Abstract
It has been demonstrated that circulating MicroRNAs (miRNAs) could be potential biomarkers for cancer diagnosis and prognosis. For pancreatic cancer (PCa), little is known about miR-221-3p biological function or its prognostic value. In the current study, we profiled miR-221-3p expression in PCa cell lines. Compared with normal pancreases ductal epithelial cells, miR-221-3p is up-regulated in all PCa cell lines analysed. In SW1990 cells, overexpression of miR-221-3p increased cell proliferation and inhibited apoptosis, while inhibition of miR-221-3p decreased cell growth rate and promoted apoptosis. Compared with adjacent non-tumour tissues, miR-221-3p was up-regulated in all 21 PCa tissues. Expression level of miR-221-3p was investigated in plasma and statistical analyses showed that circulating miR-221-3p expression level was correlated with distant metastasis and TNM stages. The receiver-operating characteristic (ROC) curves and the area under the ROC curve (AUC) suggested that the diagnostic efficacy for distant metastasis of miR-221-3p is better than CA19-9 (AUC: 0.689 vs. 0.587). To summary, we found miR-221-3p could promote cell proliferation and inhibit apoptosis in PCa cells and circulating miR-221-3p could serve as a biomarker for PCa.
Introduction
Pancreatic cancer (PCa) is one of the most deadly cancers worldwide [Citation1]. Despite the improvements achieved in surgery and chemotherapy, the prognosis of PCa is still extremely poor [Citation2]. The median survival is about 5–8 months and the 5-year survival rate of PCa is less than 5% [Citation2]. Local invasion and distant metastases usually occur at early stage of the clinical course of PCa, which is the main reason for the poor survival of PCa [Citation3]. Therefore, early detection and diagnosis of PCa at early stage is the key to improve survival rates.
Several cancer associated molecular biomarkers have been identified to improve diagnosis of PCa, and carbohydrate antigen 19-9 (CA19-9) and carcinoembryonic antigen (CEA) are two most famous circulating biomarkers [Citation4–6]. However, the sensitivity and specificity of these tumour markers are not satisfying for the early detection and dynamic monitoring of PCa. MicroRNAs (miRNAs) are a kind of small noncoding RNA molecules which are about ∼21 nt and regulate gene expression at post-transcription level [Citation7,Citation8]. miRNAs are involved in almost every aspects of cellular biological processes and deregulation of miRNAs has been reported in various diseases, especially for cancers [Citation9–11]. Recently, evidence has demonstrated that miRNA are detectable in plasma or serum, which indicates that circulating miRNAs could be sensitive biomarkers [Citation11]. It is believed that circulating miRNAs could bind with proteins, like Argonaute 2 or contained by exosomes in plasma, thus they are resistant to endogenous ribonuclease [Citation12–14]. For non-small cell lung cancer, it has been found that a signature consists of three miRNAs could efficiently predict survival of patients [Citation11].
In terms of PCa, several tumour suppressors or oncogene like miRNAs have been reported and some of these miRNAs are detectable in plasma [Citation15,Citation16]. However, these circulating miRNAs fail to predict the malignant potential of PCa.miR-221-3p, which is a member of antiangiogenic gene-regulating miRNAs family, is encoded by a gene cluster on the X chromosome and is abundant in the intima layer of endothelial cells of human atherosclerotic vessels [Citation17]. The overexpression of miR-221-3p indirectly decreases the synthesis and expression of endothelial nitric oxide synthase, which results in limited tube formation, endothelial cell proliferation, migration and angiogenesis. miR-221-3p has also been found to play a crucial role in abnormal cell proliferation, differentiation in the setting of various cancers [Citation18–20]. Such as Wu et al. [Citation21] reported that the tumour suppressive role of miR-221-3p in epithelial ovarian cancer and directly target ARF4, suggesting the miR-221-3p might be a novel potential candidate for clinical prognosis and therapeutics of epithelial ovarian cancer. However, studies about miR-221-3p are rare in PCa [Citation22–24]. In the current study, found miR-221-3p is overexpressed in PCa tissues compared with adjacent non-tumour tissues and circulating miR-221-3p level is correlated with distant metastases of PCa patients. Additionally, in vitro experiments showed that miR-221-3p could promote proliferation of PCa cell line.
