Abstract
Hepatocarcinoma is one of the most lethal malignancy haunting the Chinese population, which is partially due to the difficulties in diagnosis at an early stage. The search for a biomarker that could signify the presence and progress of hepatocarcinoma is never ended. MicroRNAs are 22-nt RNAs that could bind to 3′ UTR of target mRNAs, mediating degradation of mRNAs or inhibiting the translation. Although much has been investigated, the role of miR-124 in hepatocarcinoma remained elusive. We first detected aberrant expression level of miR-124 in HCC tissues of 112 patients. By exploring the clinical parameters, we found a significantly inverse correlation between miR-124 level and TNM stages. Consistent with this, the survival analysis indicated the association of low miR-124 with longer survival time. Subsequent forced expression miR-124 resulted in reduced cell viability of Hep3B and SMMC-7221, which cell lines have high and low background expression of miR-124, respectively. TargetScan prediction rendered a subset of target candidates, which were selected for experimental validation, KLF4 was subject to luciferase assay. Ectopic expression of KLF4 increased the sphere formation ability and CD44/133-positive cell numbers, which can be reversed by abundant expression of miR-124, suggesting that KLF4 is a functional target of miR-124 in tumourigenesis and cancer progression of HCC.
Introduction
Hepatocellular carcinoma (HCC) is one of the most lethal cancers, making up a large proportion of cancer-related death in the world [Citation1]. Most patients were diagnosed at late stage, which becomes refractory to surgery and chemotherapies [Citation2]. Though increasing evidence highlighted the genetic etiologies for cancers, the tumourigenesis of HCC can be attributed to a variety of risk factors including diabetes, obesity and chronic alcohol consumption [Citation3,Citation4]. Over 80% HCC cases are associated with infections of hepatitis B virus (HBV) or hepatitis C virus (HCV) [Citation3]. Moreover, metastasis and tumour cell dissemination deteriorate the clinical outcomes [Citation5]. Though the 5-year survival rates has doubled over the last two decades, drug therapy was not proportionately improved [Citation6]. Therefore, identifying genes and pathways underlying tumourigenesis and development of HCC is critical to researches and trials of therapeutic regimes.
MicroRNAs are a cluster of short non-transcriptional RNAs that are 22–24 nt in length, which have been characterized as critical regulators in protein translation [Citation7]. miRNAs can either initiate cleavage of mRNAs by miRNA-associated RNA-induced silencing complex (miRISC) or deter the translation of protein due to spacial impairment [Citation8]. As its short length, a miRNA could impact the function of a subset of genes, and vice versa. Therefore, miRNAs and protein coding genes form a regulatory interaction network that encompasses critical biological pathways underpinning the cellular activities and orchestration thereof [Citation7,Citation9–13]. The implication of miRNAs in a variety of cancers was extensively investigated [Citation14–16]. Recently, an increasing number of studies have revealed the association between miRNA and HCC [Citation10,Citation17–20]. For instance, miR-21 is an oncogenic miRNA that was overexpressed in liver cancer, particularly in HCC, which enhances the proliferative potency via repressing mitogen-activated protein kinase-kinase 3 (MAP2K3), a member of MAPK family that transduces extracellular signals into intracellular response [Citation21]. Wong et al. detected elevated expression of miR-222 expression in primary HCC tumours, compared to normal liver tissues, suggesting a strong relationship between high miR-222 expression and tumour progression [Citation22], which functions by suppressing PPP2R2A, an inhibitor of AKT signalling pathway [Citation23]. Ding et al. established HCC cell lines to overexpress miR-145 and observed that cell proliferation, migration and invasion were significantly inhibited [Citation24]. Subsequently, ROCK1 was identified as a novel target of miR-145, which is an important regulator of actomyosin contraction and focal adhesions, thereby increasing cell polarity and migration. In HCC metastasis, aberrant expression of miRNAs was detected by microarray analysis [Citation25]. Although the implication of miRNAs in HCC was well recognized, the precise role of miRNA in every regulatory layer remains elusive [Citation26]. miR-124 was identified as a tumour suppressor that is epigenetically silenced in HCC [Citation27]. Ectopic expression of miR-124 directly resulted in downregulation of VIQGAP1, ABCE1 and CDK6, as well as impaired cell growth, suggesting its important role in hepatocarcinogenesis [Citation27]. Given this, the further exploration of precise role of miR-124 is warranted.
