Abstract
Acute myeloid leukaemia (AML) is a genetically heterogeneous, severe and rapidly progressing disease triggered by blocking granulocyte or monocyte differentiation and maturation. Overexpression of myeloid cell leukaemia-1 (Mcl-1) and Survivin is associated with drug resistance, tumour progression and inhibition of apoptotic mechanisms in leukaemia and several cancers. In the present study, we examined the combined effect of etoposide and dual siRNA-mediated silencing of Mcl-1 and Survivin on U-937 AML cells. The AML cells were co-transfected with Mcl-1 and Survivin-specific siRNAs and genes silencing were confirmed by quantitative real-time PCR and Western blotting. Subsequently, MTT assay was used for the evaluation of cytotoxic effects by dual siRNA and etoposide on their own and in combination. For the studying of apoptosis, DNA-histone ELISA and annexin-V/FITC assays were performed. Co-transfection of Mcl-1 and Survivin siRNA significantly blocked their expression at the mRNA and protein levels, leading to the induction of apoptosis and strong inhibition of growth (p < .05). Besides, combined treatment of etoposide with Mcl-1 and Survivin siRNAs co-transfection leads to synergistically enhance etoposide-induced cytotoxic and apoptotic effects (p < .05). The results showed that Mcl-1 and Survivin play a major role in the U937 cells survival and their resistance relative to etoposide. Thus, Mcl-1 and Survivin can be considered as promising molecular targets for the treatment of AML. The combination treatment with etoposide, and siRNA-mediated silencing of corresponding genes may be a novel strategy in chemoresistance AML treatment.
Introduction
Acute myeloid leukaemia (AML) is one of the severe, rapidly progressing genetic disorders triggered by blocking granulocyte or monocyte differentiation and maturation and interfere with the production of normal blood cells [Citation1,Citation2]. It is the most common leukaemia in elderly and comprises about 25% of different leukaemia cases in western countries. AML includes a wide spectrum of malignancies which if left untreated can be seriously lethal or slow growing [Citation3]. Cytostatic drugs enter cells by a passive diffusion through the plasma membrane and exert their effects intra-cellular [Citation4]. Many of cytostatic drugs act via binding to DNA and prevent DNA processes including transcription and replication. Other cytostatic drugs can create a cross-linking with DNA strands and cause DNA strand breaks. Others can interfere with the function of topoisomerases, inhibiting the DNA synthesis and resulting in cell death [Citation5]. By the treatment of cytostatic drugs, some patients were cured; however, a number of patients have no complete response or not at all. The development of drug resistance at diagnosis or during treatment of cytostatic drugs is the major cause of these incomplete responses [Citation3,Citation6]. Therefore, the advancements that have taken place in identification of molecular basis for cancer have led to introduction of targeted therapy methods using biologic drugs [Citation7].
siRNA belongs to RNAi gene silencing technology which cleaved the target mRNA in post-transcriptional level [Citation8,Citation9]. Development and identification of such a novel effective molecule, that are able to suppress proliferation, induce cell death and cellular mobility effectively in cancer cells [Citation10,Citation11].
Myeloid cell leukaemia-1 (Mcl-1) is a tightly regulated member of anti-apoptotic Bcl-2 family [Citation12]. Mcl-1 is highly expressed in many tumour cells including AML [Citation13,Citation14]. TRAF-2 or Survivin is a member of IAP family which inhibits caspases activation and consequently apoptosis. In lymphoma and leukaemia, overexpression of the human Survivin gene may be related to cancer progression and drug resistance [Citation15–17].
Due to the fact that any lacks in Mcl-1 and Surviving function are enough to induce apoptosis in tumour cells separately. Mcl-1 and Survivin can be assumed as unique targets for anti-cancer therapies.
In the present study, we analyse the role of dual siRNA Mcl-1 and Survivin knockdown in sensitizing U937 leukaemic cells to etoposide treatment. In this regard, we assess the effects of etoposide alone and in combination with dual siRNA in promoting apoptosis and U937 cell viability.
