Abstract
CaMKIIγ in dorsal root ganglion neurons is closely related to the neuropathic pain, neuron injury induced by local anesthetics. To get great insight into the function of CaMKIIγ in dorsal root ganglion neurons, we need one cell model to specially inhibit the CaMKIIγ mRNA expression. The present study was aimed to establish one cell model to specially inhibit the CaMKIIγ mRNA expression. We designed the CaMKIIγ shRNA sequence and connected with pYr-1.1 plasmid. The ligation product of the CaMKIIγshRNA and pYr-1.1 plasmid was recombined with pAd/PL-DEST vector into pAD-CaMKIIγ-shRNA. adenovirus vector. pAD-CaMKIIγ-shRNA. adenovirus vector infected the dorsal root ganglion neuron to inhibit the CaMKIIγ mRNA expression in vitro. The pAD-CaMKIIγ-shRNA adenovirus vector was verified to be correct by the digestion, sequence. And pAD-CaMKIIγ-shRNA. adenovirus vector can infect the DRG cells to inhibit the CaMKIIγ mRNA or protein expression by the real-time polymerase chain reaction (PCR) or western blotting. Those results showed that we successfully constructed one adenovirus vector that can infect the dorsal root ganglion neuron to inhibit the CaMKIIγ mRNA and protein expression. That will supply with one cell model for the CaMKIIγ function study.
Introduction
Calcium/calmodulin-dependent protein kinase II (CaMKII) is one kind of multifunction Ser/Thr protein kinase. It is widely distributed in the central and peripheral nervous system. As the main component of the excitatory postsynaptic density, it is the key factor to regulate the synaptic transmission and the function of the neurons (Ataei et al. Citation2015, Kennedy and Greengard Citation1981, Liu and Murray Citation2012, Racioppi and Means Citation2012). In our previous study, we detected the expression of the CaMKIImRNA of dorsal root ganglion neuron (DRG) in rat. There are four subtype of CaMKII in the DRG of the rat, CaMKIIα, CaMKIIβ, CaMKIIδ, CaMKIIγ, respectively (Boric et al. Citation2013, Ferhatovic et al. Citation2014). As well as, ropivacaine hydrochloride, one kind of local anesthetics, can upregulate all four kinds of the expression of CaMKII mRNA with the concentration and exposure time of the ropivacaine hydrochloride increase.
CaMKIIγ can activate Cav3.2 T-type channel, increase the density of the Cav3.2 T-type current in vitro. Furthermore, the inhibitor of CaMKIIγ can reduce the Cav3.2 T-type calcium current (Barrett et al. Citation2000, Chemin et al. Citation2006, Iftinca and Zamponi Citation2009, Welsby et al. Citation2003, Wolfe et al. Citation2002). T-type calcium channel has been testified to be related with the local anesthetics neurotoxicity (Wen et al. Citation2013, Citation2016). To investigate the role of CaMKIIγ in the nerve block or neurotoxicity of the local anesthetics, we need to construct one cell model to specially inhibit the CaMKIIγ mRNA expression.
RNA interference (RNAi) is often used to regulate the gene expression in clinic and science research (Alagia and Eritja Citation2016, Bobbin and Rossi Citation2016, Fakhr et al. Citation2016, Prabha et al. Citation2015, Shenas et al. Citation2016). It is a biological process that mRNA special degradation induced by the highly conservation double-stranded RNA (daRNA). There are different methods for the RNAi to inhibit the mRNA expression. The shRNA, one typical method in RNAi, is a small hairpin RNA or short hairpin RNA. The shRNA molecule has a tight hairpin turn that can silence the target gene expression. As well as, the shRNA expression in cell need to be connected with the plasmids or through virus and bacterial vectors.
In this study, we construct one adenovirus vector that can infect the dorsal root ganglion neuron to inhibit the CaMKIIγ mRNA expression. We designed the CaMKIIγ shRNA sequence and connected with pYr-1.1 plasmid. The ligation product of the CaMKIIγshRNA and pYr-1.1 plasmid was recombined with pAd/PL-DEST vector into pAD-CaMKIIγ-shRNA adenovirus vector. pAD-CaMKIIγ-shRNA adenovirus vector infected the dorsal root ganglion neuron to inhibit the CaMKIIγ mRNA expression in vitro. That will supply with one cell model for the CaMKIIγ function study.