In this study, we detected aberrant expression level of miR-124 in HCC tissues of 179 patients. By exploring the clinical parameters, we found a significantly inverse correlation between miR-124 level and TNM stages. Consistent with this, the survival analysis indicated the association of low miR-124 with longer survival time. Subsequent forced expression miR-124 resulted in reduced cell viability of Hep3B and SMMC-7221, which cell lines have high and low background expression of miR-124, respectively. TargetScan prediction rendered a subset of target candidates, which were selected for experimental validation, KLF4 was subject to luciferase assay. Ectopic expression of KLF4 increased the sphere formation ability and CD44/133-positive cell numbers, which can be reversed by abundant expression of miR-124, suggesting that KLF4 is a functional target of miR-124 in tumourigenesis and cancer progression of HCC.
Materials and methods
Patients and samples
A total of 112 HCC patients who were diagnosed and ascertained at The First People's Hospital of Yancheng City during from September 2010 to November 2016 were recruited. Those patients were with an average age of 58, and had a pathological diagnosis of HCC based on histological or cytological criteria via biopsy or surgical resection. All patients had no history of anticancer treatment and metastasis before surgery or biopsy, and their clinical data were complete (). Tumour-node-metastasis (TNM) staging was classified according to the 7th edition TNM classification of the International Union Against Cancer. Healthy control individuals were selected from those who took physical examination at Healthy Physical Examination Center of The First People's Hospital of Yancheng City. The checked items include blood chemistry, baseline electrolytes, blood cell counts, CEA and AFP, c-reactive protein, type-B ultrasound at abdomen and pelvis and a chest X-ray. Individuals showed abnormality in any one of those items or have findings suggesting liver pathology or constitutional symptoms were excluded. Written informed consents were obtained from patients and healthy individuals, and study design was approved by the Ethics Committee of The First People's Hospital of Yancheng City.
Table 1. Patient characteristics.
RNA isolation and serum miRNA qRT-PCR assay
Trizol (Invitrogen Life Technologies, CA, USA) was used to extract total RNA from HCC and adjacent non-tumour liver tissues as per manufacturer’s instructions. Subsequently, total RNA was reversely transcribed to cDNA using Primescript RT reagent kit (Takara Bio. Inc., Tokyo, Japan). Real-time PCR was conducted using SYBR Premix Ex Taq (Takara). All experiments were performed triplicate. Relative amounts of miRNA were normalized to internal miRNA controls RNU6B. Ct values were measured using the fixed threshold settings, which were then corrected based on standard amplification curve of series synthetic miRNA oligonucleotides of known concentrations.
Cell culture and transfection
Four HCC cell lines, HepG2, Hep3B, SNU-182 and SMMC-7221, were procured from Shanghai Institute of Cell Biology, which were maintained in Dulbecco’s modified Eagle’s medium (DMEM; Invitrogen, CA, USA) supplemented with 10% fetal bovine serum (FBS; Invitrogen), 0.2 mM non-essential amino acids, 3 mM L-glutamine and 1% streptomycine. The culture environment was conditioned at 37 °C with 5% CO2.
After growing to 70–80% confluence, SMMC-7221 cells were transfected with miR-124 mimics, and Hep3B cell with antagomir sequences (Thermo Fisher Scientific, CA, USA) for 24 h at room temperature with Lipofactamine 2000 (Invitrogen; Thermo Fisher Scientific). Both cells transfected with scramble sequence were considered as control.
Cell viability assay
The cell viability of Hep3B and SMMC-7221 cells was measured by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay. The cells were seeded in 96-well plates at a density of 2 × 105 cells/cm2 with 100 μl cell culture medium. MTT stock solution (5 mg/ml ×20 μl, Sigma-Aldrich, Saint Louis, MO, USA) was added to each well and incubated at 37 °C for 5 h. The formazan crystals were dissolved in 200 μl of DMSO. The optical density (OD) under 570 nm was taken.
