ABSTRACT
The barrier function of epithelia and endothelia depends on tight junctions, which are formed by the polymerization of claudins on a scaffold of ZO proteins. Two differentially spliced isoforms of ZO-1 have been described, depending on the presence of the α domain, but the function of this domain is unclear. ZO-1 also contains a C-terminal ZU5 domain, which is involved in a mechano-sensitive intramolecular interaction with the central (ZPSG) region of ZO-1. Here we use immunoblotting and immunofluorescence to map the binding sites for commercially available monoclonal and polyclonal antibodies against ZO-1, and for a new polyclonal antibody (R3) that we developed against the ZO-1 C-terminus. We demonstrate that antibody R40.76 binds to the α domain, and the R3 antibody binds to the ZU5 domain. The (α+) isoform of ZO-1 shows higher expression in epithelial versus endothelial cells, and in differentiated versus undifferentiated primary keratinocytes, suggesting a link to epithelial differentiation and a potential molecular adaptation to junctions subjected to stronger mechanical forces. These results provide new tools and hypotheses to investigate the role of the α and ZU5 domains in ZO-1 mechano-sensing and dynamic interactions with the cytoskeleton and junctional ligands.
Introduction
Tight junctions (TJ) are critically important for the development and physiology of vertebrate organisms. By generating semipermeable barriers to ions, water, and solutes, they allow tissues to separate different compartments of the extracellular space, modulate absorption of nutrients, and prevent pathogen entry into the body.Citation1–Citation4 TJ are localized at the apico-lateral border of polarized epithelial cells, where they form a continuous circumferential belt, placed immediately apical to the cadherin-based zonula adhaerens .Citation5 TJ are also present in endothelial tissues and cells, although here they are molecularly intermixed with adherens junctions.Citation6,Citation7 The barrier function properties of endothelial and epithelial tissues are highly variable, depending on the physiological requirements of the tissue, and can be altered in disease states.Citation1–Citation3,Citation8,Citation9
At the molecular level, the barrier is formed by a network of intramembrane strands generated by the trans-association of cis-polymers of claudins.Citation4,Citation10–Citation12 The polymerization of claudins into strands critically requires the assembly of a cytoplasmic scaffold formed by ZO proteins.Citation13,Citation14 ZO proteins (ZO-1, ZO-2, and ZO-3) were discovered in the 80s and 90s, thanks to the development of monoclonal antibodies raised against semi-purified junctional membrane fractions of epithelial tissues, and through co-immunoprecipitation studies.Citation15–Citation18 The molecular structure of ZO proteins comprises three N-terminal PDZ domains (PDZ1, PDZ2, PDZ3), a central region that contains SH3 and GUK domains, and a C-terminal region of different length.Citation19,Citation20 In ZO-1 and ZO-2, the C-terminal domain contains an actin-binding region (ABR).Citation21,Citation22 Indeed, ZO-1 and ZO-2 are fundamentally important for the linkage of TJ transmembrane proteins to actin filaments, Citation23–Citation25 and for the organization and contractility of the cortical and junctional actomyosin cytoskeleton.Citation26–Citation30 The C-terminus of ZO-1 also contains a ≈ 100 residue ZU5 domain, that was first identified in ZO-1 and in the netrin receptor UNC-5.Citation31,Citation32 FRAP studies have shown that ZO-1 dynamically exchanges between cytoplasmic soluble and junction-associated soluble and stable pools, and its dynamics depends on interactions with the actomyosin cytoskeleton.Citation33,Citation34
Recent work from our laboratory showed that ZO-1 exists in extended and folded conformations, which show different ligand-binding properties in vitro and in cells, depending on actomyosin-generated force and heterodimerization.Citation35 In the extended/stretched conformation, the N-terminal and C-terminal ends of ZO-1 are physically separated, and the molecules are arranged in a regular array with respect to the junctional membrane. The folded/autoinhibited conformation of ZO-1 is observed in cells depleted of ZO-2, when actomyosin-dependent force has been disrupted either by drugs or by growth on soft substrates.Citation35 The folded conformation of ZO-1 results from a mechano-sensitive intra-molecular interaction between a C-terminal fragment of ZO-1, that contains the ZU5 domain, and the ZPSG (PDZ3-SH3-GUK-U6) central region. In the folded conformation, ZO-1 cannot bind to its ligands ZONAB/DbpA and occludin, leading to downstream modulation of nuclear signaling and barrier function, respectively.Citation35 Although the function of the ZU5 domain is not well understood, FRAP studies suggest that it is important for the dynamics of ZO-1 and for barrier function.Citation36 Another domain of ZO-1 whose function is not completely understood is the α domain, which is localized between the ZPSG and the ABR. This domain was identified as a differentially spliced domain, which defined two isoforms of ZO-1, α(+) and α(-),Citation37 which are differentially expressed in early mouse developmentCitation38 and in different tissues.Citation39
Monoclonal and polyclonal antibodies against ZO-1 have been described and are available from commercial providers. However, the binding site for monoclonal R40.76Citation40 is not known, and to our knowledge, no antibody has been described against the C-terminal ZU5 domain of ZO-1. Here we used immunoblotting and immunofluorescence to map the binding sites of anti-ZO-1 antibodies. We show that monoclonal antibody R40.76 binds to the α domain of ZO-1, and a new R3 antibody, that we developed, specifically recognizes the ZU5 domain. Neither domain is required for the junctional localization of ZO-1. Furthermore, we examine the expression of the (α+) and (α-) isoforms of ZO-1 in different cell lines and experimental conditions, and on the basis of our results and data from the literature we propose that the ZO-1 (α+) isoform is a marker of epithelial differentiation and is tuned to junctions subjected to higher mechanical force.
Results
R40.76 and R3 bind to the α and ZU5 domains, respectively
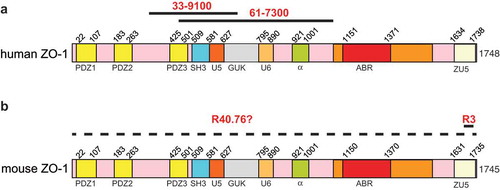
The mouse monoclonal antibody (33–9100) and the rabbit polyclonal antibody (61–7300) were raised against antigens that comprise sequences within the N-terminal half of human ZO-1 ()). Specifically, 61–7300 was generated against a 69 kD fusion protein corresponding to amino acids 463–1109 of human ZO-1,Citation19 which comprise the PDZ3, SH3, U5, GUK, U6 and α domains ()). Mouse antibody 33–9100 was raised against a human recombinant ZO-1 fusion protein encompassing amino acids 334–634, which comprises PDZ3, SH3, U5, and part of the GUK domains ()). However, no information is available regarding the localization of the epitope recognized by the rat monoclonal antibody R40.76, which was generated against a junction-enriched preparation of mouse liver canalicular membranes and was used in the early characterization of ZO-1Citation40 ()). Polyclonal antibody R3 was generated in our laboratory against a 22-residue peptide coding for amino-acids 1673–1695 of mouse ZO-1, which lies within the ZU5 domain ()). All these antibodies recognize human ZO-1, except for R40.76, which recognizes ZO-1 from mouse and dog, but not human ZO-1.