Materials and methods
Design of the shRNA
Search the interfere target of the CaMKIIγ gene (NM_133605.1) of the rats online (https://rnaidesigner.invitrogen.com/rnaiexpress/) and designed three sequence, GGATATGCCGACTTCTGAA (CaMKIIγ-shRNA-1, target of the gene locus 188 ∼ 206), GGAGCCTACGATTTCCCAT (CaMKIIγ-shRNA-2, target of the gene locus 685 ∼ 703), GGTCAACAGTGGCATCCAT(CaMKIIγ-shRNA-3, target of the gene locus 824 ∼ 842), respectively. The basic structure of the shRNA primer is BsaI + sense + loop + antisense + TTTTTT+ SacI +BsaI. The shRNA primers were synthesized by Jinsirui Ltd. (Nanjing, China) ().
Table 1. The primer of the shRNA.
Ligation of the shRNA and pYr-1.1 plasmid
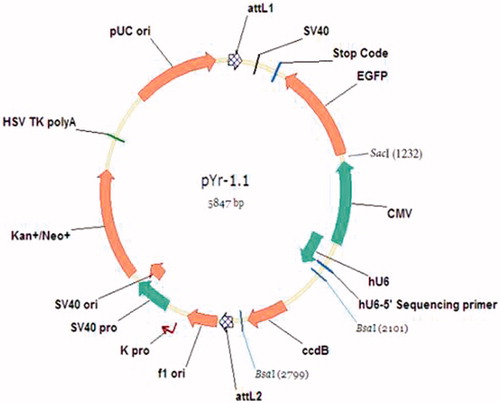
The shRNA primer (1 μg/μl), CAMK2γ-shRNA-1-A + CAMK2γ-shRNA-1-B, CAMK2γ-shRNA-2-A + CAMK2γ-shRNA-2-B, and CAMK2γ-shRNA-3-A + CAMK2γ-shRNA-3-B were annealed at 95 °C for 5 min to form the double strands. pYr-1.1 The pYr-1.1 plasmid (Wuhan Biofield biological Service Co., Ltd., Wuhan, China) was digested with BSAI (Dalian TAKARA Co., Ltd., Dalian, China) and recycled the large fragment. The map of pYr-1.1 plasmid was shown in . The mixture of pYr-1.1 plasmid recycle and annealing shRNA with the T4 DNA ligase (Dalian TAKARA Co., Ltd., Dalian, China) were stored at 4 °C overnight. The ligation product was named pYr-1.1-shRNA-1, pYr-1.1-shRNA-2 and pYr-1.1-shRNA-3.
Transformation of the pYr-1.1 plasmid and shRNA ligation product
The pYr-1.1 plasmid and shRNA ligation product 5 ul was incubated for 5 min in 65 °C to inactive the ligation enzyme. After 2-min cooling on ice, 30 μl competent cells DH5а (Dalian TAKARA Co., Ltd., Dalian, China) was mixed with the ligation product and kept on the ice for 10 min. Then, the mixture was incubated in 42 °C water bath for 30 s. After 2-min cooling, 100 μl LB medium was added into the competent cells and incubated the cells in 37 °C water bath for 30 min. Then, the competent cells were uniformly coated plates on the LB medium plates with the Kana (30 μg/ml final concentration) resistant and incubate in 37 °C constant temperature incubator overnight. Monoclonal colony was inoculated into 5 ml LB medium with Kana resistant in 37 °C shaking table overnight for the plasmid amplification. Plasmid extracted with the EZNA Plasmid Mini Kit I (Omega Bio-tek, Norcross, GA) were digested with Sac I (Dalian TAKARA Co., Ltd., Dalian, China) and sent to Jinsirui Ltd. (Nanjing, China) for the sequencing. Selected one correct sequence and named as pYr-CaMKIIγ-shRNA for the restructuring of adenovirus vector.