Western blot assay
Cells were lysed in NP-40 buffer containing 150 mM NaCl, 1.0% NP-40, 50 mM Tris-HCl and protease inhibitors (Roche, Basel, Switzerland) and 1 mM phenylmethyl sulfonylfluoride for 30 min on ice. The supernatant was subject to protein denaturation with buffer containing 2% sodium dodecylsulfate (SDS; Sigma), was subsequently analyzed using SDS-PAGE and monoclonal antibody against KLF4, PCNA and MMP2. The resultant gel was then washed and covered by appropriate horseradish peroxidase-conjugated Ig secondary antibodies. Enhanced chemiluminescence (GE Healthcare, Boston, MA, USA) was used to detect the staining.
Sphere formation assay
Cells were separated by trypsin and washed with phosphate-buffered saline (PBS). A total of 2 × 103 cells were seeded in the wells of six-well ultralow attachment plates (Corning Inc., New York, NY) in Dulbecco’s Modified Eagle’s Medium/F-12 culture medium supplemented with 10 ng/ml basic fibroblast growth factor and EGF (Invitrogen). After 10 days of culturing, mammospheres in the plate were counted under the microscope.
Luciferase assay
Plasmid carrying KLF4 3′UTR and its mutated counterpart sequence plasmids were constructed. Cells were seeded in a 24-well plate at cell density of 1 × 105 cells/well. After 24 h of culture, cells were co-transfected with firefly luciferase reporter plasmids, respectively, containing wild-type or mutant KLF4 3′UTR pRL-TK vector expressing Renilla luciferase, and miR-124 or miRNA negative control via Lipofectamine 2000 (Invitrogen). Firefly and Renilla luciferase activities were measured after 36 h by Dual Luciferase Reporter Assay (Promega, Madison, WI, USA). Each transfection was performed twice in triplicate.
Statistical analysis
The Statistical Package for the Social Sciences (SPSS) 13.0 for Windows (SPSS, Inc., Chicago, IL) was used for assessment. Each biological replicate has three technical replicates to calculate the average value of each parameter. Data are reported as mean ± SD. Kaplan–Meier analysis was applied to analyze survival data. Student’s t-test was used to test the significance of differences, and p < .05 was considered significant. Independent prognostic factors were estimated by the Cox proportional hazards stepwise regression model.
Results
miR-124 is downregulated in HCC tissues compared with peritumour tissues
To evaluate whether miR-124 is aberrantly expressed in HCC tumour tissues, we conducted RT-qPCR to measure the expression of miR-124 in HCC tumour tissue and matched peritumour tissue obtained from 112 patients. The miR-124 expression in tumour tissue is significantly lower than matched control (1.23 ± 0.24 vs. 1.89 ± 0.35, p < .01, ), indicating that miR-124 may be a tumour suppressor in HCC.
Figure 1. (A) Relative expression of miR-124 in tumour tissues and normal adjacent tissues of 112 HCC patients. (B) Overall survival rate of high miR-124 expression patients and low miR-124 expression patients. Kaplan–Meier analysis was used for the statistical assessment.
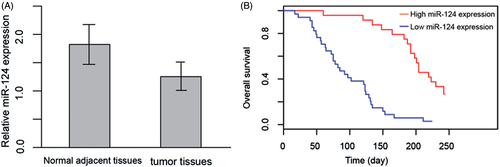
To further investigate the potential role of a biomarker for HCC, we performed Kaplan–Meier analysis to test the relationship of miR-124 expression with survival rate, as well as logistic analysis to compute the odds ratios of miR-124 in varying stages of HCC. For survival analysis, we divide the cohort into two groups, the high miR-124 expression (>1.5) and low miR-124 expression (≤1.5), which, respectively, contained 31 and 81 patients (). The high miR-124 expression group has significantly higher survival rate than its counterpart. Univariate Cox regression analysis showed that the odd ratio of miR-124 (high:low ratio) (OR: 1.92, CI: 1.67–2.43, p < .001), tumour size (OR: 1.52, CI: 1.23–1.87, p < .001) and TNM stage (OR: 2.0, CI: 1.54–2.68, p < .001) were significant prognostic factors (). Multivariate logistic analysis revealed that miR-124 expression (high:low ratio) (OR: 2.21, CI: 1.76–2.68, p = .002) were independent prognostic factors ().