Figure 1. Antigens for anti-ZO-1 antibodies.
Schematic diagrams of human (a) and mouse (b) ZO-1, with numbers indicating the position of different structural domains along the respective sequences. Continuous black lines indicate antigens used for the production of the respective antibodies ((a): 33–9100, 61–7300; (b) R40.76 and R3). The dashed line in (b) indicates that the localization of the mouse ZO-1 epitope recognized by R40.76 is not known.
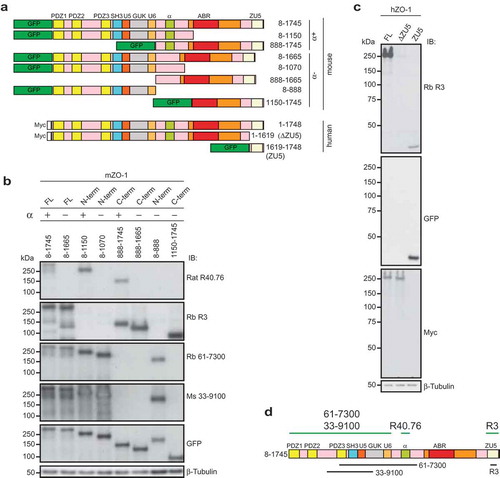
To map the antibody binding sites, we expressed in HEK cells GFP-tagged forms of either full-length mouse ZO-1 α(+) and α(-) isoforms, or C-terminal and N-terminal truncations of both isoforms, or myc-tagged constructs of human ZO-1 with and without the ZU5, domain, or a GFP-tagged construct comprising the ZU5 domain ()). The HEK lysates were analyzed by immunoblotting with the anti-ZO-1 antibodies (). Antibodies against either GFP or myc were used to verify the expression of the transgene, and antibodies against β-tubulin were used to normalize protein content (). The monoclonal R40.76 antibody labeled only the full-length ZO-1 and truncated construct that contained the 80-residue α motif (α(+) isoform) ()). In contrast, the full-length α(-) isoform and the truncated N-terminal and C-terminal fragments lacking the α motif showed no reactivity with R40.76 ()). Antibody R3 recognized both α(+) and α(-) full-length and C-terminal mouse constructs, but not the N-terminal constructs ()). The rabbit (61–7300) and mouse (33–9100) antibodies recognized both α(+) and α(-) full-length and N-terminal constructs, but not the C-terminal constructs ()). The R3 antibody labeled the myc-tagged full-length human ZO-1 and the GFP-tagged isolated ZU5 domain, but not the full-length human ZO-1 lacking the ZU5 domain (ΔZU5, 1–1619) ()). In summary, immunoblotting analysis defined the amino-acid stretches recognized by the different antibodies ()).
Figure 2. The rat monoclonal R40.76 and the rabbit polyclonal R3 bind to the α and ZU5 domains of ZO-1, respectively.
(a). Schematic diagrams of mouse and human ZO-1 constructs expressed in HEK cells, used for the mapping of antibody epitopes. The N-terminal tags, either GFP or myc, the amino-acid residues of ZO-1 included in each construct, and the presence or absence of the α domain (α+) or (α-) are indicated. (b). Antibody R40.76 binds to the α domain. Immunoblotting analysis of HEK cell lysates expressing the mouse ZO-1 constructs. FL, N-term and C-term constructs, the presence (+) or absence (-) of the α domain, and amino-acid residues of each construct are indicated on top. Numbers on the left indicate the migration of pre-stained molecular weight markers (kDa). Antibodies used for IB are indicated on the right. Antibodies against β-tubulin were used to normalize lysates. (c). Antibody R3 binds to the ZU5 domain. Immunoblotting analysis of HEK cell lysates expressing the human FL, ΔZU5, and ZU5 constructs, using antibodies against ZO-1 (R3) and GFP and myc tags. (d). Schematic diagram summarizing experimentally determined the reactivity of antibodies against indicated regions/constructs of mouse ZO-1 (green lines above the scheme), and position of antigens (black lines below the scheme)
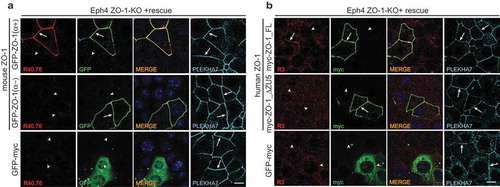
Next, we examined the reactivity of the R40.76 and R3 antibodies against full-length and mutated ZO-1 constructs expressed in the background of ZO-1-KO Eph4 cellsCitation35,Citation41 (). In agreement with the immunoblotting data, immunofluorescent signal for ZO-1 was detected by the R40.76 antibody only when KO cells were rescued with the α(+), but not the α(-) mouse ZO-1 isoform ()). Similarly, to confirm the specificity of the R3 polyclonal antiserum, we rescued ZO-1-KO cells either with full-length human ZO-1, or with a construct lacking the C-terminal ZU5 domain (ΔZU5) ()). Only cells that expressed the full-length construct, but not the cells expressing the construct lacking the ZU5 domain, showed junctional labeling ()). The localization of the α(-) and ΔZU5 constructs at junctions showed that these domains are not required for the localization of ZO-1 at junctions, in agreement with previous studies showing that sequences in the N-terminal half of ZO-1 are necessary and sufficient for ZO-1 junctional localization.Citation24
Figure 3. The α and ZU5 domains of ZO-1 are not required for the junctional localization of ZO-1.
(a–b). Immunofluorescent analysis of ZO-1-KO Eph4 cells, using either R40.76 (a) or R3 (B) antibodies, after transfection of ZO-1-KO cells either with GFP-tagged mouse constructs either with or without the α domain (a), or with myc-tagged human ZO-1 constructs either with or without the ZU5 domain (b). Control cells (bottom row) were transfected with GFP-myc alone, which was labeled with antibodies against either GFP (A, second column), or myc (B, second column). Arrows indicate junctional localization, arrowheads indicate undetectable labeling at junctions that contain the zonula adhaerens protein PLEKHA7 (A and B, 4th column), used as a reference junction marker. Merge images are shown in the third column (A, B, third column), with nuclei labeled by DAPI. Scale bar = 10 μm. Exogenous ZO-1 lacking the α domain is localized at junctions but is not labeled by R40.76, and exogenous ZO-1 lacking the ZU5 domain is localized at junctions but is not labeled by R3.