Restructuring of adenovirus vector and infection of 293 cells
The pYr-CaMKIIγ–shRNA was connected with pAd/PL-DEST vector. The ligation product was transformed with the DH5а as the above method for the pYr-CaMKIIγ-shRNA ligation products. Selected two different monoclonal colony to amplify and identify, named pAd-CaMKIIγ-shRNA-1 and pAd-CaMKIIγ-shRNA-2, respectively. pAd-CaMKIIγ-shRNA-1, and pAd-CaMKIIγ-shRNA-2 adenovirus vector was digested with xbal enzyme and sequenced to identify. Selected the correct monoclonal vector, named pAd-CaMKIIγ-shRNA, to infect 293 cells for the generation of the adenovirus. After linearized with PacI enzyme, the restructure pAd-CaMKIIγ-shRNA adenovirus vector was transfect into 293 cell with the Lipo2000 kits (Thermo Fisher Scientific Inc., Los Angeles, CA). Collected the medium of the 293 cells infected by pAd-CaMKIIγ-shRNA adenovirus to identify with PCR and sequencing (Jinsirui Ltd., Nanjing, China). The hU6 universal primer sequence was 3′-TACGATACAAGGCTGTTAGAGAG-5′ and the down primer of the CaMKIIγ was 5′-AAATGGATGCCACTGTTGACCCAAA-3′, they were synthesized by Jinsirui Ltd. (Nanjing, China). The PCR product size was 244 bp.
Infection DRG cells with pAd-CaMKIIγ-shRNA adenovirus
Neonatal rats aged one day were dislocated executed and the vertebral columns of the rats were completely separated. Carefully separate the dorsal root ganglions with the fine dissecting forceps and transferred the dorsal root ganglions cells. The cells, with 2–3 × 105/ml density, were incubated in 37 °C 5% CO2, changed the Neurobasal medium (D-glucose 4.5 g/l, L-glutamine 2 mmol/l, FBS 1%, B-27 additive 2%, NGF 10 μg/l, penicillin 100 U/l, and streptomycin 100 μg/l) every 3 days. Recombinant pAd-CaMKIIγ-shRNA adenovirus was added to the cells with the 3 × 109 TU/ml titer. The infection efficiency was observed with the fluorescence microscope.
qRT-PCR detected the CaMKIIγ mRNA expression in DRG cells
After infection with recombinant pAd-CaMKIIγ-shRNA adenovirus, the DRG cells were collected to detect the expression of the CaMKIIγ mRNA with the real-time quantity PCR with the real-time PCR kits (Invitrogen Ltd., Carlsbad, CA). Theβ-actin primer was 5′-CACGATGGAGGGGCCGGACTCATC-3′, 5′-TAAAGACCTCTATGCAACACAGT-3′, and the product size was 240 bp. The CaMKIIγ primer was 5′-TTGCTGCTGGCGAGTAAATG-3′, 5′-TAGGGATCTTTCCTCAAGACCTCA-3′, and the product size was 149 bp. The primers were synthesized by Jinsirui Ltd. (Nanjing, China). The absolute amount of corresponding gene mRNA in each cell was obtained from the corresponding gene curve. The data from the real-time PCR was converted to 2−△△Ct, where Ct represented the threshold cycle.
Western blotting detected the CaMKIIγ protein expression in DRG cells
After infection with recombinant pAd-CaMKIIγ-shRNA adenovirus, the DRG cells were collected to detect the expression of the CaMKIIγ protein with western blotting. In brief, after total protein extracted, protein concentration determined and 7.5% SDS-PAGE electrophoresis, the CaMKIIγ protein or β-actin primary antibody (1 300, Santa Cruz Biotechnology, Inc., Dallas, TX) incubated the PVDF film for 24 h 4 °C overnight. Then, the horseradish peroxidase conjugated rabbit antisheep antibody (1:500, Boster bio-engineering Ltd. Co., Wuhan, China) incubated for 1 h at room temperature and electrochemiluminescence coloration. The CaMKIIγ protein expression quantity was normalized with the β-actin protein.