Materials and methods
Cell culture conditions
U-937, the human leukaemic monocyte lymphoma cell line was purchased from Pasteur Institute Cell Culture Collection (Tehran, Iran) and cultured in RPMI-1640 medium containing 10% heat-inactivated foetal bovine serum (FBS), and 100 IU/ml penicillin, 100 μg/ml streptomycin (Gibco/Life Technologies, Grand Island, NY). The cells maintained at 37 °C with a 95% humidified atmosphere containing 5% CO2. The cells were sub-cultured after 24–48 h and all the experiments performed in the log phase of cell growth.
siRNA transfection
The Mcl-1 siGENOME siRNA (5′-AGAACGAAUUGAUGUGUAA-3′), Survivin siGENOME siRNA (5′-CAAAGGAAACCAACAAUAA-3′) and a negative control (NC) siRNA (Scrambled siGENOME siRNA had no known homology with any human genes) were ordered from Dharmacon (Lafayette, CO). To evaluate the effects of siRNAs on gene silencing, transfections were performed in six-well cell culture plates for 24, 48 and 72 h. In brief, U-937 cells were plated (1 × 106 cells/well) and allowed to grow overnight. On the day of the transfection, the cells were cultivated in RPMI-1640 medium free of serum and antibiotics. Transfection of siRNA (at a final concentration of 50 or 100 nM) was performed using Lipofectamine™ 2000 (Invitrogen, Carlsbad, CA) transfection reagent. According to the manufacturer’s recommendations, briefly, siRNA and Lipofectamine diluted in OptiMEM reduced-serum medium separately and incubated for 10 min at 18–22 °C. The siRNA dilution directly added into Lipofectamine solution and incubated for 20 min at 18–22 °C. Afterward, the siRNA/Lipofectamine mixtures added to each well-containing cells and OptiMEM medium. The cells which treated with individual lipofectamine were considered as a blank control. The cells incubated for 6 h at 37 °C in a cell culture incubator. Subsequently, we added the equal volume of RPMI-1640 medium containing 20% FBS into each well and incubated for further 24–72 h. The silencing of Mcl-1 and Survivin genes expression was then assessed by quantitative real-time PCR (qRT-PCR) and Western blot analysis.
RNA isolation, cDNA synthesis and qRT-PCR
Total RNA was extracted by the RiboEX reagent (GeneAll Biotechnology, Seoul, Korea) according to the manufacturer’s protocol. Complementary DNAs were synthesized using the RevertAid First Strand cDNA Synthesis kit (Thermo Fisher Scientific, Waltham, MA). Two micrograms of total RNA was reverse-transcribed by Moloney murine leukaemia virus transcriptase using oligo-dT primers according to the manufacturer’s instructions. qRT-PCR was applied by the Corbett Rotor-Gene™ 6000 system (Corbett Life Science, Concorde, Australia) using SYBR Premix Ex Taq (TakaRa, Ostu, Japan ). The reaction system of qRT-PCR was 3 μl of cDNA template, 1.2 μl of MgCl2, 1 μM (10 pmol) of each of the primers, 2 μl of reaction buffer, 2 μl of 10 mM dNTP mix, 0.3 μl of Taq DNA polymerase and 9.5 μl of nuclease-free water. The primer sequences is illustrated in .
Table 1. The primer sequences.
Reaction parameters were 95 °C for 3 min followed by 35 cycles consisting of 30 s at 95 °C, 40 s at a gradient temperature of 54–62 °C and 30 s at 72 °C. Final extension eventually was performed by heating at 72 °C for 5 min. Relative gene expression of Mcl-1 was calculated with the 2-(ΔΔCT) method [Citation18], using β-actin as the reference gene.