Table 2. The relationship between clinicopathological characteristics and overall survival of patients.
miR-124 expression inhibit invasion and tumourigenesis of HCC
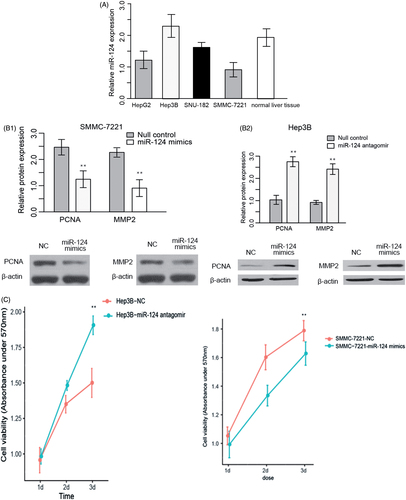
Given that we observed downregulation of miR-124 is associated with poorer survival and demonstrated that the ratio of miR-124 expression in normal tissue versus tumour tissue was an independent prognostic factor, we next sought to examine the role of tumour suppressor of miR-124 in HCC cell lines. HepG2, Hep3B, SNU-182 and SMMC-7221 were chosen for testing this hypothesis. miR-124 expression varied between cell lines. To account for differences between cell lines, we chose cell lines that express the most and least miR-124 for further experiments. Result showed that Hep3B had significantly higher miR-124 levels and SMMC-7221 displayed much lower miR-124 expression ().
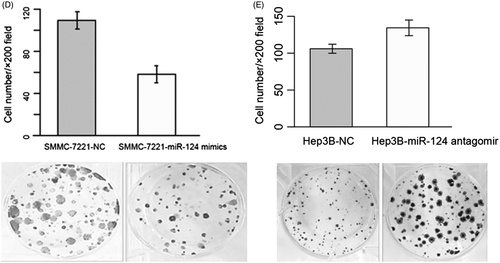
Figure 2. (A) Relative miR-124 expression in four HCC cell lines. (B) Western blot and quantization of PCNA and MMP2 in SMMC-7221 and Hep3B, respectively, transfected with miR-124 mimics and miR-124 antagomir. (C) Cell growth of Hep3B and SMMC-7221, respectively, transfected with miR-124 antagomir and miR-124 mimics. Cells transfected with scramble sequencing were used as control. Absorbance under 570 nm was taken as a measurement of living cell numbers. **: strong statistical significance. (D) Matrigel invasion assay of SMMC-7221 transfected with miR-124 mimics. (E) Matrigel invasion assay of Hep3B transfected with miR-124 antagomir.
SMMC-7221 overexpressing miR-124 mimics and Hep3B overexpressing miR-124 antagomirs were established, and PCNA and MMP2 were measured to evaluate the growth and invasive ability. Overexpression of miR-124 mimics in SMMC-7221 decreased the PCNA and MMP2 levels, while overexpression of miR-124 antagomirs in Hep3B was found to increase PCNA and MMP2 (). MTT assay demonstrated that the growth rate of miR-124 overexpressing SMMC-7221 was inhibited, whereas miR-124 silenced Hep3B displayed elevated growth (). In addition, matrigel invasion assay was performed to evaluate the metastatic status of HCC cell lines. Similar to MTT assay, the number of cell penetrating through the membrane after three days of transfection was higher in miR-124 silenced Hep3B and reduced in miR-124 overexpressing SMMC-7221 (). Taken together, these results indicated that miR-124 could inhibit cell proliferation and invasion of HCC.
KLF4 is a functional targets of miR-124 in HCC
miRNAs function through suppressing or degradating mRNA of target genes. To acquire a more in-depth understanding of mechanisms underlying the anti-proliferative function of miR-124, we employed TargetScan 7.0 to predict the target genes of miR-124. We obtained a list of candidate targets of miR-124 (Supplementary Table 1), and among them KLF4, RAB11FIP4 and RAF1 had been implicated in pathology of cancer. We next performed RT-qPCR and western blot to measure the mRNA and protein level of KLF4, RAB11FIP4 and RAF1 in miR-124 silenced Hep3B. Results showed that both mRNA and protein level of KLF4 were reduced compared with control, and RAB11FIP4 and RAF1 were not perturbed, indicating that KLF4 is a potential target of miR-124 ().
