Abstract
MicroRNAs (miRNAs) play an important role in the development and progression of human malignancy. miR-613, as a tumour suppressor, was reported to decrease in several tumours. However, the expression levels and role of miR-613 in gastric cancer remain unknown. In this study, we found that miR-613 was evidently downregulated in gastric cancer tissue and cell. The functional analysis showed that miR-613 suppressed cell proliferation and migration in gastric cancer. Next, the dual-luciferase reporter system supported CDK9 as a direct target gene of miR-613. miR-613 mimics evidently repressed CDK9 expression in gastric cancer cells. Furthermore, we found that CDK9 in upregulated in gastric cancer and the CDK9 expression levels were inversely correlated with that of miR-613 in gastric cancer tissues. Overall, the results revealed that miR-613, as a tumour suppressor, involves in gastric cancer progression and metastasis by targeting CDK9, implying a novel potential therapeutic target for the treatment of gastric cancer.
Introduction
Gastric cancer is one of the most common cancer in the world and ranks as the third leading cause of cancer mortality around the world [Citation1,Citation2]. Despite the improvement of the prognosis and treatment of gastric cancer, and the overall 5-year survival rate of this cancer remains nearly 40% [Citation3,Citation4]. Therefore, understanding the tumorigenesis and progression of gastric cancer is critical and may identify novel diagnostic and prognostic biomarkers for improving the clinical outcome of gastric cancer patients.
MicroRNAs (miRNAs), a class of small non-coding RNA species, regulate or degrade its target gene by binding to the untranslated regions (3′UTR) [Citation5]. miRNAs play critical roles in the regulation of diverse biological processes including embryogenesis, development, cell maintenance, proliferation, apoptosis and so on. In recent years, accumulating studies have reported that miRNAs involved in the development and progression in various human cancers including gastric cancer [Citation6–9]. Studies showed that miRNAs were dysregulated and acts as a prognostic biomarker in gastric cancer [Citation10,Citation11]. Furthermore, the mechanism of miRNAs involved in gastric cancer progression might help to develop strategies for its diagnosis, treatment and prognosis in the future.
MiR-613 was downregulated and serve as a potential cancer suppressor in various kinds of cancer including ovarian cancer [Citation12], oesophageal squamous cell carcinoma [Citation13], colorectal cancer [Citation14], non-small cell lung cancer [Citation15], osteosarcoma [Citation16]. MiR-613 was also reported to repress cell proliferation and migration in prostate cancer [Citation17], breast cancer [Citation18] and retinoblastoma [Citation19]. It has been reported that miR-613 can suppress cell migration and migration by targeting its target genes [Citation20]. However, miR-613 expression and role in gastric cancer remains unclear.
In this study, we found that miR-613 expression was downregulated and miR-613 suppressed the gastric cancer proliferation and migration via targeting CDK9. Our results demonstrated the mechanism of miR-613 in gastric cancer progression, suggesting a potential therapeutic target for the treatment of gastric cancer.
Material and methods
Clinical tissues samples
A total of 52 pairs of human gastric cancer tissues and matched with adjacent non-cancerous bone tissues were obtained from patients in our department. This study was approved by the Research Ethics Committee of XiangyaHospital, Central South University, Changsha, China.
Cell culture and transfection
Seven gastric cancer cell lines, AGS, BGC-823, HGC-27, MGC-803, NCI-N87 and one normal gastric epithelial cell line (GES-1) were grown in RPMI-1640 medium (Gibco, USA) supplemented with 10% (vol/vol) foetal bovine serum (FBS). Cultures were maintained at 37 °C in a humidified atmosphere with 5% CO2.
Cells were seeded in 12-well plates and transfected using Lipofectamine 2000 (Invitrogen) and Opti-MEM (Gibco, USA) 24 h later according to the manufacturer’s instructions.
RNA extraction and qRT-PCR
Total RNA was extracted from cells and tissues using Trizol Reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's protocol. The first strand of cDNA was synthesized by reverse transcription (Takara Bio Inc., Shiga, Japan), and the expression of miRNA was detected by real-time quantitative RT-PCR (qRT-PCR) analysis using the SYBR Green detection system (Roche Applied Science). U6 and GAPDH mRNA were used as the internal controls. All tests were run in triplicate. The levels of mRNA expression were normalized to the level of the β-actin mRNA expression and using the 2−ΔΔCt cycle threshold method.