Cytotoxicity assay
The effects of siRNAs co-transfection on a sensitivity of U-937 cells to etoposide were done using 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay. U-937 cells (2 × 104 cells/well) were incubated in 96-well plates each containing 200 μl of medium. The cells were divided into three groups: etoposide, NC siRNA, co-transfection (Mcl-1 and Survivin) and etoposide. siRNA transfection performed as the protocol which described above. Six hours after the siRNA transfection, the cells were treated with different concentrations of chemotherapeutic agent etoposide (0.001, 0.2, 0.5, 1, 2 and 4 μM). After 48 h, the cytotoxicities of the treatments were measured using MTT assay. Briefly, the cells treated with MTT solution (Sigma-Aldrich, Saint Louis, MO) (2 mg/ml) for 4 h, subsequently, dissolve the insoluble purple formazan product into a coloured solution via 200 µl of DMSO and 25 µl of Sorensen buffer. Finally, the absorbances of the solubilized formazan dyes were measured using an ELISA plate reader (Awareness Technology, Palm City, FL) at 570 nm. The survival rate (SR) was calculated by the following formula: SR (%) = (A Treatment/A Control) × 100%. The half inhibitory concentration (IC50) calculated using GraphPad Prism 6.01 software (GraphPad Software Inc., San Diego, CA).
Combination effect analysis
The combination effect between siRNAs (Mcl-1 and Survivin) cotransfection and etoposide was measured, based on the principles described by Chou and Talalay [Citation19]. The value of combination index (CI) was determined using the following formula: CI = SAB/(SA × SB), where SA and SB are the survival rate of etoposide and combined siRNAs relative to the corresponding controls, SAB is the survival rate of the combination treatment of siRNAs in combination to etoposide relative to the control. CI <1, CI =1 and CI >1 indicate synergistic, additive and antagonistic effects, respectively.
Cell death detection
The U-937 cells were plated at a density of 1.5 × 104 cells/well in 96-well plate and then treated with specific siRNAs, NC siRNA, the IC50 concentration of etoposide and the combination of them, as described previously. Forty-eight hours after transfection, cells were harvested and assayed for apoptosis using a Cell Death Detection ELISA Kit (Roche, Mannheim, Germany) following the manufacturer’s instructions. Briefly, the lysates were harvested and immunoreagent containing antihistone-biotin and anti-DNA-peroxidase were then transferred to each well of a streptavidin-coated plate. Subsequently, the wells were washed with incubation buffer and then 2,2-azino-bis(3-ethylbenzthiazoline-6-sulfonic acid) (ABTS) solution was added. Finally, the reactions were stopped with ABTS stop solution and the absorbance was measured using an ELISA plate reader at 405 nm. The results were expressed as the fold change in the absorbance.
Flow cytometry for cell apoptosis detection
U-937 cells were seeded at a density of 2 × 105 cell/well in six-well plates. The experiment was divided into the five groups as mentioned above. Forty-eight hours after co-treatments, the cells were stained with Annexin-V-FITC and propidium iodide (PI) using the Annexin-V-FLUOS staining kit (Roche Diagnostics GmbH). Briefly, cells were detached and washed with phosphate buffered saline. The density of U937 cells was adjusted to 5 × 105–5 × 106 cells. The cells re-suspended with 100 μl of pre-chilled binding buffer, and then 5 μl of Annexin V-FITC and 5 μl of PI were added to each sample and incubated for a limited time in the dark on ice. Then, the apoptosis evaluated by using BD FACSCalibur (BD Biosciences, Franklin Lakes, NJ) flow cytometer.
Statistical analysis
All the results of this study were presented as the mean ± standard deviation (SD). Statistical significance of differences between groups was evaluated by using analysis of variance (ANOVA) followed by Bonferroni’s and Sidak’s multiple comparisons. p Values less than .05 were considered significant. Three independent experiments were performed for each assay. All the statistical analyses were performed using GraphPad Prism software (La Jolla, CA; http://www.graphpad.com). The value of p less than .05 was considered significant.