Methods
Patients and tissue samples
Ethical approval for this study was obtained (Ethics committee, Qilu Hospital), informed written consent was obtained from each patient. PCa tissues and adjacent non-tumour tissues were collected from 21 PCa patients undergone resection between 2011 and 2014. Once resected, tissue samples were snap frozen and stored in liquid nitrogen till RNA extraction.
Plasma samples
Blood samples were collected with sodium EDTA-Na tubes from each patients and controls. To separate plasma, the blood samples were centrifuged at 3500 rpm for 10 min at room temperature. After centrifuge, the plasma supernatant was collected and stored at −80 °C until RNA extraction.
PCa cell lines and cell culture
Pancreatic cancer cell lines SW1990, CFPAC-1, PANC1, MIA, and PaCa-2, and two normal pancreatic ductal epithelial cells H6C7 and HPDE were purchased from the Institute of Biochemistry and cell biology of Chinese academy of science (Shanghai, China). Cells were cultured with DMEM medium (GIBCO, NY, USA) supplemented with 10% foetal bovine serum, 100 U/ml penicillin, and 100 mg/ml streptomycin in humidified air at 37 °C with 5% CO2.
Transfection of cells
Mimics and inhibitors of miR-211-3p were transfected using Lipofectamine 2000 (Invitrogen, Shanghai, China) according to the manufacturer’s instructions. Cells were harvested after 24 h of transfection for other experiments.
Cell count kit-8 assay
SW1990 cells were seeded into 96-well plates (2 × 104/well) and cultured for 96 h. The Cell Counting Kit-8 (CCK8) assay was used to determine the relative proliferation rate according to the manufacturer’s instructions. The absorbance at 450 nm was measured.
Flow cytometry analysis
For the analysis of apoptosis, SW1990 cells were double stained with fluorescein isothiocyanate (FITC)-Annexin V and propidium iodide by the FITC Annexin V Apoptosis Detection Kit (BD Biosciences, CY, USA) according to the manufacturer’s instructions. The cells were analysed by a flow cytometry (FACScan; BD Biosciences, CY, USA) with the Cell Quest software (BD Biosciences, CY, USA). The stained cells were categorised into viable cells, dead cells, early apoptotic cells, and apoptotic cells.
RNA extraction
The mirVana PARIS Kit (Ambion, TX, USA) was used to extract total RNA from cell lines and plasma and the pellet was eluted into 100 ml of preheated (95 °C) Elution Solution according to the manufacturer’s directions. Total RNA of tissues samples were extracted with Trizol Reagent according to manufacturer’s protocol.
Quantitative reverse transcription polymerase chain reaction (qRT-PCR)
The human TaqMan MicroRNA Assay Kit (Applied Biosystems, Foster City, CA, USA) was used to quantify the amount of miRNAs. The synthesised cDNA was subjected to RT-PCR reaction with specific probes on a 7300 Realtime PCR system (Applied Biosystems, Foster City, CA, USA). The RT-PCR cycles were as follows: incubation at 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s, and 60 °C for 1 min. Cycle threshold (Ct) values were calculated. To calculate the amounts of miRNAs in plasma, the standard curve method was used by constructed with the mirVana miRNA Reference Panel (Ambion, Austin, TX, USA). For the expression of miRNAs in cell lines and tissue samples, the 2−DDCt method relative to U6 small nuclear RNA (RNU6B) was utilised.
Statistical analyses
The differences expression levels of miR-221-3p in plasma between cancer patients and healthy volunteers were analysed by the Mann–Whitney U-test. To evaluate the correlations between plasma miR-221-3p expression levels and clinical characteristics of PCa patients, χ2 test or Fisher’s exact probability test was used. The diagnostic accuracy of miR-221-3p and CA19-9 was measured by the receiver-operating characteristic (ROC) curves and the area under the ROC curve (AUC). A p <.05 was considered statistically significant.
Results
Study design
shows the flow chart of the current study. The oncogenic function of miR-221-3p has been reported in many types of cancer, while report about miR-221-3p is rare in PCa. Therefore, we first profiled the expression of miR-221-3p in PCa cell lines and analysed its biological function with in vitro experiments. After that, we investigated miR-221-3p expression in PCa tissues and plasma with qRT-PCR and evaluated the association between miR-221-3p level and clinical characteristics.