Figure 3. (A) Western blot and quantization of KLF4, RAB11FIP4 and RAF1 in SMMC-7221 transfected with miR-124 mimics. (B) Luciferase assay was used to confirm the targetability of KLF4 3′ UTR and miR-124. Wild type represent bona fide sequence of KLF4 3′ UTR; Mutant represents a sequence mimicking the KLF4 3′ UTR, but introduced with four single nucleotide variant.
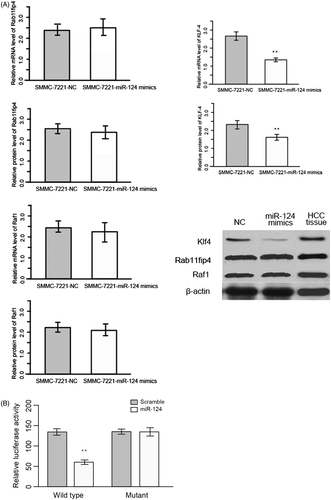
To validate those candidate targets, we perform luciferase assay. Fragments of 3′-UTR of KLF4, RAB11FIP4 and RAF1 which contain binding site of miR-124 were cloned into the pMIR-Report luciferase vector with firefly luciferase reporter gene. Luciferase activity of KLF4 was lower than control (). These data suggested that KLF4 is a target of miR-124.
The promotion of KLF4 on stemness of HCC can be reversed by miR-124
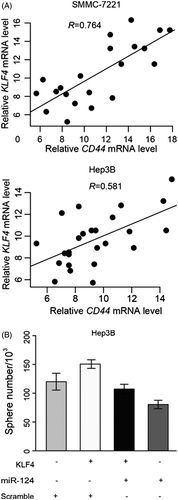
Cell stemness was considered as an outstanding characteristic of malignancy, which enhances the oncogenic property of tumour. Since the KLF4 was identified as target of miR-124, the relationship of KLF4 and miR-124 with cell stemness of HCC was not explored. To this end, we first determined the mRNA level of CD44 and CD133 to evaluate the impact of miR-124 on stemness of HCC. We detected a negative correlation between miR-124 and mRNA of CD44 and CD133 Hep3B and SMMC-7221 cells (). Further, sphere formation assay was conducted. We observed that the overexpression of KLF4 led to increase of the sphere formation ability and CD44/133-positive cell numbers () in Hep3B cell lines. These data implied that miR-124 antagonizes the stemness enhanced by KLF4.
Discussion
miRNAs are critical regulators in various biological activities, such as proliferation, differentiation, by mediating degradation or inhibiting translation at 3′ UTR. miRNAs are ubiquitous that more than 60% of human genes are regulated by miRNAs and they are involved in multiple cellular pathways implicated in tumourigenesis and cancer development [Citation28]. Those pathways affect many aspects of tumour biology including stemness and epithelial-to-mesenchymal transition (EMT). Multiple lines of evidence have implicated abnormal expression of miRNAs in a wide variety of cancers, including HCC [Citation29]. Over 300 miRNAs are expressed in healthy liver tissues, and dysregulation of them are reported as relevant to different aspects of hepatocarcinogenesis and HCC cell biology [Citation30,Citation31]. For example, hepatitis B and hepatitis C virus (HBV and HCV) could alter the miRNA profile and exploit host miRNAs to promote viral replication and tumour-associated activities [Citation32,Citation33]. Suppression of miR-122 regulates hepatocyte differentiation, thereby enhancing the metastatic potential of HCC cells [Citation34]. miR-21, a regulator renowned for its implications in cancer, targets PTEN and repress its activity, leading to elevated cell growth, invasion and metastasis [Citation35].