Results
Mcl-1 and Survivin mRNA and protein down-regulation in U-937 cells after siRNAs transfection
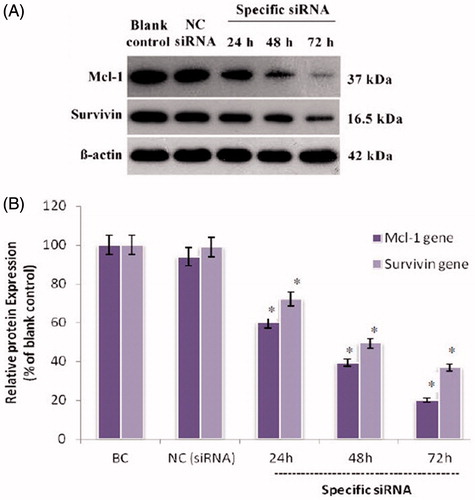
We initially explored the effect of specific siRNA on Mcl-1 and Survivin expression by qRT-PCR and Western blot analysis at 24, 48 and 72 h after transfection. As shown in , Mcl-1 and Survivin siRNA transfection separately reduced the amount of Mcl-1 and Survivin mRNA. Results of untreated group (blank control) were considered as 100% and relative gene expression was calculated in relation to blank control. The level of Mcl-1 mRNA in the siRNA-treated group at 24, 48 and 72 h post-transfection was about 43.7%, 21.1% and 17.2%, and for Survivin was 43.7%, 21.1% and 17.2%, respectively (p < .05; ). There was no significant difference between NC siRNA group and untreated cells. The Mcl-1 and Survivin siRNA was also effective in knocking down Mcl-1 and Survivin protein expression. As shown in , while the β-actin internal control showed equal loading among all groups, the level of Mcl-1 protein at 24, 48 and 72 h post-transfection was 51.4%, 20.5%, and 13.2% and for Survivin was 51.4%, 20.5%, and 13.2%, respectively. Meanwhile, NC siRNA had an insignificant effect on Mcl-1 and Survivin mRNA and protein expression compared to the blank control group.
Figure 1. The Mcl-1 and Survivin proteins expression levels in treated human myelomonocytic leukaemia cells. (A) Representative western blot bands of β-actin, Mcl-1 and Survivin proteins from U937 cells transfected with siRNAs. (B) The density of each band was quantified by the Image J software (NIH, Bethesda, MD) and the expression of each Mcl-1 and Survivin normalized to the corresponding β-actin. Results are expressed in relation to the blank control. The data represent mean ± SD (n = 3); *p < .05 versus blank control.
Synergistic chemosensitization effect of simultaneous Mcl-1 and Survivin suppression
To compare the chemosensitization effect of concomitant gene suppression with the single gene suppression, the cells were exposed to a mixture of etoposide and co-transfection of siRNAs, then 24 h later analysed by MTT assay. The results showed that the combination of dual siRNAs markedly decreased the cell survival rate to 66.11%, compared with the blank control cells (p < .05; ).
Figure 2. (A) Effect of siRNAs co-transfection on etoposide sensitivity of U-937 cells. Six hours after transfection of siRNAs or NC siRNA, cells were exposed to etoposide at indicated doses. After 18 h, cytotoxicity of treatments was determined by MTT assay as described above. The data represent mean ± SD (n = 4); *p < .05 versus etoposide or Mcl-1 siRNA alone; #p < .05 versus blank control. (B) Dose–response curves of treatments in MTT assay. A dose–response curve was created with the familiar symmetrical sigmoidal shape model which fit a dose–response curve to determine the IC50 of the etoposide. The data represent mean ± SD (n = 4).
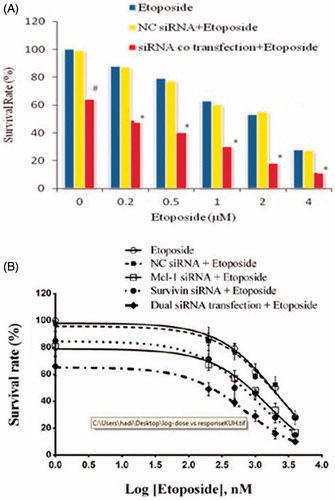
Moreover, transfection of dual siRNAs synergistically enhanced chemosensitivity of the cell and further lowered the IC50 value of etoposide relative to single siRNA transfection (from 2.06 to 0.70 μM) (p < .05; and ). NC siRNA (100 nM) had also a minimal effect on the chemosensitivity of the cells relative to the etoposide (p > .05; and ).