Statistical analysis
Data are expressed as means ± standard error. The comparisons of mRNA and protein expression of the CaMKIIγ in the dorsal root ganglions neurons were performed using one-way analysis of variance (ANOVA). P < 0.05 was considered to indicate a statistically significant difference.
Results
pYr-1.1 plasmid digest with BASI enzyme
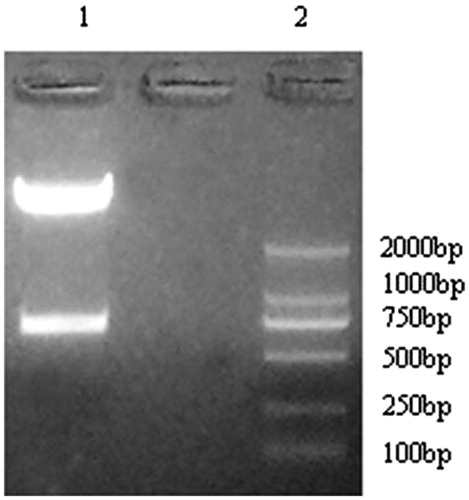
There were two BASI enzyme cutting sites in the pYr-1.1 plasmid, 2799 and 2101, respectively. After digest with the BASI enzyme, there was one 698 bp band ().
pYr-1.1–CaMKIIγ-shRNA plasmid digest with BASI enzyme and sequencing
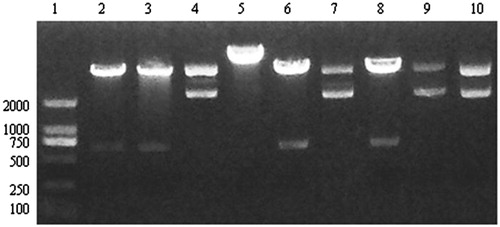
The pYr-1.1-CaMKIIγ-shRNA plasmids were digest with BASI enzyme to identify the fragment size. We selected three different bacterial colons in the pYr-1.1-CaMKIIγ-shRNA-1, the pYr-1.1-CaMKIIγ-shRNA-2, and the pYr-1.1-CaMKIIγ-shRNA-3. From the , we can find that the bacterial colons in the lane 2 and lane 3 (from pYr-1.1-CaMKIIγ-shRNA-1) and lane 6 (from pYr-1.1-CaMKIIγ-shRNA-2) and lane 8 (pYr-1.1-CaMKIIγ-shRNA-3) can be digest with BASI enzyme and one 698 size band can be found. Those suggested that the band size was correct. We send the lane 2 plasmid from pYr-1.1-CaMKIIγ-shRNA-1, lane 6 plasmid from pYr-1.1-CaMKIIγ-shRNA-2, and lane 8 from the pYr-1.1-CaMKIIγ-shRNA-3 to Jinsirui Ltd., (Nanjing, China) for the sequencing. The data from indicated that the sequence of the pYr-1.1-CaMKIIγ-shRNA-1, pYr-1.1-CaMKIIγ-shRNA-2, and pYr-1.1-CaMKIIγ-shRNA-3 were all correct.
pAd-CaMKIIγ-shRNA digested and sequencing
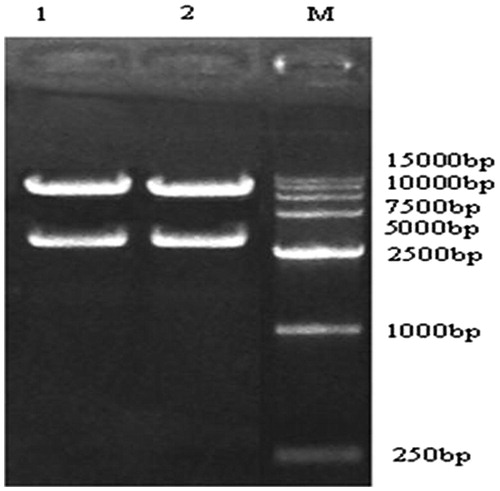
The pAd-CaMKIIγ-shRNA digested with xbal enzyme. From , we can find that there was one 3645 bp band. And sent the pAd-CaMKIIγ-shRNA to sequence, all the data suggested that the CaMKIIγ-shRNA had been inserted into pAd/PL-DEST vector.