Luciferase reporter assay
The 3′ UTR of the human CDK9 with predicted miR-613-binding site was amplified from cDNA library of AGS cells and cloned to pGL3-Control vector. The binding site mutant of CDK9/pGL3 was also generated by a Site-Directed Mutagenesis Kit (SBS Genetech, Beijing, China). The cells were seeded in 24-well plates. AGS cells were cotransfected with the CDK9/pGL3 vectors and miR-613 or the miRNA control. Luciferase activity values were determined using the Dual-Luciferase Reporter Assay System (Promega, Madison, WI).
Cell proliferation assay
Cell proliferation was measured using the 3–(4,5-dimethylthiazole-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay. In brief, the cells were plated in 96-well plates at 5 × 103 per well after transfection. MTT assay was conducted. Finally, the optical density was determined at 570 nm using the ELISA plate reader (Model 550; Bio-Rad, Hercules, CA).
Cell migration assay
The wound-healing assay was used to detect cell migration capability as previous work [Citation21].
Western blot assay
Total proteins were extracted from AGS cells with a RIPA buffer with 0.5% sodium dodecyl sulphate (SDS) and 3% proteinase inhibitor cocktail (Sigma) for 30 min on ice. The concentration of protein was determined using the BCA protein assay kit (Santa Cruz, CA). For western blotting, proteins were separated by 10% SDS–PAGE and transferred onto PVDF membranes (Invitrogen, Carlsbad, CA). After blocking with 5% bovine serum albumin at room temperature for 1 h, membranes were incubated with primary antibody against CDK9 (1:200, Abcam, Hong Kong, China) at 4 °C overnight. Membranes were then incubated with secondary antibody for 2 h at room temperature. The intensities of the bands were quantified using an image analysis system.
Statistical analysis
All experiments were repeated at least three times with similar results. Representative data are shown. Statistical analyses were performed using paired Student’s t-tests. A value of p < .05 was considered statistically significant.
Results
miR-613 was frequently downregulated in gastric cancer
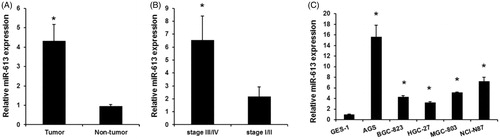
We detected the miR-613 expression in a total of 52 patients with a median age of 60 years. And 30 patients were at stage III/IV gastric cancer and 22 were at stage I/II. We found that the expression of miR-613 was evidently downregulated in tumour samples compared with adjacent non-tumour tissues. (). Moreover, miR-613 expression in patients with stage-III/stage-IV gastric cancer was significantly higher than in patients with stage-I/stage-II gastric cancer (). Finally, we detected the expression of miR-613 in gastric cancer cells and normal gastric epithelial GES-1 cells, miR-613 expression was down-regulated in AGS, BGC-823, HGC-27, MGC-803, NCI-N87 (). The results indicated that miR-613 may function as a tumour suppressor in gastric cancer.
Figure 1. miR-613 expression levels in gastric cancers. (A), qPCR assay detected the expression levels of miR-613 in gastric cancer samples and adjacent non-tumour tissues. Data are presented as means ± SD from three independent experiments. *p < .01 versus non-tumour group. (B), qPCR assay detected the expression levels of miR-613 in III/IV gastric cancer and stage-I/stage-II gastric cancer. (C), the expression levels of miR-613 in normal gastric epithelial GES-1 cells and gastric cancer cells (AGS, BGC-823, HGC-27, MGC-803, NCI-N87). Data are presented as means ± SD from three independent experiments. *p < .01 versus GES-1 group.