Figure 3. Effect of combination of Mcl-1 and Survivin suppression on etoposide-induced apoptosis. After 6 h of siRNAs co-transfection, the cells were exposed to etoposide (IC50) for 18 h. After 24 h of siRNA transfection, ELISA cell death assay was used to measure apoptosis induction. The results are expressed as mean ± SD (n = 3); *p < .05 versus blank control; #p < .05.
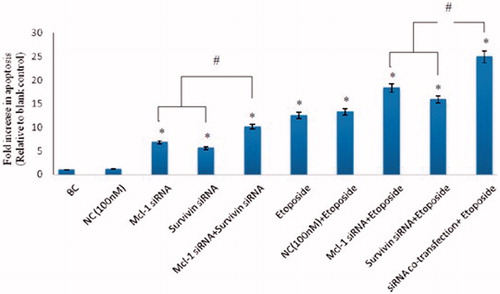
Table 2. Summarized inhibitory concentration (IC50) values of etoposide in MTT assay.
Mcl-1 and Survivin silencing enhancedetoposide-induced apoptosis in U937 cells
To confirm the synergistic cytotoxic effects between siRNAs co-transfection and etoposide could induce the apoptosis in human myelomonocytic leukaemia cell (U937), the ELISA-based cell death detection was examined. The results demonstrated that treatment of U937 cells with the combination of Mcl-1 and Survivin siRNA significantly added to the apoptotic effect caused by etoposide (compare to the single siRNA transfection) (p < .05; ). However, treatment with scrambled siRNA (NC) individual or in combination with the chemotherapeutic agent (etoposide) showed no distinct changed in the apoptosis induction compared to the blank control group or etoposide mono-treatment group, respectively (p > .05; ). Therefore, it can be concluded that the chemosensitization effect of dual Mcl-1 and Survivin suppression was partially attributed to the enhancement of apoptosis alone and synergically by etoposide.
Induction of apoptosis by siRNAs and etoposide treatments was also confirmed by flow cytometry
In the flow cytometric assay, annexin V-FITC and PI double staining were performed to quantify the percentage of apoptotic cells. Lower left (LL) quadrant (annexin V−/PI−) was considered as the population of live cells, lower right quadrant (LR) (annexin V+/PI−) was regarded as the cell population in the early stage of apoptosis, upper right (UR) quadrant (annexin V+/PI+) is considered as the population of cell at late apoptosis and early necrosis stages and upper left (UL) quadrant (annexin V−/PI+) represents the population of cells in the late necrosis stage.
As shown in and , the percentage of annexin V + cells were 27.55 and 27.81 for etoposide (IC50), dual siRNA-treated groups, respectively, after 24 h of incubation. On the other hand, interestingly, when cells were subjected to Survivin and Mcl-1 knockdown and concurrent etoposide exposure, the population of Annexin V + cells was increased significantly relative to the etoposide mono treatment (, ).
Figure 4. Flow cytometric apoptosis analysis of U-937 cells. Cells were stained with annexin-V-FITC and PI after treatment with blank control, 100 nM of NC siRNA, etoposide (IC50), 50 nM of Mcl-1 siRNA +50 nM of survivin siRNA and 50 nM of Mcl-1 siRNA +50 nM of survivin siRNA + etoposide (IC50). Dot plots are representative of three experiments.