ZO-1 α(+) expression is higher in epithelial than endothelial cells, and is induced by keratinocyte differentiation
To assess the expression of the α(+) and α(-) isoforms of ZO-1, we examined by immunoblotting lysates from different epithelial and non-epithelial cells from dog, mouse, and human origin (). The two isoforms of ZO-1 can be identified through their slightly different electrophoretic mobility.Citation37 Both α(+) and α(-) isoforms were detected by antibodies R3, 61–7300, and 33–9100 in lysates of mouse kidney collecting duct (mCCD), human umbilical vein endothelial cells (HUVEC) mouse brain microvascular cells (bEnd.3), mouse endothelial cells (H5V), mouse aortic endothelium-derived cells (meEC), human lung carcinoma cells (Calu-I), and mouse mammary epithelium cells (Eph4) ()). Monoclonal R40.76 only labeled the more slowly migrating, upper polypeptide, in agreement with the observation that R40.76 only binds to the isoform that contains the α(+) motif ()). In MDCK lysates all the antibodies labeled one major polypeptide, which was also recognized by the R40.76 antibody, demonstrating that MDCK cells only or predominantly express the α(+) isoform ()). In lysates of human lung carcinoma cells (A427), human myeloblastic-derived cells (Hap1) and human keratinocytes (HaCaT) there was very low or undetectable labeling for the α(+) isoform, indicating that these cells mostly express the α(-) isoform (). In contrast, the signal for the α(-) isoform was stronger than the signal for the α(+) isoform in lysates of endothelial cells that express VE-cadherinCitation42 (HUVEC, bEnd.3, H5V) ()). Significantly, roughly equal labeling for both isoforms was detected in lysates of meEC cells, which are derived from endothelium, but have lost VE-cadherin expression and acquired E-cadherin expression, and contain a continuous junctional circumferential beltCitation42 (). In summary, these observations indicate that the α(-) isoform is predominant in endothelial cells (see alsoCitation39) whereas epithelial cells express roughly the same amount of α(+) and α(-) isoforms, except for MDCKII cells, where the α(-) isoform is very low or undetectable.
Figure 4. Increased expression of the ZO-1 (α+) isoform correlates with epithelial differentiation.
(a). Immunoblotting analysis of lysates from the indicated cell lines, using antibodies against ZO-1 indicated on the right. (b) Quantification or relative expression of (α+) and (α-) IB signals in the indicated cell lines, following densitometric analysis (Materials and Methods). (c) Immunoblotting analysis of lysates from primary mouse keratinocytes either from WT or CGN-KO cells, either cultured in the absence of 1.2 mM calcium chloride (-), or after calcium-induced differentiation (+), using antibodies indicated on the right. Anti-cingulin antibody (C532) was used to verify genotype. Lysates of mouse mCCD cells were used as a control for the migration and expression of the two ZO-1 isoforms. (d). Quantification of relative expression of (α+) and (α-) in mouse primary keratinocytes, and mCCD cells, based on densitometric analysis of the data shown in (c).
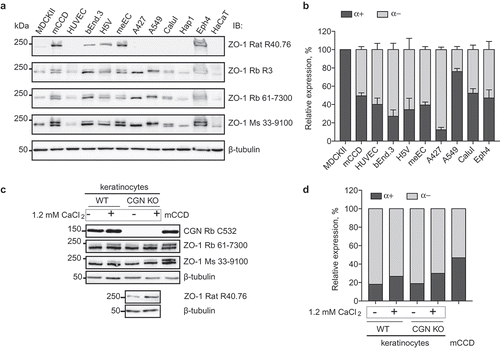
Differentiating agents such as histone deacetylase inhibitors up-regulate the expression of TJ proteins,Citation43 and increased extracellular calcium concentration drives terminal differentiation of keratinocytes and junction formation both in vitro and in vivo.Citation44–Citation47 Thus, we examined the expression of ZO-1 α(+) and α(-) isoforms in primary cultures of undifferentiated and calcium-differentiated primary keratinocyte cultures from wild-type and CGN-KO mice.Citation48,Citation49 Immunoblotting analysis with antibodies 61–7300 and 33–9100 showed that undifferentiated keratinocytes express both isoforms ()), but the α(+) isoform was expressed at low levels ()). In contrast, upon calcium-induced differentiation, there was an increase of the expression of the α(+) isoform (), which was also detected using the R40.76 antibody ()). The increase in the immunoblotting signal for the α(+) isoform was similar in CGN WT and KO keratinocytes (), indicating that cingulin, which is recruited to TJ by ZO-1Citation41,Citation50 does not influence the relative levels of expression of ZO-1 isoforms.
Discussion
Antibodies are essential tools to characterize the cellular and subcellular distribution of proteins and their isoforms, and can also be used to identify and modulate intramolecular interactions in vitro and in vivo.Citation35,Citation51–Citation53 Recently we showed that antibody labeling can be used in combination with high-resolution microscopy, to define the spatial relationship between N-terminal and C-terminal ends of ZO-1, and to study the effect of mechanical force on ZO-1 conformation.Citation35 Here we report that the epitope recognized by the monoclonal antibody R40.76 lies within the ZO-1 α domain, and we describe a new polyclonal antibody (R3) against the ZU5 domain of ZO-1. These results are useful to advance our knowledge about the expression and functions of the α(+) and α(-) ZO-1 isoforms and to investigate the spatial localization and function of the ZU5 domain in different experimental contexts.