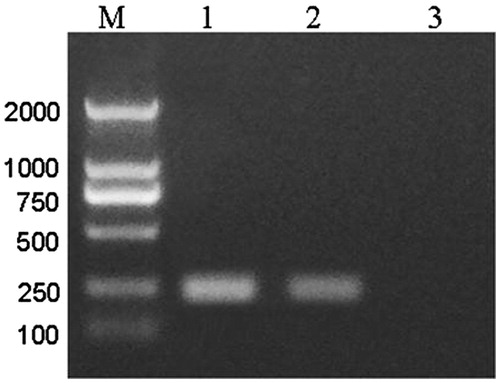
PCR verify the production of recombinant adenovirus in 293 cells
shows the positive control with the pAd/PL-DEST vector (lane 1) and the pAd-CaMKIIγ-shRNA adenovirus had one 244 bp band, however, the negative control without vector had no band. It indicated that the hu6 promoter and CaMKIIγ shRNA were expressed in the pAd-CaMKIIγ-shRNA adenovirus.
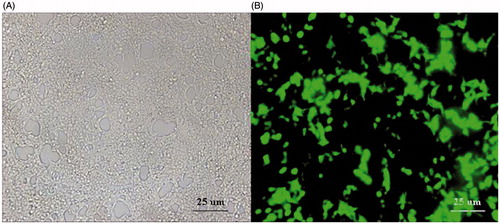
DRG cells infected pAd-CaMKIIγ-shRNA adenovirus under fluorescence microscope
After infected 24 h with the pAd-CaMKIIγ-shRNA adenovirus, the DRG cells were observed under the fluorescence microscope (Leica M165 FC, Wetzlar, Germany), because that the pY-1.1 plasmid contain the EGFP, the infection efficiency was estimated with the green fluorescence. The green fluorescence reflected the pAd-CaMKIIγ-shRNA adenovirus DRG cells ().
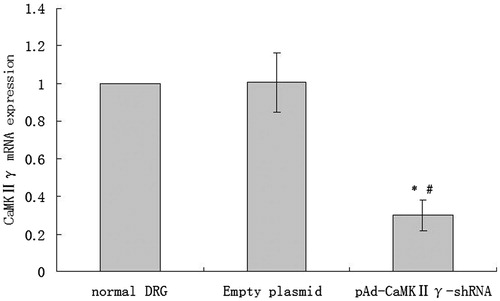
CaMKIIγ mRNA expression of DRG cells infected pAd-CaMKIIγ-shRNA adenovirus detected by real-time PCR
From , compared with the normal DRG cells, the empty plasmid without the CaMKIIγ-shRNA had no effect on the DRG cells CaMKIIγ mRNA expression. However, compared with the normal DRG cells and empty plasmid cells, the CaMKIIγ mRNA expression of the DRG cells infected with pAd-CaMKIIγ-shRNA adenovirus significantly reduced.
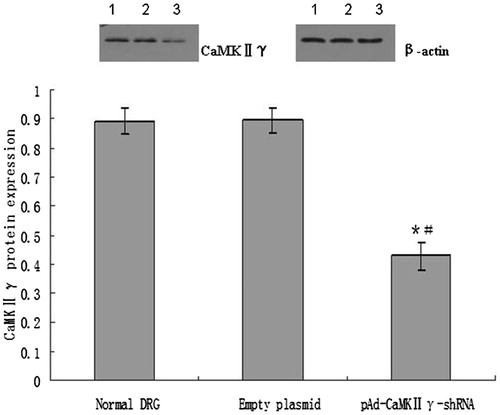
CaMKIIγ protein expression of DRG cells infected pAd-CaMKIIγ-shRNA adenovirus by western blotting
From , it was observed that there was no difference of the CaMKIIγ protein expression between the normal DRG cells and empty plasmid DRG cells. Compared with normal DRG cells and empty plasmid cells, the CaMKIIγ protein expression of the DRG cells infected with pAd-CaMKIIγ-shRNA adenovirus significantly reduced.