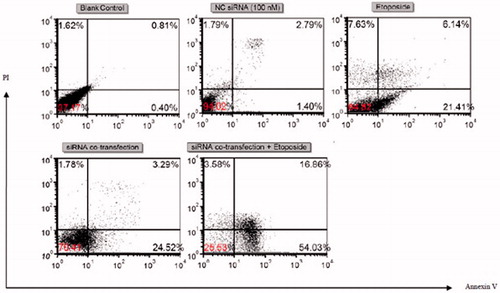
Table 3. Flow cytometric results of U-937 cells treated with anticancer agents.
Nevertheless, no significant changes in the apoptosis were displayed in scramble (NC) siRNA group or NC siRNA in combination to etoposide groups compared to the blank control group or individual etoposide, respectively (p > .05; and ).
Discussion
AML is a malignant disease characterized by an arrest in cell differentiation and uncontrolled proliferation whose standard chemotherapeutic treatment consists mainly of cytarabine and anthracyclines [Citation20], but resistance to therapy is a common clinical obstacle in AML [Citation21].
Mcl-1 is a member of Bcl-2 anti-apoptotic family of proteins and is overexpressed in a variety of human malignancies including colon, breast, CNS, lung, renal, melanoma, ovarian, prostate and leukaemia [Citation22]. Mcl-1 upregulation in tumour cells enabled them to evade apoptosis, to survive and as a mechanism to develop drug resistance [Citation23,Citation24].
Survivin as a member of IAP protein family is normally expressed in foetal period but its overexpression incompletely differentiated cells is frequently reported in malignant and cancerous cells [Citation25].
According to the literature, the overexpression of Mcl-1 and Survivin genes is directly associated with poor prognosis, tumour development and resistance to chemotherapy drugs in many cancers such as AML. In addition, some reports indicate that Mcl-1 and Survivin down-regulation can sensitize tumour cells to anticancer agents [Citation24,Citation26,Citation27].
In this regards, the present study to investigate whether dual siRNA-mediated silencing of Mcl-1 and Survivin would affect drug resistance and chemosensitivity of leukaemic cells to etoposide, we examine dual siRNA in combination to etoposide and alone in U-937 AML cells.
The results of qRT-PCR and western blot analysis showed that co-transfection with Mcl-1 and Survivin siRNAs could significantly reduce the mRNAs levels of Mcl-1 and Survivin and their translated protein, suggesting that Mcl-1 and Survivin siRNA could effectively degrade Mcl-1 and Survivin mRNA and inhibited its translation to protein.
The results of cell viability assay revealed that Mcl-1 and Survivin suppression had a significant effect on decreasing U-937 cell growth and proliferation, suggesting that these proteins may have an important role in cell proliferation.
Furthermore, the results of MTT assay revealed that pre-treatment with dual siRNA could synergistically reduce the IC50 of leukaemic cells to etoposide, demonstrating that Mcl-1 and Survivin downregulation could sensitize U-937 AML cells to etoposide synergistically. In order to better understand the effect of Survivin on drug resistance in leukaemic cells, we evaluated the effects of Mcl-1 and Survivin siRNA on the apoptotic effect of etoposide. The results of this study showed the amount of apoptosis by two distinct methods to determine whether the decrease in cell viability is because of reduced cell proliferation or increased apoptosis. The results of both assays showed that single treatment with etoposide remarkably increased apoptosis in U-937 cells.
Furthermore, Mcl-1 and Survivin siRNAs co-transfection in combination with etoposide were tested and indicated that dual gene silencing was more effective than single gene silencing in promoting etoposide-induced apoptosis.
However, NC siRNA nor Lipofectamine 2000 could not change the chemosensitivity of etoposide, indicating the important role of Mcl-1 and Survivin siRNAs in chemosensitization.
Etoposide activates both intrinsic and extrinsic pathways of apoptosis [Citation27,Citation28]. This anti-cancer agent disturbed DNA topoisomerase II by interfering DNA re-ligation, this causes in numerous errors in DNA synthesis at the premitotic stage of cell division. Consequently, a larger number of errors in DNA synthesis of malignant cells caused by etoposide which results in apoptosis. However, by inhibition of caspase activities and other possible mechanisms, Survivin is associated with microtubules and the mitotic spindle protein and blocks the both intrinsic and extrinsic pathways and leads to chemo- or radiotherapy resistance [Citation29–34].