ZO-1 is a key protein of vertebrate TJ, because of its role in the clustering and polymerization of claudin-based strands, and in the organization and junctional anchoring of the actomyosin cytoskeleton. Determining the role of the different structural domains of ZO-1 is crucially important to understand its mechanisms of functioning in epithelial and endothelial cells and tissues. Recently we showed that ZO-1 is a mechano-sensor, since its conformation in cells can either be extended/stretched or folded/autoinhibited, depending on mechanical force and heterodimerization with ZO-2.Citation35 The autoinhibited conformation is mediated by an intramolecular interaction between the ZU5 domain (C-terCitation35), and a central region of ZO-1, comprising the PDZ3, SH3 and GUK domains (ZPSG,Citation35). Little information is available on the function of the ZU5 domain. FRAP studies indicate that the ZU5 stabilizes ZO-1 and contributes to the regulation of barrier function.Citation36 The ZU5 domain of ZO-1 was also reported to interact with MRCKβ, a Cdc42 effector kinase that modulates cell protrusion and migration at the leading edge of migrating cells.Citation54 The activation of Cdc42 primes the kinase to be in a conformation that allows interaction with the ZU5 domain, and this interaction promotes the accumulation of the kinase at the leading edge of migrating cells.Citation54 The ZPSG region of ZO-1 binds to several ligands, including DbpA, occludin, afadin, and α-catenin,Citation14 and we showed that when ZO-1 is in the folded/autoinhibited conformation, the binding of DbpA and occludin is blocked by the intramolecular interaction between the ZU5 and the ZPSG.Citation35 In the extended conformation, a tag downstream from the ZU5 domain can be resolved from an N-terminal tag of ZO-1.Citation35 Thus, an antibody specific for the ZU5 domain can be useful to study the mechano-regulation of endogenous ZO-1, by establishing its position relative to the N-terminal region and/or its ligands. Previous studies established that the molecular environment of the N-terminal and C-terminal ends of ZO-1 is distinct,Citation55 in agreement with our observations about the spatial separation of the two ends of ZO-1 when it is in the extended/stretched conformation.Citation35
Here we also report the mapping of the epitope of the R40.76 rat monoclonal antibodyCitation40 within the α domain of ZO-1. The α domain is an 80-residue stretch that was detected in some, but not all initial ZO-1 cDNAs isolates,Citation37 resulting in two isoforms can be distinguished on the basis of their electrophoretic migration.Citation37,Citation39,Citation40,Citation56 Based on the abundant literature where the use of R40.76 is described,Citation35,Citation37,Citation40,Citation57–Citation62 it appears that essentially all cultured and/or native epithelial cells, for example, from intestine, liver, lung, mammary gland, and skin, express the α(+) isoform. However, the relative ratios for the two isoforms vary depending on cell types, from 0.5:1 to 20:1 (α(-)/α(+)).Citation37 Using affinity-purified antibodies that reacted exclusively with either the α(+) or the α(-) isoforms of ZO-1, it was indeed shown that the two isoforms are differentially expressed in cultured cells and mouse embryos and tissues.Citation38,Citation39,Citation56 For example, using an affinity purified polyclonal antibody specific for the α domain, the signal for the α(+) isoform was missing from the filtration slits diaphragms of rat kidney glomeruli, but the same structure was labeled by antibodies that recognize both α(+) and α(-) isoforms,Citation56,Citation63 suggesting that only ZO-1 α(-) is expressed in glomerular slits. Using affinity purified antibodies against the two isoforms, the α(+) isoform was detected in stomach epithelial cells, and the α(-) isoform in capillary endothelial junctions, Sertoli cells, and glomerular slits.Citation39 Maternal mRNA coding for the ZO-1 α(-) isoform was detected in unfertilized mouse oocytes and in all preimplantation stages, whereas α(+) transcripts were first detected after the 8-cell stage.Citation38 ZO-1 α(+) protein was detectable at TJ at the 16–32-cell stage, immediately before TJ become functional, and blastocoele fluid accumulation begins.Citation38 Here we show that in MDCK cells the α(-) isoform is essentially undetectable, suggesting that it is either not expressed or expressed at very low levels, whereas in Hap1 and HaCaT cells the α(+) isoform is not detectable. We also show that epithelial and endothelial cells show a lower α(+)/α(-) ratio, in agreement with previous results.Citation39 Interestingly, meEC cells originated from endothelial aortic cells, but have differentiated into an epithelial phenotype, as detected from the switch in expression from VE-cadherin to E-cadherin.Citation42 In these cells, there is a higher α(+) to α(-) ratio than in endothelial cells (bEnd.3, H5V, HUVEC) that have retained expression of VE-cadherin (bEnd.3, H5V, HUVEC).Citation42 These results indicate that epithelial differentiation and expression of E-cadherin correlate with increased expression of the α(+) isoform, in agreement with previous studies on Caco2 cells.Citation64
The function of the α domain is not known. The α domain is located immediately downstream of the U6 ZO-1 domain, which has been implicated in the modulation of the interaction of the U5-GUK domain with occludin, and the TJ localization of ZO-1Citation65 (). An early hypothesis was that the α domain contributes to modulating TJ barrier function and that the expression of the α(-) isoform reflects a “leaky” TJ.Citation56 However, the observation that the α(+) isoform is detected in both leaky and tight epithelia was not consistent with this idea.Citation56 It is now known that TJ leakiness to ions depends on the tissue-specific expression of claudinsCitation4,Citation66 and that the actomyosin cytoskeleton modulates the paracellular permeability barrier both to ions and solutes (reviewed inCitation1,Citation67). Interestingly, it was recently demonstrated that TJ positioning in the stratified skin epithelium depends on EGFR- and E-cadherin- integrated mechano-transduction in the uppermost layer (SG2) of the stratum granulosum.Citation68 In this latter study ZO-1 was labeled for immunofluorescence with the R26.4C antibody, which also recognizes only one polypeptide in lysates of MDCK cells and mouse tissues,Citation40 indicating that, as R40.76, it is specific for the α(+) isoform. It was found that ZO-1 is expressed in all skin layers, in a punctate pattern, but is highly enriched at circumferential cell-cell contacts of the SG2 layer.Citation68 This same layer also displayed highly enriched staining for vinculin, and for the mechano-sensitive epitope of α-catenin, indicating higher mechanical tension.Citation68 Knock-out of E-cadherin resulted in a reduction in tension across intercellular contacts and reduced junctional ZO-1 labeling. Importantly, the formation of circumferential zonulae adhaerentes and TJ is associated with the establishment of a highly contractile peri-junctional ring of actomyosin that connects the cytoskeletons of adjacent cells.Citation69 Here we show that primary keratinocyte differentiation, which recapitulates differentiation from the lower to upper layers of the epidermis, correlates with increased levels of ZO-1 α(+) isoform, as detected by immunoblotting. Collectively, these observations and the correlation between increased E-cadherin-mediated junctional tension in the apical layers and junctional accumulation of ZO-1 α(+) suggest that the α(+) isoform is specifically adapted to junctions subjected to stronger mechanical forces, such as those generated in the upper skin layer. Similarly, the expression of the α(+) isoform just prior to blastocoele formationCitation38 may be related to the increased hydrostatic pressure resulting from the accumulation of apical fluid in the blastocoele. Finally, when comparing ZO-1 isoform expression in cells derived either from the proximal kidney tubule (MDCK) or from kidney collecting ducts (mCCD), the higher expression of the α(+) isoform in MDCK cells, with respect to mCCD cells, may be related to the fact that intratubular fluid hydrostatic pressure and flow rate are much higher in the proximal segments of the nephron .Citation70 Taken together, these observations indicate that the α(+) domain is required for ZO-1 to function at junctions that are subjected to higher mechanical stresses. Whether this is through modulation of intramolecular ZO-1 interactions, or through modulation of interactions with other junctional ligands remains to be established. It will be very interesting to test these ideas, and explore the role of the α domain in the mechano-sensing, dynamics, and molecular interactions of ZO-1.