MiR-221-3p expression level in PCa cell lines
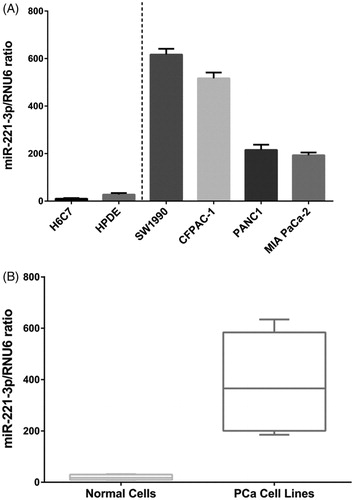
We first analysed the expression profile of miR-221-3p in PCa cell lines and normal pancreatic ductal epithelial cells, H6C7 and HPDE. As shown in , compared with H6C7 and HPDE, miR-221-3p is significantly up-regulated in the 4 PCa cell lines analysed and miR-221-3p expression level is highest in SW1990 cell line.
Biological role of miR-221-3p in PCa cell line
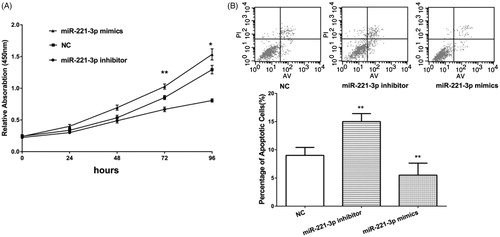
Then, we try to identify the biological role of miR-221-3p with in vitro experiments. The SW1990 cell was used as a cell model and mimics and inhibitor of miR-221-3p were transfected to overexpress or inhibit miR-221-3p, respectively. CCK-8 assay shows that overexpression of miR-221-3p increases the proliferation rate of SW1990 cells, while cell proliferation rate was significantly decreased by miR-221-3p inhibitor (). Apoptosis was also analysed by PI staining followed by flow cytometry analysis. As shown, the percentage of apoptotic cells significantly increased after inhibition of miR-221-3p, while after overexpression of miR-221-3p the percentage of apoptotic cells decreased (). These results suggested that miR-221-3p could prove PCa cell proliferation and inhibit apoptosis.
Figure 3. Compared with negative control (NC), mimics of miR-221-3p increased cell proliferation rate in SW1990 cell line, while inhibitor of miR-221-3p reduced cell proliferation (A). For cell apoptosis analysis, miR-221-3p inhibitor increased the percentage of apoptotic cells, while miR-221-3p mimics decreased the percentage of apoptotic cells (B). *p < .05, **p < .01.
Expression of miR-221-3p in PCa patients
Then we analysed the expression profiles and clinical utility of miR-221-3p in PCa tissues and blood samples, respectively. We first detected miR-221-3p expression in 21 pairs of PCa tissues and adjacent non-tumour tissues. As shown in , the expression level of miR-221-3p was significantly higher in PCa tissues. Then, miR-221-3p expression was analysed in blood samples of 48 healthy controls and 87 PCa patients, and miR-221-3p level was also significantly higher in plasma of PCa patients than that in controls. Further statistical analyses revealed that miR-221-3p level in plasma was associated with clinical characteristics of PCa patients and the results were presented in . We found miR-221-3p expression level was higher in patients with distant metastasis and those with IIb–IV stages. Then we compared the diagnostic efficiency of miR-221-3p for detecting distant metastasis with conventional biomarker CA19-9. The ROC assay indicated that miR-221-3p has better diagnostic (AUC = 0.687) than CA19–9 (AUC = 0.589, ).
Figure 4. Expression miR-221-3p was analysed in 21 pairs of PCa tissues and adjacent non-tumour tissues (A), and miR-221-3p expression is significantly higher in PCa tissues (B). ROC curve suggested that the AUC of miR-221-3p (0.687) is larger than that of CA19-9 (0.589).
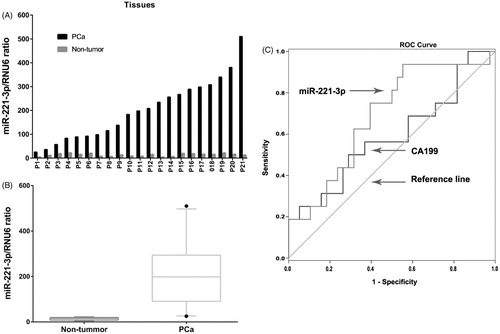
Table 1. Association between plasma miR-221-3p expression level and clinical characteristics of PCa patients.