Figure 9. The expression of CaMKIIγ protein (mean ± sd, n = 6). *P < 0.01 versus normal DRG cells, #P < 0.01 versus empty plasmid DRG cells. Lane 1, the expression of CaMKIIγ protein in normal DRG cells. Lane 2, the expression of CaMKIIγ protein in empty DRG cells. Lane 3, the expression of CaMKIIγ protein in pAd-CaMKIIγ-shRNA adenovirus DRG cells.
Discussion
Dorsal root ganglion neurons are the primary neurons. Nerve impulse conduction from the peripheral neurons through DRG transmit to the central nervous system and vice versa. DRG plays a very important role in the nerve impulse conduction (Van Steenwinckel et al. Citation2009, Walwyn et al. Citation2009). At the same time, it is the target cells of the local anesthetic in nerve block. In fact, the role of nerve block or neurotoxicity of the local anesthetics in spinal anesthesia was all involved with the DRG cell (Yin et al. Citation2016, Zhang et al. Citation2016). Our previous study found that there was CaMKIIγ mRNA expression in DRG cells, and ropivacaine hydrochloride can upregulate the CaMKIIγ mRNA expression with the concentration or exposure time increase. Those data indicated that the CaMKIIγ may be involved with the nerve block or neurotoxicity of the local anesthetics. Therefore, we constructed pAd-CaMKIIγ-shRNA adenovirus to infect the DRG cells to in inhibit the CaMKIIγ mRNA expression of the DRG cells.
In this study, we designed two BASL restriction enzyme cutting site in the shRNA sequence. Meanwhile, the BASL restriction enzyme cutting site in the pYr-1.1 plasmid matched with the shRNA. After digested with BASL enzyme, pYr-1.1 plasmid connected with shRNA. According to the sequencing result, the shRNA can be correctly inserted into the pYr-1.1 plasmid. Because the pYr-1.1 plasmid contains EGFP, we can observe the restructure pAd-CaMKIIγ-shRNA adenovirus vector and the infection efficiency of the pAd-CaMKIIγ-shRNA adenovirus to the DRG cells under fluorescence microscope. In this study, we found that CaMKIIγ mRNA and protein expression in the DRG cells infected with pAd-CaMKIIγ-shRNA adenovirus significantly reduced by real-time PCR and western blotting method. Those data indicated that we constructed pAd-CaMKIIγ-shRNA adenovirus can inhibit CaMKIIγ mRNA expression of the DRG cells.
In conclusion, we successfully constructed the pAd-CaMKIIγ-shRNA adenovirus, which can infect the DRG cells and inhibit the CaMKIIγ mRNA expression.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
References
- Alagia A, Eritja R. 2016. siRNA and RNAi optimization. Wiley Interdiscip Rev RNA. 7:316–329.
- Ataei N, Sabzghabaee AM, Movahedian A. 2015. Calcium/calmodulin-dependent protein kinase II is a ubiquitous molecule in human long-term memory synaptic plasticity: a systematic review. Int J Prev Med. 6:88.
- Barrett PQ, Lu HK, Colbran R, Czernik A, Pancrazio JJ. 2000. Stimulation of unitary T-type Ca(2+) channel currents by calmodulin-dependent protein kinase II. Am J Physiol Cell Physiol. 279:C1694–C1703.
- Bobbin ML, Rossi JJ. 2016. RNA interference (RNAi)-based therapeutics: delivering on the promise? Annu Rev Pharmacol Toxicol. 56:103–122.
- Boric M, Kadic AJ, Ferhatovic L, Sapunar D, Puljak L. 2013. Calcium/calmodulin-dependent protein kinase II in dorsal horn neurons in long-term diabetes. Neuroreport. 24:992–996.