Materials and methods
Cell culture
MDCKII, mCCD, bEnd.3, H5V, meEC, A427, A549, and HaCaT were cultured as previously described.Citation42 Calu-I (human non-small-cell lung carcinoma, a kind gift of Marco Paggi, Regina Elena National Cancer Institute, Italy)Citation71 was cultured in RPMI (Gibco) supplemented with 10% inactivated fetal bovine serum (FBS, PAN Biotech), 1x minimal essential medium non-essential amino acids (MEM NEAA, PAN Biotech), 100 units/ml penicillin and 100 μg/ml streptomycin (PAN Biotech). Eph4 and Hap1 cells were cultured as previously describedCitation35,Citation72,Citation73. HUVEC cell line (a kind gift of Beat Imhof, University of Geneva, Switzerland) was cultured in M199 medium (Gibco) supplemented with 20% inactivated fetal bovine serum (FBS, PAN Biotech), 15 µg/ml Endothelial Cell Growth Supplement (ECGS), 10 µg/ml heparin (Sigma H-3149), 100 nM hydrocortisone, 10 µg/ml vitamin C, 1x penicillin-streptomycin-glutamine (Thermo Fisher Scientific) on tissue culture plates pre-coated with 10 µg/ml collagen G and 0.2% gelatin (15–30 min at 37°C). Mouse primary keratinocytes were isolated from WT and CGN-KO newborn mice as described.Citation49 Differentiation was obtained by allowing cells to reach confluence, and then incubating them in medium containing 1.2 mM CaCl2 for 48 h. Cells were plated on collagen I-coated tissue culture dishes at 1 × 105 cells/cm2. HEK293T cells were cultured in DMEM, supplemented with 10% FBS, and antibiotics (100 U/ml penicillin and 100 µg/ml streptomycin (PAN Biotech)).
Antibodies
Primary antibodies were: monoclonal rat anti-ZO-1 (R40.76, 1:500 IB, 1:100 IF a gift from Prof. Daniel Goodenough, Harvard Medical School), polyclonal rabbit anti-ZO-1 (Thermo Fischer Scientific, 61–7300, 1:2000 IB) and monoclonal mouse anti-ZO-1 (Thermo Fischer Scientific, 33–9100, 1:2000 IB), rabbit anti-cingulin C532 (1:5000 IB),Citation74 monoclonal mouse anti-GFP (Roche Applied Science, 11814460001, 1:2000 IB, 1:200 IF), mouse anti-myc 9E10 (1:200, IF), guinea pig anti-PLEKHA7 GP2737 (1:300 IF), monoclonal mouse anti-β-tubulin (Invitrogen, 32–2600, 1:5000 IB). The polyclonal rabbit anti-ZO-1 R3 was obtained by immunizing rabbits with a peptide (CRDNSILPPLDKEKGETLLSPLV) corresponding to residues 1673–1695 within the ZU5 domain of mouse ZO-1 (Covalab, Lyon, France), and was used at a dilution of 1:1000 for IB, and 1:100 for IF. Secondary antibodies for IF were anti-mouse and anti-rabbit Alexa Fluor 488, anti-mouse, anti-rabbit, and anti-rat Cy3, anti-guinea pig Alexa Fluor 647 (Jackson ImmunoResearch Europe, Newmarket, UK, 1:300). Anti-mouse and anti-rabbit (1:20000, Promega, W402B and W401B, respectively), and anti-rat (1:10000, Thermo Fisher Scientific, 62–9520) IgG, HRP-conjugated antibodies were used for immunoblotting.
Plasmids and transfections
cDNAs coding for either the α (+) (residues 8–1745) or α(-) isoforms (residues 8–1665) of mouse ZO-1 were obtained from sequences in different clones obtained by screening λgt11 expression libraries.Citation18,Citation23 After PCR amplification, the indicated sequences were cloned into the NotI and Acc65I sites of a pCDNA3.1myc vector backbone, engineered to contain GFP between the BamHI and NotI sites.Citation75 To generate N-terminal and C-terminal fragments of the two ZO-1 isoforms, cDNAs corresponding to the indicated residues were amplified by PCR, and similarly inserted between the NotI and Acc65I sites of pCDNA-GFP-BamH1-Not1-Acc65I. Constructs of human full-length ZO-1 (residues 1–1748) were described previously.Citation35 PCR amplification was performed on myc-ZO-1 (human) full-length with appropriate oligonucleotides for generation of depleted ZO-1 constructs, i.e., myc-ZO-1-ΔZU5 (residues 1–1619) subsequently cloned into the BamHI-XhoI sites of previously prepared pCDNA3.1 vector. PCR amplification was performed on myc-ZO-1 full-length with appropriate oligonucleotides for generation of ZU5 constructs (residues 1619–1748) subsequently cloned into the KpnI-NotI sites of previously prepared pCDNA3.1 vector. All constructs obtained by PCR were verified by sequencing the entire insert. Transfections of HEK cells were performed using Lipofectamine2000, following the manufacturer’s guidelines (Thermo Fisher Scientific). HEK cells (2x106) plated in 100 mm2 dish were transfected with 20 μg of different ZO-1 constructs, lysed 2 days after transfection. Transfections of Eph4 cells were carried out using jetOPTIMUS DNA transfection reagent (Polyplus), following the manufacturer’s guidelines.
Preparation of lysates, SDS-PAGE and immunoblotting
HEK cells, used for expression of ZO-1 constructs, were washed with cold PBS, lysed with CO-IP buffer (150 mM NaCl, 20 mM Tris-HCl, pH 7.5, 1% Nonidet P-40, 1 mM EDTA, 5 mg/ml antipain, 5 mg/ml leupeptin, 5 mg/ml pepstatin, 1 mM PMSF) with Roche inhibitor (1x) at 4°, and incubated for 15 min with gentle agitation. Lysates were sonicated 5 s at 66% power (3 bursts), centrifuged 20 min at 13,000 rpm, and supernatants were recovered. For the preparation of lysates and immunoblotting from other cultured cell lines, the procedure was as described in,Citation42 using RIPA buffer. Mouse primary keratinocytes were lysed with 250 µl RIPA buffer (150 mM NaCl, 40 mM Tris-HCl, pH 7.5, 2 mM EDTA, 10% glycerol, 1% Triton X-100, 0.5% sodium deoxycholate, 0.2% SDS, protease inhibitors) for 10 min at 4°C, followed by sonication (5 s at 33% amplitude with a Branson sonifier). Proteins were mixed with SDS sample buffer and incubated at 95°C for 5 min. Protein lysates were analyzed on SDS gels (8% acrylamide, 100 V). For immunoblots, gels were transferred onto nitrocellulose (0.45 μm) (100 V for 80 min at 4°C), and blots were incubated with primary antibody, followed by secondary HRP-labeled antibody, and visualized with the ECL (Amersham). Quantification of IB signal was performed using Image Studio Lite (LI-COR). The sum of signal intensities of upper band (corresponding to α+) and lower band (corresponding to α-) of immunoblots using anti-ZO-1 antibodies was taken as 100%. Then, the relative expression of each isoform was calculated, in relation to the sum of their signal intensities. Data in ) represent the mean of three densitometric analyses using antibodies recognizing both ZO-1 isoforms, carried out on two independent experimental repeats (n = 2). Each blot was repeated two times, and data were pooled from blots using the three different antibodies. Data are represented as a mean with the bars indicating standard deviation. Graphs were generated using Prism 7 (GraphPad).