miR-613 repressed gastric cancer cells proliferation and migration
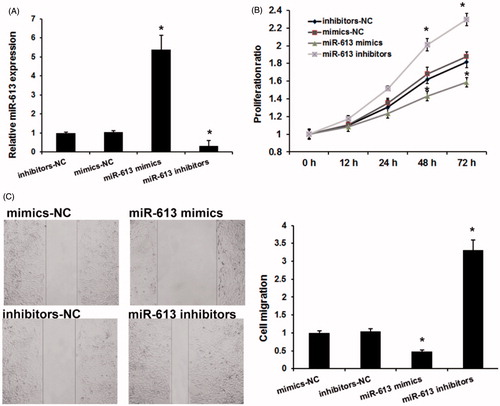
We next detected the role of miR-613 on gastric cancer progression by using MTT and wound healing analysis. We used miR-613 mimics to induce miR-613 expression and miR-613 inhibitors to repress miR-613 expression (). Consistent with previous studies, MTT assay revealed that miR-613 decreased the proliferation (). Moreover, the wound-healing assay showed that AGS cells treated with miR-613 mimics had a significantly reduced cell migration (). These results indicated that miR-613 involved gastric cancer progression by repressing cells proliferation and migration.
Figure 2. miR-613 repressed gastric cancer cells proliferation and migration. (A), The effects of miR-613 mimics and miR-613 inhibitors on miR-613 expression. (B), MTT assay detected the effects of miR-613 on gastric cancer cell proliferation. (C), Wound-healing assay detected the effects of miR-613 on gastric cancer cell migration. Data are presented as means ± SD from three independent experiments. *p < .01 versus NC group.
CDK9 is a direct target of miR-613 in gastric cancer
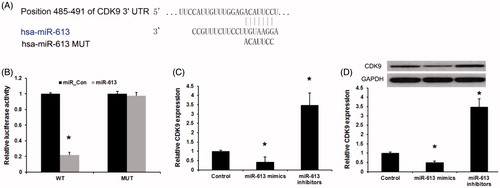
In this study, miRanda and Targetscan were used to predict the target gene of miR-613 in gastric cancer. As shown in , miR-613 could directly bind to 3′ UTR of CDK9 mRNA. To verify the prediction, we constructed wild-type CDK9 3′ UTR and mutated luciferase reporter plasmids. The luciferase reporter assays showed that miR-613 mimics evidently inhibited the luciferase activity of the wild type but had no effect on that of the mutant CDK9 3′-UTR (). Then, we detected the affection of miR-613 on the CDK9 expression in gastric cancer. We found that, miR-613 mimics repressed and miR-613 inhibitors induced endogenous CDK9 mRNA in AGS cells (). Meanwhile, miR-613 also inhibits CDK9 protein level in AGS cells (). Taken together, these results suggest that miR-613 directly targets and modulates the expression of CDK9 in gastric cancer cells.
Figure 3. CDK9 is a direct target of miR-613 in gastric cancer. (A), Sequence alignment of miR-613 and 3′ UTR of CDK9 using mirco-RNA.org. (B), Luciferase reporter assay revealed the target role of miR-613 on CDK9. (C), qPCR assay and (D), Western blot assay revealed the effects of miR-613 on the expression levels of CDK9 in gastric cancers. Data are presented as means ± SD from three independent experiments. *p < .01 versus control group.
CDK9 expression levels in gastric cancer tissues and cell lines
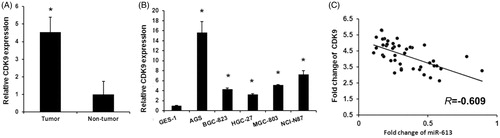
To validate the role of CDK9 in gastric cancer, we detected the expression levels of CDK9 in gastric cancer tissues and cell lines using qRT-PCR. As shown in , upregulated CDK9 expression were detected in gastric cancer tissues compared with adjacent non-cancerous tissues. CDK9 expression was also upregulated in gastric cancer cell lines (AGS, BGC-823, HGC-27, MGC-803, NCI-N87) compared with normal gastric epithelial GES-1 (). Then, we assessed the correlation between miR-613 and CDK9 in gastric cancer. As expected, we found that the levels of miR-613 exhibited a significant negative correlation with the levels of CDK9 mRNA (Pearson's correlation coefficient of r = −0.609, p < .01) (). Overall, our finding indicates that the expression level of CDK9 was upregulated and negatively associated with those of miR-613 in clinical gastric cancer tissues.
Figure 4. miR-613 expression is negatively correlated with CDK9 expression in gastric cancer. (A), qPCR assay revealed the expression levels of miR-613 in gastric cancer tissues and adjacent non-tumour tissues. Data are presented as means ± SD from three independent experiments. *p < .01 versus non-tumour group. (B), qPCR assay revealed the expression levels of miR-613 in normal gastric epithelial GES-1 cells and gastric cancer cells (AGS, BGC-823, HGC-27, MGC-803, NCI-N87). Data are presented as means ± SD from three independent experiments. *p < .01 versus GES-1 group. (C), Correlation of miR-613 levels with CDK9 mRNA levels was examined by qRT-PCR in 30 cases of glioma tissues.