On the other hand, Mcl-1 localized to mitochondrial outer membrane and hetero-dimerize with and neutralize pro-apoptotic Bcl-2 family proteins, leading to inhibition of cytochrome c release from mitochondria which is needed for apoptosis induction and eventually cell survival would be increased [Citation13]. In addition, Mcl-1 mainly blocks the apoptosis through intrinsic pathway [Citation13].
The results of our study showed that dual siRNA-mediated downregulation of Mcl-1 and Survivin led to the increased apoptotic effect of etoposide, suggesting the possibility of that Mcl-1 and Survivin sensitizes AML cells in the Bcl2 family especially Bak and Bax and a caspase 3-dependent mechanism.
Taking together, the results from this study demonstrated that Mcl-1 and Survivin have a vital role in cell survival and sensitivity of U-937 cells to etoposide. Downregulation of Mcl-1 and Survivin by specific siRNAs and co-exposure to etoposide led to a significant synergistic inhibition of cell proliferation and enhanced apoptosis and anti-tumour effect. Therefore, these can be considered as attractive targets for AML treatment and a novel strategy to overcome drug resistance and sensitize leukaemic cells to chemotherapeutics.
Acknowledgments
We acknowledge the Iran National Science Foundation (INSF) for funding this research.
Disclosure statement
Authors declare no competing financial interests by the results presented in this manuscript.
References
- Löwenberg B, Rowe JM. Introduction to the review series on advances in acute myeloid leukemia (AML). Blood. 2016;127:1.
- Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2006. CA Cancer J Clin. 2006;56:106–130.
- Kupsa T, Milos Horacek J, Jebavy L. The role of cytokines in acute myeloid leukemia: a systematic review. Biomed Pap. 2012;156:291–301.
- Frézard F, Garnier-Suillerot A. Permeability of lipid bilayer to anthracycline derivatives. Role of the bilayer composition and of the temperature. Biochim Biophys Acta. 1998;1389:13–22.
- Rabbani A, Finn RM, Ausio J. The anthracycline antibiotics: antitumor drugs that alter chromatin structure. Bioessays. 2005;27:50–56.
- Smits EL, Berneman ZN, Van Tendeloo VF. Immunotherapy of acute myeloid leukemia: current approaches. Oncologist. 2009;14:240–252.
- Le Tourneau C, Delord J-P, Gonçalves A, et al. Molecularly targeted therapy based on tumour molecular profiling versus conventional therapy for advanced cancer (SHIVA): a multicentre, open-label, proof-of-concept, randomised, controlled phase 2 trial. Lancet Oncol. 2015;16:1324–1334.
- Mansoori B, Mohammadi A, Shir Jang S, et al. Mechanisms of immune system activation in mammalians by small interfering RNA (siRNA). Artif Cells Nanomed Biotechnol. 2016;44:1589–1596.
- Prabha S, Vyas R, Gupta N, et al. RNA interference technology with emphasis on delivery vehicles—prospects and limitations. Artif Cells Nanomed Biotechnol. 2016;44:1391–1399.
- Shajari N, Davudian S, Kazemi T, et al. Silencing of BACH1 inhibits invasion and migration of prostate cancer cells by altering metastasis-related gene expression. Artif Cells Nanomed Biotechnol. 2017 [cited Sep 11]; [10 p.]. DOI:10.1080/21691401.2017.1374284
- Musavi Shenas SMH, Mansoori B, Mohammadi A, et al. SiRNA-mediated silencing of Snail-1 induces apoptosis and alters micro RNA expression in human urinary bladder cancer cell line. Artif Cells Nanomed Biotechnol. 2017;45:969–974.
- Cai M, Chen Q, Chen C, et al. Activation of triggering receptor expressed on myeloid cells-1 protects monocyte from apoptosis through regulation of myeloid cell leukemia-1. Anesthesiology. 2013;118:1140–1149.