Immunofluorescence
Eph4 ZO-1-KO cells were seeded on glass coverslips in 24 well plates at a density of 100,000 cells/well and were transfected the next day. Two days after transfection monolayers were fixed with cold methanol for 10 min at −20°C, incubated with primary antibodies overnight at 4°C, and then incubated with fluorophore conjugated secondary antibodies for 30 min at 37°C. Coverslips were mounted with Vectashield containing DAPI (Reactolab) and observed with Zeiss LSM800 confocal microscope using a 63x objective.
Author contribution
FR and EV prepared constructs, carried out cell culture, transfections, immunoblotting and immunofluorescence analyses, and prepared Figures. DS and ST provided essential constructs and cDNAs. FR, EV, and ST provided comments on the manuscript SC initiated, coordinated and supervised the project, wrote the manuscript and finalized Figures.
Acknowedgements
The Citi laboratory is supported by the Swiss National Foundation (31003A_172809) and by the State of Geneva. We thank Christina Van Itallie, James Anderson and Eric Feraille for discussions, and Lionel Jond for technical assistance.
Additional information
Funding
References
- Anderson JM, Van Itallie CM. Physiology and function of the tight junction. Cold Spring Harb Perspect Biol. 2009;1(a002584):1–16. doi:10.1101/cshperspect.a002584.
- Buckley A, Turner JR. Cell biology of tight junction barrier regulation and mucosal disease. Cold Spring Harb Perspect Biol. 2018;10(1) doi: 10.1101/cshperspect.a029314.
- Citi S. Intestinal barriers protect against disease. Science. 2018;359(6380):1097–1098. doi:10.1126/science.aat0835.
- Tsukita S, Tanaka H, Tamura A. The claudins: from tight junctions to biological systems. Trends Biochem Sci. 2019;44(2):141–152. doi:10.1016/j.tibs.2018.09.008.
- Zihni C, Mills C, Matter K, Balda MS. Tight junctions: from simple barriers to multifunctional molecular gates. Nat Rev Mol Cell Biol. 2016;17(9):564–580. doi:10.1038/nrm.2016.80.
- Dejana E. Endothelial cell-cell junctions: happy together. Nat Rev Mol Cell Biol. 2004;5(4):261–270. doi:10.1038/nrm1357.
- Dejana E, Orsenigo F. Endothelial adherens junctions at a glance. J Cell Sci. 2013;126(Pt 12):2545–2549. doi:10.1242/jcs.124529.
- Gonzalez-Mariscal L, Lechuga S, Garay E. Role of tight junctions in cell proliferation and cancer. Prog Histochem Cytochem. 2007;42(1):1–57. doi:10.1016/j.proghi.2007.01.001.
- Krug SM, Schulzke JD, Fromm M. Tight junction, selective permeability, and related diseases. Semin Cell Dev Biol. 2014;36:166–176. doi:10.1016/j.semcdb.2014.09.002.
- Tsukita S, Furuse M, Itoh M. Multifunctional strands in tight junctions. Nat Rev Mol Cell Biol. 2001;2(4):285–293. doi:10.1038/35067088.
- Suzuki H, Nishizawa T, Tani K, Yamazaki Y, Tamura A, Ishitani R, Dohmae N, Tsukita S, Nureki O, Fujiyoshi Y. Crystal structure of a claudin provides insight into the architecture of tight junctions. Science. 2014;344(6181):304–307. doi:10.1126/science.1248571.
- Suzuki H, Tani K, Tamura A, Tsukita S, Fujiyoshi Y. Model for the architecture of claudin-based paracellular ion channels through tight junctions. J Mol Biol. 2015;427(2):291–297. doi:10.1016/j.jmb.2014.10.020.
- Umeda K, Ikenouchi J, Katahira-Tayama S, Furuse K, Sasaki H, Nakayama M, Matsui T, Tsukita S, Furuse M, Tsukita S. ZO-1 and ZO-2 independently determine where claudins are polymerized in tight-junction strand formation. Cell. 2006;126(4):741–754. doi:10.1016/j.cell.2006.06.043.
- Fanning AS, Anderson JM. Zonula occludens-1 and −2 are cytosolic scaffolds that regulate the assembly of cellular junctions. Ann N Y Acad Sci. 2009;1165:113–120. doi:10.1111/j.1749-6632.2009.04440.x.
- Stevenson BR, Siliciano JD, Mooseker MS, Goodenough DA. Identification of ZO-1: a high molecular weight polypeptide associated with the tight junction (zonula occludens) in a variety of epithelia. J Cell Biol. 1986;103:755–766. doi:10.1083/jcb.103.3.755.
- Gumbiner B, Lowenkopf T, Apatira D. Identification of a 160 kDa polypeptide that binds to the tight junction protein ZO-1. Proc Natl Acad Sci USA. 1991;88:3460–3464. doi:10.1073/pnas.88.8.3460.
- Balda MS, Gonzalez-Mariscal L, Matter K, Cereijido M, Anderson JM. Assembly of the tight junction: the role of diacylglycerol. J Cell Biol. 1993;123:293–302. doi:10.1083/jcb.123.2.293.
- Itoh M, Nagafuchi A, Yonemura S, Yasuda-Kitani T, Tsukita S, Tsukita S. The 220kD protein colocalizing with cadherins in non-epithelial cells is identical to ZO-1 a tight junction-associated protein in epithelial cells: cDNA cloning and immunoelectron microscopy. J Cell Biol. 1993;121:491–502.
- Willott E, Balda MS, Fanning AS, Jameson B, Van Itallie C, Anderson JM. The tight junction protein ZO-1 is homologous to the drosophila discs-large tumor suppressor protein of septate junctions. Proc Natl Acad Sci USA. 1993;90:7834–7838. doi:10.1073/pnas.90.16.7834.
- Stevenson BR, Haskins J, Hibbard J, Tamber M, Weber D. P130 is homologous to ZO-1 and ZO-2 and is a novel member of the MAGUK family of proteins. Mol Biol Cell. 1996;7:605a. (Abstr.).
- Wittchen ES, Haskins J, Stevenson BR. Protein interactions at the tight junction. Actin has multiple binding partners, and zo-1 forms independent complexes with zo-2 and zo-3. J Biol Chem. 1999;274(49):35179–35185. doi:10.1074/jbc.274.49.35179.
- Fanning AS, Ma TY, Anderson JM. Isolation and functional characterization of the actin binding region in the tight junction protein ZO-1. Faseb J. 2002;16(13):1835–1837. doi:10.1096/fj.02-0121fje.
- Itoh M, Nagafuchi A, Moroi S, Tsukita S. Involvement of ZO-1 in cadherin-based cell adhesion through its direct binding to alpha catenin and actin filaments. J Cell Biol. 1997;138(1):181–192. doi:10.1083/jcb.138.1.181.
- Fanning AS, Jameson BJ, Jesaitis LA, Anderson JM. The tight junction protein ZO-1 establishes a link between the transmembrane protein occludin and the actin cytoskeleton. J Biol Chem. 1998;273(45):29745–29753. doi:10.1074/jbc.273.45.29745.