In the present study, we detected downregulation of miR-124 in HCC tissues, and Kaplan–Meier analysis showed that high miR-124 expression are associated with longer survival time, and Univariate cox regression exhibited significant association between miR-124 and N:T ratio, indicating that miR-124 is a promising prognostic factor, which was supported in multivariate logistic analysis. Ex vivo experiments performed in two HCC cell lines with different level of base line expression of miR-124 revealed that overexpression of miR-124 inhibits the proliferation and invasion of SMMC-7221 and Hep3B, and PCNA and MMP2 which signify the activity of proliferation and invasion were also increased. Together, the antiproliferative function of miR-124 was characterized.
Target gene prediction of miR-124 by TargetScan resulted in over one thousand genes, from which we selected top ten predictions ranked by “Cumulative weighted context++ score” in ascending order. This model has improved predictive accuracy than other published models and was comparable with in vivo crosslinking approaches [Citation36]. Among the candidates, the targetability of KLF4 was verified by luciferase experiment, which has been implicated as a tumour suppressor in a variety of cancers. Subsequently, we observed miR-124 overexpression could reduce the stemness at the presence of KLF4. These results amount to verify that KLF4 is a functional target of miR-124.
KLF4 was initially characterized as an important regulator of cell proliferation, which suppresses the tumour growth in a wide range of carcinomas, including medulloblastoma, colorectal, bladder and cervical cancers [Citation37–40]. However, its dual role as a tumour suppressor and oncogene are both highlighted. KLF4 induces CDK inhibitors CDKN1A and CDKN1B, both of which are crucial switch molecules in cell cycles, and decreases cyclin D1 and FOXM1 expression, thereby inhibiting cell proliferation in pancreatic cancers [Citation40–42]. Canonical WNT signalling, commonly recognized as a mediator of cancer cell proliferation, was found to be a target of KLF4. KLF4 interacts with TCF4 and β-catenin to prohibit the binding of β-catenin and TCF4, and disable the trigger of WNT pathway [Citation43,Citation44]. In gastric cancer cell lines, KLF4 suppresses CTNNB1 to reduce proliferative potent. EMT underlying invasive properties of tumours is an important stage for metastasis [Citation45]. In murine models, KLF4 exerts significant antagonizing effects on lung and liver metastasis. Noteworthily, Liu et al. showed that KLF4 expression is significantly correlated with downregulation of SLUG, and deficiency in KLF4 is sufficient to trigger SLUG-dependent EMT [Citation46]. In human HCC tumours, aberrantly high KLF4 expression levels were associated with aggressive tumour behaviours, particularly in vascular invasion, tumour differentiation and survival rates [Citation47]. Consistent with the HCC research, the present study demonstrated that ectopic expression of KLF4 promoted the sphere formation ability in Hep3B cells, and overexpression of miR-124 alleviated this effect.
In conclusion, we first detected aberrant expression level of miR-124 in HCC tissues of 112 patients. By exploring the clinical parameters, we found a significantly inverse correlation between miR-124 level and TNM stages. Consistent with this, the survival analysis indicated the association of low miR-124 with longer survival time. Forced expression miR-124 resulted in reduced cell viability of Hep3B and SMMC-7221, which cell lines have high and low background expression of miR-124, respectively. TargetScan prediction rendered a subset of target candidates, which were selected for experimental validation, KLF4 was subject to luciferase assay. Ectopic expression of KLF4 increased the sphere formation ability and CD44/133-positive cell numbers, which can be reversed by abundant expression of miR-124, suggesting that KLF4 is a functional target of miR-124 in tumourigenesis and cancer progression of HCC.
Supplementary_table_1.xlsx
Download MS Excel (121.3 KB)Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Jemal A, Bray F, Ferlay J. Global cancer statistics: 2011 [Internet]. CA Cancer J Clin. 1999;49:33–64.
- Singal AG, Pillai A, Tiro J. Early detection, curative treatment, and survival rates for hepatocellular carcinoma surveillance in patients with cirrhosis: a meta-analysis. PLoS Med. 2014;11:e1001624.
- Elserag HB. Epidemiology of viral hepatitis and hepatocellular carcinoma. Gastroenterology. 2012;142:1264.
- Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557–2576.
- Bruix J, Reig M, Sherman M. Evidence-based diagnosis, staging, and treatment of patients with hepatocellular carcinoma [Internet]. Gastroenterology. 2016;150:836–853.