Discussion
Gastric cancer is the fourth most frequently occurring aggressive cancer and the second leading cause of death from cancer worldwide, particularly in East Asia. Both inherited genetic alterations and environmental factors may contribute to cancer development. The underlying mechanism of gastric cancer carcinogenesis is critical for predicting prognosis and developing therapeutic strategy. Therefore, it is urgent to investigate the function of deregulated molecules in gastric cancer progression. In the present study, we indicated miR-613, as a tumour suppressor, regulated gastric cancer cell proliferation and migration by directly targeting CDK9.
Downregulated miR-613 was commonly observed in various kinds of cancers [Citation15,Citation22]. However, the expression and role of miR-613 in gastric cancer carcinogenesis remains unclear. Here, we confirmed downregulated miR-613 involved cell proliferation and migration in gastric cancer, suggesting that miR-613 may be a novel therapeutic strategy for gastric cancer treatment.
Subsequently, we were interested in the underlying mechanism of how miR-613 exerted its effect on cancer cells. CDK9 is a CDC2-related kinase and the catalytic subunit of the positive-transcription elongation factor b and the Tat-activating kinase [Citation23]. CDK9 regulates AR transcriptional activity and plays important role on prostate cancer cell growth [Citation24]. The bioinformatic databases predicted that miR-613 has a putative binding CDK9 sites in the 3′UTR. Then, the luciferase activity reporter assay and western blot analysis showed that miR-613 might be a novel negative regulator for CDK9. Moreover, the CDK9 expression levels were upregulated and negatively relationship with miR-613 expression in clinical gastric cancer tissues. Studies showed that deregulation of CDK9 has been associated with many cancer types [Citation25–27] and inhibition of CDK9 may represent an interesting approach as a cancer therapeutic target [Citation28,Citation29]. All together, we speculated that CDK9 is a target gene of miR-613 and plays important roles in tumorigenesis of gastric cancer.
In conclusion, the present study provides new insights into the mechanism of miR-613 in gastric cancer proliferation and migration, and suggests that targeting of miR-613/CDK9 axis may provide a potential therapeutic strategy for gastric cancer.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Yan X, Song X, Wang Z. Construction of specific magnetic resonance imaging/optical dual-modality molecular probe used for imaging angiogenesis of gastric cancer. Artif Cells Nanomed Biotechnol. 2017;45:399–403.
- Mohammadian F, Pilehvar-Soltanahmadi Y, Mofarrah M, et al. Down regulation of miR-18a, miR-21 and miR-221 genes in gastric cancer cell line by chrysin-loaded PLGA-PEG nanoparticles. Artif Nanomed Biotechnol. 2016;44:1972–1978.
- Musavi Shenas SMH, Mansoori B, Mohammadi A, et al. SiRNA-mediated silencing of Snail-1 induces apoptosis and alters microRNA expression in human urinary bladder cancer cell line. Artif Nanomed Biotechnol. 2017;45:969–974.
- Liang SH, Yan XZ, Wang BL, et al. Increased expression of FOXQ1 is a prognostic marker for patients with gastric cancer. Tumor Biol. 2013;34:2605–2609.
- Sun K, Lai EC. Adult-specific functions of animal microRNAs. Nat Rev Genet. 2013;14:535–548.
- Li X, Yang H, Tian Q, et al. Upregulation of microRNA-17-92 cluster associates with tumor progression and prognosis in osteosarcoma. NEO. 2014;61:453–460.
- Wang G, Shen N, Cheng L, et al. Downregulation of miR-22 acts as an unfavorable prognostic biomarker in osteosarcoma. Tumor Biol. 2015;36:7891–7895.
- Han K, Zhao T, Chen X, et al. microRNA-194 suppresses osteosarcoma cell proliferation and metastasis in vitro and in vivo by targeting CDH2 and IGF1R. Int J Oncol. 2014;45:1437–1449.