- Akgul C. Mcl-1 is a potential therapeutic target in multiple types of cancer. Cell Mol Life Sci. 2009;66:1326–1336.
- Bose P, Grant S. Mcl-1 as a therapeutic target in acute myelogenous leukemia (AML). Leuk Res Rep. 2013;2:12–14.
- Kang MH, Reynolds CP. Bcl-2 inhibitors: targeting mitochondrial apoptotic pathways in cancer therapy. Clin Cancer Res. 2009;15:1126–1132.
- Ghanbari P, Mohseni M, Tabasinezhad M, et al. Inhibition of Survivin restores the sensitivity of breast cancer cells to docetaxel and vinblastine. Appl Biochem Biotechnol. 2014;174:667–681.
- Hosseini S, Hashemzadeh S, Estiar MA, et al. Expression analysis of aurora-C and Survivin, two testis-specific genes, in patients with colorectal cancer. Clin Lab. 2015;61(5–6):475–480.
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–408.
- Chou T-C. Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res. 2010;70:440–446.
- Fathi AT, Grant S, Karp JE. Exploiting cellular pathways to develop new treatment strategies for AML. Cancer Treat Rev. 2010;36:142–150.
- Åström M, Bodin L, Nilsson I, et al. Treatment, long-term outcome and prognostic variables in 214 unselected AML patients in Sweden. Br J Cancer. 2000;82:1387.
- Shore GC, Warr MR. Unique biology of Mcl-1: therapeutic opportunities in cancer. Curr Mol Med. 2008;8:138–147.
- Chetoui N, Sylla K, Gagnon-Houde J-V, et al. Down-regulation of mcl-1 by small interfering RNA sensitizes resistant melanoma cells to fas-mediated apoptosis. Mol Cancer Res. 2008;6:42–52.
- Wei S-H, Dong K, Lin F, et al. Inducing apoptosis and enhancing chemosensitivity to gemcitabine via RNA interference targeting Mcl-1 gene in pancreatic carcinoma cell. Cancer Chemother Pharmacol. 2008;62:1055–1064.
- Zaffaroni N, Daidone MG. Survivin expression and resistance to anticancer treatments: perspectives for new therapeutic interventions. Drug Resist Updat. 2002;5:65–72.
- Fukuda S, Pelus LM. Survivin, a cancer target with an emerging role in normal adult tissues. Mol Cancer Ther. 2006;5:1087–1098.
- Adida C, Recher C, Raffoux E, et al. Expression and prognostic significance of survivin in de novo acute myeloid leukaemia. Br J Haematol. 2000;111:196–203.
- Montecucco A, Biamonti G. Cellular response to etoposide treatment. Cancer Lett. 2007;252:9–18.
- Zaffaroni N, Pannati M, Diadone MG. Survivin as a target for new anticancer interventions. J Cell Mol Med. 2005;9:360–372.
- Church DN, Talbot DC. Survivin in solid tumors: rationale for development of inhibitors. Curr Oncol Rep. 2012;14:120–128.
- Rodel F, Sprenger T, Kaina B, et al. Survivin as a prognostic/predictive marker and molecular target in cancer therapy. Curr Med Chem. 2012;19:3679–3688.
- Mansoori B, Mohammadi A, Shirjang S, et al. HMGI-C suppressing induces P53/caspase9 axis to regulate apoptosis in breast adenocarcinoma cells. Cell Cycle. 2016;15:2585–2592.
- Kachalaki S, Baradaran B, Majidi J, et al. Reversal of chemoresistance with small interference RNA (siRNA) in etoposide resistant acute myeloid leukemia cells (HL-60). Biomed Pharmacother. 2015;75:100–104.
- Mansoori B, Mohammadi A, Goldar S. Silencing of high mobility group isoform IC (HMGI-C) enhances paclitaxel chemosensitivity in breast adenocarcinoma cells (MDA-MB-468). Adv Pharm Bull. 2016;6:171.