- Simard EP, Ward EM, Siegel R, et al. Cancers with increasing incidence trends in the United States: 1999 through 2008 [Internet]. CA Cancer J Clin. 2012;62:118–128.
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function [Internet]. Cell. 2014;116:281–297.
- Zamore PD, Haley B. Ribo-gnome: the big world of small RNAs. Science. 2005;309:1519–1524.
- Lee EJ, Gusev Y, Jiang J, et al. Expression profiling identifies microRNA signature in pancreatic cancer. Int J Cancer. 2006;120:1046–1054.
- Meng F, Henson R, Wehbejanek H, et al. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133:647–658.
- Takamizawa J, Konishi H, Yanagisawa K, et al. Reduced expression of the let-7 MicroRNAs in human lung cancers in association with shortened postoperative survival [Internet]. Cancer Res. 2004;64:3753–3756.
- Michael MZ, O’Connor SM, van Holst Pellekaan NG, et al. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol Cancer Res. 2003;1:882–891.
- Zhao X, Yang B, Ma L, et al. MicroRNA-1 effectively induces differentiation of myocardial cells from mouse bone marrow mesenchymal stem cells. Artif Cells Nanomed Biotechnol. 2016;44:1665–1670.
- Jinling W, Sijing S, Jie Z, et al. Prognostic value of circulating microRNA-21 for breast cancer: a systematic review and meta-analysis. Artif Cells Nanomed Biotechnol. 2017;45:1216–1221.
- Cong J, Liu R, Wang X, et al. MiR-634 decreases cell proliferation and induces apoptosis by targeting mTOR signaling pathway in cervical cancer cells. Artif Cells Nanomed Biotechnol. 2016;44:1694–1701.
- Wang M, Xie R, Si H, et al. Integrated bioinformatics analysis of miRNA expression in osteosarcoma. Artif Cells Nanomed Biotechnol. 2017;45:1.
- Gramantieri L, Ferracin M, Fornari F, et al. Cyclin G1 is a target of miR-122a, a MicroRNA frequently down-regulated in human hepatocellular carcinoma. Cancer Res. 2007;67:6092–6099.
- Wong QWL, Lung RWM, Lai PBS, et al. MicroRNA-223 is commonly repressed in hepatocellular carcinoma and potentiates expression of stathmin1. Gastroenterology. 2008;135:257–269.
- Varnholt H, Drebber U, Schulze F, et al. MicroRNA gene expression profile of hepatitis C virus-associated hepatocellular carcinoma. Hepatology. 2007;47:1223–1232.
- Huang X, Wang Q, Chen J, et al. Bead-based microarray analysis of microRNA expression in hepatocellular carcinoma: miR-338 is downregulated . Hepatol Res. 2009;39:786–794.
- Xu G, Zhang Y, Wei J, et al. MicroRNA-21 promotes hepatocellular carcinoma HepG2 cell proliferation through repression of mitogen-activated protein kinase-kinase 3. BMC Cancer. 2013;13:469.
- Wong QWL, Ching AKK, Chan AWH, et al. MiR-222 overexpression confers cell migratory advantages in hepatocellular carcinoma through enhancing AKT signaling. Clin Cancer Res. 2010;16:867–875.
- Zeng L, Hu Z, Li K, et al. miR-222 attenuates cisplatin-induced cell death by targeting the PPP2R2A/Akt/mTOR Axis in bladder cancer cells. J Cell Mol Med. 2016;20:559–567.
- Ding W, Tan H, Zhao C, et al. MiR-145 suppresses cell proliferation and motility by inhibiting ROCK1 in hepatocellular carcinoma. Tumour Biol. 2016;37:6255–6260.
- Sun J, Lu H, Wang X, et al. MicroRNAs in hepatocellular carcinoma: regulation, function, and clinical implications. Sci World J. 2013;2013:924206.
- Huang Y, Chen H, Chiang C, et al. Identification of a two-layer regulatory network of proliferation-related microRNAs in hepatoma cells. Nucleic Acids Res. 2012;40:10478–10493.