- Liu HT, Xing AY, Chen X, et al. MicroRNA-27b, microRNA-101 and microRNA-128 inhibit angiogenesis by down-regulating vascular endothelial growth factor C expression in gastric cancers. Oncotarget. 2015;6:37458–37470.
- Li C, Song L, Zhang Z, et al. MicroRNA-21 promotes TGF-beta1-induced epithelial-mesenchymal transition in gastric cancer through up-regulating PTEN expression. Oncotarget. 2016;7:66989–67003.
- Cao C, Sun D, Zhang L, et al. miR-186 affects the proliferation, invasion and migration of human gastric cancer by inhibition of Twist1. Oncotarget. 2016;7:79956–79963.
- Fu X, Cui Y, Yang S, et al. MicroRNA-613 inhibited ovarian cancer cell proliferation and invasion by regulating KRAS. Tumor Biol. 2016;37:6477–6483.
- Guan S, Wang C, Chen X, et al. MiR-613: a novel diagnostic and prognostic biomarker for patients with esophageal squamous cell carcinoma. Tumor Biol. 2016;37:4383–4391.
- Li B, Xie Z, Li Z, et al. MicroRNA-613 targets FMNL2 and suppresses progression of colorectal cancer. Am J Transl Res. 2016;8:5475–5484.
- Li D, Li DQ, Liu D, et al. MiR-613 induces cell cycle arrest by targeting CDK4 in non-small cell lung cancer. Cell Oncol (Dordr). 2016;39:139–147.
- Li X, Sun X, Wu J, et al. MicroRNA-613 suppresses proliferation, migration and invasion of osteosarcoma by targeting c-MET. Am J Cancer Res. 2016;6:2869–2879.
- Ren W, Li C, Duan W, et al. MicroRNA-613 represses prostate cancer cell proliferation and invasion through targeting Frizzled7. Biochem Biophys Res Commun. 2016;469:633–638.
- Wu J, Yuan P, Mao Q, et al. miR-613 inhibits proliferation and invasion of breast cancer cell via VEGFA. Biochem Biophys Res Commun. 2016;478:274–278.
- Zhang Y, Zhu X, Zhu X, et al. MiR-613 suppresses retinoblastoma cell proliferation, invasion, and tumor formation by targeting E2F5. Tumour Biol. 2017;39. doi: 1010428317691674
- Jiang B, Li Z, Zhang W, et al. miR-874 Inhibits cell proliferation, migration and invasion through targeting aquaporin-3 in gastric cancer. J Gastroenterol. 2014;49:1011–1025.
- Zhang LQ, Sun SL, Li WY, et al. Decreased expression of tumor suppressive miR-874 and its clinical significance in human osteosarcoma. Genet Mol Res. 2015;14:18315–18324.
- Nohata N, Hanazawa T, Kikkawa N, et al. Tumour suppressive microRNA-874 regulates novel cancer networks in maxillary sinus squamous cell carcinoma. Br J Cancer. 2011;105:833–841.
- Garriga J, Bhattacharya S, Calbo J, et al. CDK9 is constitutively expressed throughout the cell cycle, and its steady-state expression is independent of SKP2. Mol Cell Biol. 2003;23:5165–5173.
- Gordon V, Bhadel S, Wunderlich W, et al. CDK9 regulates AR promoter selectivity and cell growth through serine 81 phosphorylation. Mol Endocrinol. 2010;24:2267–2280.
- Krystof V, Baumli S, Furst R. Perspective of cyclin-dependent kinase 9 (CDK9) as a drug target. CPD. 2012;18:2883–2890.
- Mitra P, Yang RM, Sutton J, et al. CDK9 inhibitors selectively target estrogen receptor-positive breast cancer cells through combined inhibition of MYB and MCL-1 expression. Oncotarget. 2016;7:9069–9083.
- Morales F, Giordano A. Overview of CDK9 as a target in cancer research. Cell Cycle. 2016;15:519–527.
- Yin T, Lallena MJ, Kreklau EL, et al. A novel CDK9 inhibitor shows potent antitumor efficacy in preclinical hematologic tumor models. Mol Cancer Ther. 2014;13:1442–1456.
- Liu X, Shi S, Lam F, et al. CDKI-71, a novel CDK9 inhibitor, is preferentially cytotoxic to cancer cells compared to flavopiridol. Int J Cancer. 2012;130:1216–1226.