- Van Itallie CM, Fanning AS, Bridges A, Anderson JM. ZO-1 stabilizes the tight junction solute barrier through coupling to the perijunctional cytoskeleton. Mol Biol Cell. 2009;20(17):3930–3940. doi:10.1091/mbc.e09-04-0320.
- Fanning AS, Van Itallie CM, Anderson JM. Zonula occludens-1 and −2 regulate apical cell structure and the zonula adherens cytoskeleton in polarized epithelia. Mol Biol Cell. 2012;23(4):577–590. doi:10.1091/mbc.E11-09-0791.
- Choi W, Acharya BR, Peyret G, Fardin M-A, Mège R-M, Ladoux B, Yap AS, Fanning AS, Peifer M. Remodeling the zonula adherens in response to tension and the role of afadin in this response. J Cell Biol. 2016;213(2):243–260. doi:10.1083/jcb.201506115.
- Odenwald MA, Choi W, Kuo W-T, Singh G, Sailer A, Wang Y, Shen L, Fanning AS, Turner JR. The scaffolding protein ZO-1 coordinates actomyosin and epithelial apical specializations in vitro and in vivo. J Biol Chem. 2018;293:17317–17335. doi:10.1074/jbc.RA118.003908.
- Odenwald MA, Choi W, Buckley A, Shashikanth N, Joseph NE, Wang Y, Warren MH, Buschmann MM, Pavlyuk R, Hildebrand J, et al. ZO-1 interactions with F-actin and occludin direct epithelial polarization and single lumen specification in 3D culture. J Cell Sci. 2017;130(1):243–259. doi:10.1242/jcs.188185.
- Yamazaki Y, Umeda K, Wada M, Nada S, Okada M, Tsukita S, Tsukita S. ZO-1- and ZO-2-dependent integration of myosin-2 to epithelial zonula adherens. Mol Biol Cell. 2008;19(9):3801–3811. doi:10.1091/mbc.e08-04-0352.
- Ackerman SL, Kozak LP, Przyborski SA, Rund LA, Boyer BB, Knowles BB. The mouse rostral cerebellar malformation gene encodes an UNC-5-like protein. Nature. 1997;386(6627):838–842. doi:10.1038/386838a0.
- Leonardo ED, Hinck L, Masu M, Keino-Masu K, Ackerman SL, Tessier-Lavigne M. Vertebrate homologues of C. elegans UNC-5 are candidate netrin receptors. Nature. 1997;386(6627):833–838. doi:10.1038/386833a0.
- Shen L, Weber CR, Turner JR. The tight junction protein complex undergoes rapid and continuous molecular remodeling at steady state. J Cell Biol. 2008;181(4):683–695. doi:10.1083/jcb.200711165.
- Yu D, Marchiando AM, Weber CR, Raleigh DR, Wang Y, Shen L, Turner JR. MLCK-dependent exchange and actin binding region-dependent anchoring of ZO-1 regulate tight junction barrier function. Proc Natl Acad Sci. 2010;107:8237–8241. doi:10.1073/pnas.0908869107.
- Spadaro D, Le S, Laroche T, Mean I, Jond L, Yan J, Citi S. Tension-dependent stretching activates ZO-1 to control the junctional localization of its interactors. Curr Biol. 2017;27(24):3783–3795. e8. doi:10.1016/j.cub.2017.11.014.
- King JM, Tan CJP, Thomason NC, White AR, Shen L, Turner JR. Zonula occludens-1 ZU5 domain contributes essential stabilizing interactions at the tight junction. Faseb J. 2016;30:1250.7.
- Willott E, Balda MS, Heintzelman M, Jameson B, Anderson JM. Localization and differential expression of two isoforms of the tight junction protein ZO-1. Am J Physiol. 1992;262:C1119–24. doi:10.1152/ajpcell.1992.262.5.C1119.
- Sheth B, Fesenko I, Collins JE, Moran B, Wild AE, Anderson JM, Fleming TP. Tight junction assembly during mouse blastocyst formation is regulated by late expression of ZO-1 alpha+ isoform. Development. 1997;124:2027–2037.
- Balda MS, Anderson JM. Two classes of tight junctions are revealed by ZO-1 isoforms. Am J Physiol. 1993;264:C918–24. doi:10.1152/ajpcell.1993.264.4.C918.
- Anderson JM, Stevenson BR, Jesaitis LA, Goodenough DA, Mooseker MS. Characterization of ZO-1, a protein component of the tight junction from mouse liver and madin-darby canine kidney cells. J Cell Biol. 1988;106:1141–1149. doi:10.1083/jcb.106.4.1141.
- Umeda K, Matsui T, Nakayama M, Furuse K, Sasaki H, Furuse M, Tsukita S. Establishment and characterization of cultured epithelial cells lacking expression of ZO-1. J Biol Chem. 2004;279(43):44785–44794. doi:10.1074/jbc.M406563200.
- Vasileva E, Sluysmans S, Bochaton-Piallat M-L, Citi S. Cell-specific diversity in the expression and organization of cytoplasmic plaque proteins of apical junctions. Ann NY Acad Sci. 2017;1405:160–176. doi:10.1111/nyas.13391.
- Bordin M, D’Atri F, Guillemot L, Citi S. Histone deacetylase inhibitors up-regulate the expression of tight junction proteins. Mol Cancer Res. 2004;2:692–701.
- Rice RH, Green H. Presence in human epidermal cells of a soluble protein precursor of the cross-linked envelope: activation of the cross-linking by calcium ions. Cell. 1979;18(3):681–694. doi:10.1016/0092-8674(79)90123-5.
- Duden R, Franke WW. Organization of desmosomal plaque proteins in cells growing at low calcium concentrations. J Cell Biol. 1988;107:1049–1063. doi:10.1083/jcb.107.3.1049.
- Watt FM, Mattey DL, Garrod DR. Calcium-induced reorganization of desmosomal components in cultured human keratinocytes. J Cell Biol. 1984;99:2211–2215. doi:10.1083/jcb.99.6.2211.
- Fuchs E. Epidermal differentiation and keratin gene expression. J Cell Sci Suppl. 1993;17:197–208.
- Guillemot L, Schneider Y, Brun P, Castagliuolo I, Pizzuti D, Martines D, Jond L, Bongiovanni M, Citi S. Cingulin is dispensable for epithelial barrier function and tight junction structure, and plays a role in the control of claudin-2 expression and response to duodenal mucosa injury. J Cell Sci. 2012;125:5005–5012. doi:10.1242/jcs.101261.
- Guillemot L, Guerrera D, Spadaro D, Tapia R, Jond L, Citi S. MgcRacGAP interacts with cingulin and paracingulin to regulate Rac1 activation and development of the tight junction barrier during epithelial junction assembly. Mol Biol Cell. 2014;25(13):1995–2005. doi:10.1091/mbc.E13-11-0680.
- D’Atri F, Nadalutti F, Citi S. Evidence for a functional interaction between cingulin and ZO-1 in cultured cells. J Biol Chem. 2002;277(31):27757–27764. doi:10.1074/jbc.M203717200.
