e318281f54e.
- Colgan SP, Curtis VF, Campbell EL. The inflammatory tissue microenvironment in ibd. Inflamm Bowel Dis. 2013;19(10):2238–2244. PMID:3749286. doi:10.1097/MIB.0b013e31828dcaaf.
- Lambeth JD, Neish AS. Nox enzymes and new thinking on reactive oxygen: A double-edged sword revisited. Annu Rev Pathol-Mech. 2014;9:119–145. doi:10.1146/annurev-pathol-012513-104651.
- Mangoni ML, McDermott AM, Zasloff M. Antimicrobial peptides and wound healing: biological and therapeutic considerations. Exp Dermatol. 2016;25(3):167–173. doi:10.1111/exd.12929.
- Okai S, Usui F, Ohta M, Mori H, Kurokawa K, Matsumoto S, Kato T, Miyauchi E, Ohno H, Shinkura R. Intestinal iga as a modulator of the gut microbiota. Gut Microbes. 2017;8(5):486–492. doi:10.1080/19490976.2017.1310357.
- Smithson JE, Campbell A, Andrews JM, Milton JD, Pigott R, Jewell DP. Altered expression of mucins throughout the colon in ulcerative colitis. Gut. 1997;40(2):234–240. doi:10.1136/gut.40.2.234.
- Everard A, Belzer C, Geurts L, Ouwerkerk JP, Druart C, Bindels LB, Guiot Y, Derrien M, Muccioli GG, Delzenne NM, et al. Cross-talk between akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc Natl Acad Sci U S A. 2013;110(22):9066–9071. PMID:3670398. doi:10.1073/pnas.1219451110.
- Derrien M, Van Baarlen P, Hooiveld G, Norin E, Muller M, de Vos WM. Modulation of mucosal immune response, tolerance, and proliferation in mice colonized by the mucin-degrader akkermansia muciniphila. Front Microbiol. 2011;2:166. PMID:3153965. doi:10.3389/fmicb.2011.00166.
- Plovier H, Everard A, Druart C, Depommier C, Van Hul M, Geurts L, Chilloux J, Ottman N, Duparc T, Lichtenstein L, et al. A purified membrane protein from akkermansia muciniphila or the pasteurized bacterium improves metabolism in obese and diabetic mice. Nat Med. 2017;23(1):107–113. doi:10.1038/nm.4236.