- Chemin J, Traboulsie A, Lory P. 2006. Molecular pathways underlying the modulation of T-type calcium channels by neurotransmitters and hormones. Cell Calcium. 40:121–134.
- Fakhr E, Zare F, Teimoori-Toolabi L. 2016. Precise and efficient siRNA design: a key point in competent gene silencing. Cancer Gene Ther. 23:73–82.
- Ferhatovic L, Jelicic Kadic A, Boric M, Puljak L. 2014. Changes of calcium/calmodulin-dependent protein kinase II expression in dorsal root ganglia during maturation in long-term diabetes. Histol Histopathol. 29:649–658.
- Iftinca MC, Zamponi GW. 2009. Regulation of neuronal T-type calcium channels. Trends Pharmacol Sci. 30:32–40.
- Kennedy MB, Greengard P. 1981. Two calcium/calmodulin-dependent protein kinases, which are highly concentrated in brain, phosphorylate protein I at distinct sites. Proc Natl Acad Sci USA. 78:1293–1297.
- Liu XB, Murray KD. 2012. Neuronal excitability and calcium/calmodulin dependent protein kinase type II: location, location, location. Epilepsia. 53:45–52.
- Prabha S, Vyas R, Gupta N, Ahmed B, Chandra R, Nimesh S. 2015. RNA interference technology with emphasis on delivery vehicles-prospects and limitations. Artif Cells Nanomed Biotechnol. [Epub ahead of print]. doi: 10.3109/21691401.2015.1058808.
- Racioppi L, Means AR. 2012. Calcium/calmodulin-dependent protein kinase kinase 2: roles in signaling and pathophysiology. J Biol Chem. 287:31658–31665.
- Shenas SMM, Mansoori B, Mohammadi A, Salehi S, Kaffash B, Talebi B, et al. 2016. SiRNA-mediated silencing of Snail-1 induces apoptosis and alters micro RNA expression in human urinary bladder cancer cell line. Artif Cells Nanomed Biotechnol. [Epub ahead of print]. doi: 10.1080/21691401.2016.1198361.
- Van Steenwinckel J, Noghero A, Thibault K, Brisorgueil MJ, Fischer J, Conrath M. 2009. The 5-HT2A receptor is mainly expressed in nociceptive sensory neurons in rat lumbar dorsal root ganglia. Neuroscience. 161:838–846.
- Walwyn W, John S, Maga M, Evans CJ, Hales TG. 2009. Delta receptors are required for full inhibitory coupling of mu-receptors to voltage-dependent Ca(2+) channels in dorsal root ganglion neurons. Mol Pharmacol. 76:134–143.
- Welsby PJ, Wang H, Wolfe JT, Colbran RJ, Johnson ML, Barrett PQ. 2003. A mechanism for the direct regulation of T-type calcium channels by Ca2+/calmodulin-dependent kinase II. J Neurosci. 23:10116–10121.
- Wen X, Xu S, Liu H, Zhang Q, Liang H, Yang C, Wang H. 2013. Neurotoxicity induced by bupivacaine via T-type calcium channels in SH-SY5Y cells. PLoS One. 8:e62942.
- Wen X, Xu S, Zhang Q, Li X, Liang H, Yang C, Wang H, Liu H. 2016. Inhibitory gene expression of the Cav3.1 T-type calcium channel to improve neuronal injury induced by lidocaine hydrochloride. Eur J Pharmacol. 775:43–49.
- Wolfe JT, Wang H, Perez-Reyes E, Barrett PQ. 2002. Stimulation of recombinant Ca(v)3.2, T-type, Ca(2+) channel currents by CaMKIIgamma(C). J Physiol (Lond). 538:343–355.
- Yin K, Baillie GJ, Vetter I. 2016. Neuronal cell lines as model dorsal root ganglion neurons: A transcriptomic comparison. Mol Pain. 12. doi: 10.1177/1744806916646111.
- Zhang Y, Yan L, Cao Y, Kong G, Lin C. 2016. Long noncoding RNA BDNF-AS protects local anesthetic induced neurotoxicity in dorsal root ganglion neurons. Biomed Pharmacother. 80:207–212.