- Furuta M, Kozaki KI, Tanaka S, et al. miR-124 and miR-203 are epigenetically silenced tumor-suppressive microRNAs in hepatocellular carcinoma. Carcinogenesis. 2009;31:766–776.
- Garzon R, Calin GA, Croce CM. MicroRNAs in cancer. Annu Rev Med. 2001;60:167–179.
- Ziebarth JD, Bhattacharya A, Cui Y. Integrative analysis of somatic mutations altering MicroRNA targeting in cancer genomes. PLoS One. 2012;7:1–10.
- González-Vallinas M, Breuhahn K. MicroRNAs are key regulators of hepatocellular carcinoma (HCC) cell dissemination – what we learned from microRNA-494. HepatoBiliary Surg Nutr. 2016;5:372–376.
- Gamazon ER, Innocenti F, Wei R, et al. A genome-wide integrative study of microRNAs in human liver. BMC Genomics. 2013;14:395.
- Li H, Jiang J, Peng Z. MicroRNA-mediated interactions between host and hepatitis C virus. World J Gastroenterol. 2016;22:1487.
- Sun X, Zhang S, Ma X. Prognostic value of MicroRNA-125 in various human malignant neoplasms: a meta-analysis. Clin Lab. 2015;61:1667.
- Coulouarn C, Factor VM, Andersen JB, et al. Loss of miR-122 expression in liver cancer correlates with suppression of the hepatic phenotype and gain of metastatic properties. Oncogene. 2009;28:3526–3536.
- Wang L, Yue Y, Wang X, et al. Function and clinical potential of microRNAs in hepatocellular carcinoma (Review). Oncol Lett. 2015;10:3345–3353.
- Agarwal V, Bell GW, Nam J, et al. Predicting effective microRNA target sites in mammalian mRNAs. eLife. 2015;4:e05005.doi: https://doi.org/10.7554/eLife.05005
- Yang W, Zheng P. Krüppel-like factor 4 functions as a tumor suppressor in cervical carcinoma. Cancer. 2012;118:3691–3702.
- Ohnishi S, Ohnami S, Laub F, et al. Downregulation and growth inhibitory effect of epithelial-type Krüppel-like transcription factor KLF4, but not KLF5, in bladder cancer. Biochem Biophys Res Commun. 2003;308:251–256.
- Chen ZY, Shie J, Tseng C. Gut-enriched Kruppel-like factor represses ornithine decarboxylase gene expression and functions as checkpoint regulator in colonic cancer cells. J Biol Chem. 2002;277:46831–46839.
- Nakahara Y, Northcott PA, Li M, et al. Genetic and epigenetic inactivation of Kruppel-like factor 4 in medulloblastoma. Neoplasia. 2010;12:20–27.
- Zammarchi F, Morelli M, Menicagli M, et al. KLF4 is a novel candidate tumor suppressor gene in pancreatic ductal carcinoma. Am J Pathol. 2011;178:361–372.
- Kong X, Li L, Li Z, et al. Dysregulated expression of FOXM1 isoforms drives progression of pancreatic cancer. Cancer Res. 2013;73:3987–3996.
- Sellak H, Wu S, Lincoln TM. KLF4 and SOX9 transcription factors antagonize β-catenin and inhibit TCF-activity in cancer cells. Biochim Biophys Acta. 2012;1823:1666–1675.
- Evans PM, Chen X, Zhang W, et al. KLF4 interacts with beta-catenin/TCF4 and blocks p300/CBP recruitment by beta-catenin. Mol Cell Biol. 2010;30:372–381.
- Wei D, Gong W, Kanai M, et al. Drastic down-regulation of Krüppel-Like Factor 4 expression is critical in human gastric cancer development and progression. Cancer Res. 2005;65:2746–2754.
- Liu YN, Aboukheir W, Yin JJ, et al. Critical and reciprocal regulation of KLF4 and SLUG in transforming growth factor β-initiated prostate cancer epithelial-mesenchymal transition. Mol Cell Biol. 2012;32:941–953.
- Yin X, Li Y, Jin J, et al. The clinical and prognostic implications of pluripotent stem cell gene expression in hepatocellular carcinoma. Oncol Lett. 2013;5:1155–1162.