Discussion
Evidence has proved that miRNAs are critical regulators in the pathological processes of cancer [Citation8]. Aberrant expression of miRNAs is found in almost all kinds of cancer [Citation25]. For PCa, many miRNAs have been reported to regulate the proliferation, apoptosis, metastasis, and drug resistance of PCa cells, such as miR [Citation26–28]. For miR-221-3p, it is reported that miR-221-3p could induce the 5-fluorouracil resistance in human PCa by directly binding to the 3′UTR of RB1 and decreased its expression [Citation29]. This report indicates miR-221-3p may play oncogenic role in PCa. However, the biological role of miR-221-3p is elusive in PCa and it has not been fully elucidated in other cancers.
In the current study, we first profiled miR-221-3p expression in PCa cell lines and normal pancreatic ductal epithelial cells. High expression of miR-221-3p was observed in all 4 PCa cell lines analysed, and miR-221-3p showed highest expression level in SW1990 cell line. With miR-221-3p mimics and inhibitor, we overexpressed and inhibited miR-221-3p in PCa cells. CCK-8 assay found that miR-221-3p overexpression promoted cell proliferation rate, while miR-221-3p inhibition decreased cell proliferation rate. Additionally, PI staining assay also confirmed that miR-221-3p overexpression reduced the percentage of apoptotic cells, while miR-221-3p inhibition increased the percentage of apoptotic cells. These results suggest miR-221-3p could promote cell proliferation and inhibit cell apoptosis in PCa.miRNAs are small in size, often bind with proteins, and miRNAs is contained in exosomes in plasmas. Due to these features, miRNAs are relatively stable in plasma, making it suitable biomarkers. Many efforts have been made to develop the diagnostic or prognostic value of circulating miRNAs. Hu et al developed a signature of 3 miRNAs could precisely predict lung cancer survival [Citation11]. For PCa, various miRNAs have been investigated [Citation30], such as miR-744 [Citation31] and miR-484-3p [Citation32].
In the current study, we found miR-221-3p is overexpressed in PCa tissues compared with adjacent non tumour tissues. And miR-221-3p expression level is correlated with distant metastasis and TNM stages. Compared with previous studies, we analysed miR-221-3p expression level in both tumour tissues and plasma, suggesting the high level of miR-221-3p in plasma may or from PCa tissues. In addition, we compared the diagnostic efficacy between miR-221-3p and CA19-9 with ROC and miR-221-3p showed better diagnostic efficacy.
In the current, we found miR-221-3p is over expressed in PCa tissues compared with adjacent non-tumour tissues. Plasma miR-221-3p expression level is correlated with distant metastasis and TNM stages. In vivo experiments demonstrated that miR-221-3p promoted proliferation and inhibit apoptosis in PCa cells.
Disclosure statement
The authors have declared no conflict of interest.
References
- Siegel R, Ma J, Zou Z, et al. Cancer statistics, 2014. CA Cancer J Clin. 2014;64:9–29.
- Vincent A, Herman J, Schulick R, et al. Pancreatic cancer. Lancet. 2011;378:607–620.
- Von Hoff DD, Ramanathan RK, Borad MJ, et al. Gemcitabine plus nab-paclitaxel is an active regimen in patients with advanced pancreatic cancer: a phase I/II trial. J Clin Oncol. 2011;29:4548–4554.
- Satake K, Kanazawa G, Kho I, et al. A clinical evaluation of carbohydrate antigen 19-9 and carcinoembryonic antigen in patients with pancreatic carcinoma. J Surg Oncol. 1985;29:15–21.
- Bauer TM, El-Rayes BF, Li X, et al. Carbohydrate antigen 19-9 is a prognostic and predictive biomarker in patients with advanced pancreatic cancer who receive gemcitabine-containing chemotherapy: a pooled analysis of 6 prospective trials. Cancer. 2013;119:285–292.
- Thornton GD, McPhail MJ, Nayagam S, et al. Endoscopic ultrasound guided fine needle aspiration for the diagnosis of pancreatic cystic neoplasms: a meta-analysis. Pancreatology. 2013;13:48–57.
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233.
- Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861–874.
- Lin S, Gregory RI. MicroRNA biogenesis pathways in cancer. Nat Rev Cancer. 2015;15:321–333.
- Song S, Ajani JA. The role of microRNAs in cancers of the upper gastrointestinal tract. Nat Rev. 2013;10:109–118.
- Hu Z, Chen X, Zhao Y, et al. Serum microRNA signatures identified in a genome-wide serum microRNA expression profiling predict survival of non-small-cell lung cancer. J Clin Oncol. 2010;28:1721–1726.