- Citi S, Cross RA, Bagshaw CR, Kendrick-Jones J. Parallel modulation of brush border myosin conformation and enzyme activity induced by monoclonal antibodies. J Cell Biol. 1989;109(2):549–556. doi:10.1083/jcb.109.2.549.
- Hoener B, Citi S, Kendrick-Jones J, Jockusch BM. Modulation of cellular morphology and locomotory activity by antibodies against myosin. J Cell Biol. 1988;107:2181–2189. doi:10.1083/jcb.107.6.2181.
- Citi S, Kendrick-Jones J. Brush border myosin filament assembly and interaction with actin investigated with monoclonal antibodies. J Muscle Res Cell Motil. 1988;9:306–319.
- Huo L, Wen W, Wang R, Kam C, Xia J, Feng W, Zhang M. Cdc42-dependent formation of the ZO-1/MRCKbeta complex at the leading edge controls cell migration. Embo J. 2011;30(4):665–678. doi:10.1038/emboj.2010.353.
- Van Itallie CM, Aponte A, Tietgens AJ, Gucek M, Fredriksson K, Anderson JM. The N and C termini of ZO-1 are surrounded by distinct proteins and functional protein networks. J Biol Chem. 2013;288(19):13775–13788. doi:10.1074/jbc.M113.466193.
- Kurihara H, Anderson JM, Farquhar MG. Diversity among tight junctions in rat kidney: glomerular slit diaphragms and endothelial junctions express only one isoform of the tight junction protein ZO-1. Proc Natl Acad Sci USA. 1992;89:7075–7079. doi:10.1073/pnas.89.15.7075.
- Jesaitis LA, Goodenough DA. Molecular characterization and tissue distribution of ZO-2, a tight junction protein homologous to ZO-1 and the Drosophila discs-large tumor suppressor protein. J Cell Biol. 1994;124:949–961. doi:10.1083/jcb.124.6.949.
- Miragall F, Krause D, de Vries U, Dermietzel R. Expression of the tight junction protein ZO-1 in the olfactory system: presence of ZO-1 on olfactory sensory neurons and glial cells. J Comp Neurol. 1994;341:433–448. doi:10.1002/cne.903410402.
- Li C, Poznansky MJ. Characterization of the ZO-1 protein in endothelial and other cell lines. J Cell Sci. 1990;97:231–237.
- Howarth AG, Hughes MR, Stevenson BR. Detection of the tight junction-associated protein ZO-1 in astrocytes and other nonepithelial cell types. Am J Physiol. 1992;262:C461–C469. doi:10.1152/ajpcell.1992.262.2.C461.
- Howarth AG, Stevenson BR. Molecular environment of ZO-1 in epithelial and non-epithelial cells. Cell Motil Cytosk. 1995;31:323–332. doi:10.1002/cm.970310408.
- Petrov T, Howarth AG, Krukoff TL, Stevenson BR. Distribution of the tight junction-associated protein ZO-1 in circumventricular organs of the CNS. Molec Brain Res. 1994;21:235–246. doi:10.1016/0169-328X(94)90254-2.
- Schnabel E, Anderson JM, Farquhar MG. The tight junction protein ZO-1 is concentrated along slit diaphragms of the glomerular epithelium. J Cell Biol. 1990;111:1255–1263. doi:10.1083/jcb.111.3.1255.
- Ciana A, Meier K, Daum N, Gerbes S, Veith M, Lehr C-M, Minetti G. A dynamic ratio of the alpha+ and alpha- isoforms of the tight junction protein ZO-1 is characteristic of Caco-2 cells and correlates with their degree of differentiation. Cell Biol Int. 2010;34(6):669–678. doi:10.1042/CBI20090067.
- Fanning AS, Little BP, Rahner C, Utepbergenov D, Walther Z, Anderson JM. The unique-5 and −6 motifs of ZO-1 regulate tight junction strand localization and scaffolding properties. Mol Biol Cell. 2007;18(3):721–731. doi:10.1091/mbc.e06-08-0764.
- Gunzel D, Yu AS. Claudins and the modulation of tight junction permeability. Physiol Rev. 2013;93(2):525–569. doi:10.1152/physrev.00019.2012.
- Shen L, Weber CR, Raleigh DR, Yu D, Turner JR. Tight junction pore and leak pathways: a dynamic duo. Annu Rev Physiol. 2011;73:283–309. doi:10.1146/annurev-physiol-012110-142150.
- Rübsam M, Mertz AF, Kubo A, Marg S, Jüngst C, Goranci-Buzhala G, Schauss AC, Horsley V, Dufresne ER, Moser M, et al. E-cadherin integrates mechanotransduction and EGFR signaling to control junctional tissue polarization and tight junction positioning. Nat Commun. 2017;8(1):1250. doi:10.1038/s41467-017-01170-7.
- Ebrahim S, Kachar B. Myosin transcellular networks regulate epithelial apical geometry. Cell Cycle. 2013;12(18):2931–2932. doi:10.4161/cc.26229.
- Arendshorst WJ, Finn WF, Gottschalk CW. Pathogenesis of acute renal failure following temporary renal ischemia in the rat. Circ Res. 1975;37(5):558–568. doi:10.1161/01.res.37.5.558.
- Fogh J, Fogh JM, Orfeo T. One hundred and twenty-seven cultured human tumor cell lines producing tumors in nude mice. J Natl Cancer Inst. 1977;59(1):221–226. doi:10.1093/jnci/59.1.221.
- Popov LM, Marceau CD, Starkl PM, Lumb JH, Shah J, Guerrera D, Cooper RL, Merakou C, Bouley DM, Meng W, et al. The adherens junctions control susceptibility to Staphylococcus aureus α-toxin. Proc Natl Acad Sci U S A. 2015;112(46):14337–14342. doi:10.1073/pnas.1510265112.
- Shah J, Rouaud F, Guerrera D, Vasileva E, Popov LM, Kelley WL, Rubinstein E, Carette JE, Amieva MR, Citi S. A dock-and-lock mechanism clusters ADAM10 at cell-cell junctions to promote alpha-toxin cytotoxicity. Cell Rep. 2018;25(8):2132–2147. e7. doi:10.1016/j.celrep.2018.10.088.
- Cardellini P, Davanzo G, Citi S. Tight junctions in early amphibian development: detection of junctional cingulin from the 2-cell stage and its localization at the boundary of distinct membrane domains in dividing blastomeres in low calcium. Dev Dyn. 1996;207(1):104–113. doi:10.1002/(SICI)1097-0177(199609)207:1<104::AID-AJA10>3.0.CO;2-0.
- Paschoud S, Guillemot L, Citi S. Distinct domains of paracingulin are involved in its targeting to the actin cytoskeleton and regulation of apical junction assembly. J Biol Chem. 2012;287(16):13159–13169. doi:10.1074/jbc.M111.315622.