- Arroyo JD, Chevillet JR, Kroh EM, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci USA. 2011;108:5003–5008.
- Vickers KC, Palmisano BT, Shoucri BM, et al. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol. 2011;13:423–433.
- Kosaka N, Iguchi H, Yoshioka Y, et al. Secretory mechanisms and intercellular transfer of microRNAs in living cells. J Biol Chem. 2010;285:17442–17452.
- Yu X, Koenig MR, Zhu Y. Plasma miRNA, an emerging biomarker for pancreatic cancer. Ann Transl Med. 2015;3:297.
- Xu JW, Wang TX, You L, et al. Insulin-like growth factor 1 receptor (IGF-1R) as a target of MiR-497 and plasma IGF-1R levels associated with TNM stage of pancreatic cancer. PLoS One. 2014;9:e92847.
- Zhang X, Shao S, Geng H, et al. Expression profiles of six circulating microRNAs critical to atherosclerosis in patients with subclinical hypothyroidism: a clinical study. J Clin Endocrinol Metab. 2014;99:766–774.
- Xue Y, Wei Z, Ding H, et al. MicroRNA-19b/221/222 induces endothelial cell dysfunction via suppression of PGC-1α in the progression of atherosclerosis. Atherosclerosis. 2015;241:671–681.
- Ergun S, Tayeb TS, Arslan A, et al. The investigation of miR-221-3p and PAK1 gene expressions in breast cancer cell lines. Gene. 2015;25:377–381.
- Tao K, Yang J, Guo Z, et al. Prognostic value of miR-221-3p, miR-342-3p and miR-491-5p expression in colon cancer. Am J Transl Res. 2014;18:391–401. doi: 10.1016/j.bbrc.2017.01.002
- Wu Q, Ren X, Zhang Y, et al. MiR-221-3p targets ARF4 and inhibits the proliferation and migration of epithelial ovarian cancer cells. Biochem Biophys Res Commun. 2017.
- Shi J, Zhang Y, Jin N, Li Y, Wu S, Xu L. MicroRNA-221-3p plays an oncogenic role in gastric carcinoma by inhibiting PTEN expression. Oncol Res. 2016. [Epub ahead of print]. doi: 10.3727/096504016X14756282819385
- Coskunpinar E, Cakmak HA, Kalkan AK, et al. Circulating miR-221-3p as a novel marker for early prediction of acute myocardial infarction. Gene. 2016;591:90–96.
- Kristensen H, Thomsen AR, Haldrup C, et al. Novel diagnostic and prognostic classifiers for prostate cancer identified by genome-wide microRNA profiling. Oncotarget. 2016;7:30760–30771.
- Roberts TC, Wood MJ. Therapeutic targeting of non-coding RNAs. Essays Biochem. 2013;54:127–145.
- Kuninty PR, Bojmar L, Tjomsland V, et al. MicroRNA-199a and -214 as potential therapeutic targets in pancreatic stellate cells in pancreatic tumor. Oncotarget. 2016;7:16396–16408.
- Wei X, Wang W, Wang L, et al. MicroRNA-21 induces 5-fluorouracil resistance in human pancreatic cancer cells by regulating PTEN and PDCD4. Cancer Med. 2016;5:693–702.
- Zhou X, Zhang L, Zheng B, et al. MicroRNA-761 is upregulated in hepatocellular carcinoma and regulates tumorigenesis by targeting Mitofusin-2. Cancer Sci. 2016;107:424–432.
- Zhao L, Zou D, Wei X, et al. MiRNA-221-3p desensitizes pancreatic cancer cells to 5-fluorouracil by targeting RB1. Tumour Biol. 2016. doi: 10.1007/s13277-016-5445-8
- Permuth JB, Choi J, Balarunathan Y, et al. Combining radiomic features with a miRNA classifier may improve prediction of malignant pathology for pancreatic intraductal papillary mucinous neoplasms. Oncotarget. 2016;7:85785–85797.
- Miyamae M, Komatsu S, Ichikawa D, et al. Plasma microRNA profiles: identification of miR-744 as a novel diagnostic and prognostic biomarker in pancreatic cancer. Br J Cancer. 2015;113:1467–1476.
- Abue M, Yokoyama M, Shibuya R, et al. Circulating miR-483-3p and miR-21 is highly expressed in plasma of pancreatic cancer. Int J Oncol. 2015;46:539–547.