Abstract
Renal carcinoma (RCC) is widely accepted as a malignant tumour of urinary system. Long intergenic non-coding RNA 1939 (LINC01939) is a novel lncRNA which was found to be down-regulated in RCC. Thus, we set out to explore the effect and regulation mechanism of LINC01939 in RCC. LINC01939 and miR-154 in RCC tissues and cell lines were detected using qRT-PCR assay. To examine cellular viability of ACHN and CAKI-1 cells, cell counting kit-8 (CCK-8) assay was exploited here. Flow cytometric analysis was conducted to examine apoptosis. Cell mobility was valued through wound healing assays. Western blotting was applied for examination of proteins related to proliferation, apoptosis, migration and Wnt/β-catenin/Notch. LINC01939 was down-regulated in RCC tissues. LINC01939 overexpression impeded proliferation and migration, and induced apoptosis. Further study found that the overexpression of LINC01939 strongly suppressed miR-154 expression. Then, the inhibiting effect of overexpressed LINC01939 on proliferation and mobility and the promoting role of LINC01939 in apoptosis were abolished by the combination of miR-154 mimic. Finally, we found that overexpressed LINC01939 inactivated Wnt/β-catenin and Notch through suppressing miR-154. Up-regulation of LINC01939 inhibited proliferation and migration of RCC cells by down-regulating miR-154.
Introduction
Renal cell carcinoma (RCC) is a common chemotherapeutic drug-resistant disease. The global annual incidence of RCC is about 20,000 cases, with 102,000 deaths [Citation1]. Among them, 80–90% patients die of clear cell renal cell carcinoma (ccRCC) which has distinctive features such as local invasion, distant metastasis and high lethal rate [Citation2]. Thus, the prognosis of patients with ccRCC is worse and the incidence of ccRCC is gradually increasing with a mortality rate about 40% [Citation3]. At present, the main treatment of RCC is surgical treatment. However, even after early radical surgery, there are still many tumours recurrence and metastasis cases. Thus, it is of great significance to investigate the molecular mechanisms of RCC with multi-gene and multi-level diagnostic and therapeutic tools, aiming to get in-depth understanding of RCC.
Long non-coding RNA (lncRNA), defined as RNAs with no protein coding potential, is longer than 200 nt. LncRNA was initially thought to be merely transcriptional noise, which has no biological function [Citation4]. But due to the growing depth of sequencing technology, more and more new lncRNAs have been found to be involved in various physiological and pathological processes [Citation5]. Studies have shown that a number of lncRNAs are abnormally expressed in many tumours and may be involved in multiple processes of tumorigenesis and development, including cell differentiation and proliferation, cell cycle regulation, invasion and metastasis [Citation6–8]. Thus, lncRNAs have been regarded as new biomarkers in various cancers including RCC. Recently, Chen et al. detected a decrease of long intergenic non-coding RNA 1939 (LINC01939) in gastric cancer tissues, which has been associated with clinical stage of gastric cancer patients [Citation9]. However, the biological function in RCC has never been explored as well as the underlying mechanisms.
microRNAs (miRNAs) are recognised as the small non-coding RNAs essentially with a length of 18–25 nt, which are widely distributed in eukaryotic organisms. They are mainly responsible for regulating gene and protein expression and participating in many physiological and pathological processes, for instance, cell growth, differentiation and apoptosis [Citation10]. These single-stranded mature miRNAs mediate protein coding by binding to 3′-untranslated region (3′-UTR) of mRNA which shares the homologous sequence [Citation11]. miRNAs are very important regulator in urinary tumours and the expression and function of miRNAs can be observed in most urinary tumours [Citation12,Citation13]. Recently, an increasing number of studies reported that miR-154 modulated by lncRNAs is included in human cancer diseases [Citation14,Citation15]. However, the regulating effects between miR-154 and lncRNAs have never been explored in RCC before.
In our present study, we noticed the down-regulation of LINC01939 in RCC tissues. Further study revealed that overexpressed LINC01939 weakened proliferative activities and inhibited motile phenotypes, as well as induced apoptotic process by down-regulating miR-154 with bluntness of Wnt/β-catenin and Notch, providing novel diagnostic and therapeutic targets for RCC.
Materials and methods
Clinical specimens
Kidney cancer tissue specimens were acquired from patients (n = 18) who had tumour resection at the First Affiliated Hospital of Soochow University. Para-carcinoma tissues were obtained from the resected tissues. Prior written and informed consents were procured from all volunteer patients. Our study had the approval from the Medical Ethics Committee of the First Affiliated Hospital of Soochow University, and the process was performed in compliance with the guidelines.
Cell culture
Human renal cell adenocarcinoma cell line ACHN was obtained from Cell Lines Service (Item No. 300117; Eppelheim, Germany). ACHN cells were grown in Eagle’s essential medium (Sigma-Aldrich, St Louis, MO, USA) with addition of 10% foetal bovine serum (FBS) (Sigma-Aldrich). Human kidney carcinoma CAKI-1 cell line was provided by DSMZ (Item No. ACC731; Braunschweig, Germany). CAKI-1 cells were maintained in McCoy’s 5a medium (Gibco, Gaithersburg, MD, USA) in supplementation with 20% FBS. Both ACHN and CAKI-1 cells were grown in an atmosphere consisting of 95% air and 5% CO2 at 37 °C.
Cell transfection
The full-length of LINC01939 sequences were constructed in pcDNA3.1 plasmid (Invitrogen, Carlsbad, CA, USA) to construct the recombinant plasmid pcDNA3.1-LINC01939. Empty vector-transfected ACHN and CAKI-1 cells served as the control cells. Following the manufacturer’s instructions, pcDNA3.1-LINC01939 or empty plasmid was introduced into ACHN and CAKI-1 cells in the presence of lipofectamine 3000 reagent (Life Technologies, Carlsbad, CA, USA). Subsequently, 0.5 mg/mL G418 (Sigma-Aldrich) conditioned medium was used to select the transfectants. Synthesised by Life Technologies, miR-154 mimic and the negative control mimic were transfected into ACHN and CAKI-1 cells. LINC01939 and miR-154 were quantified by qRT-PCR.
Cell viability
To detect the viable cells, 5,000 of ACHN or CAKI-1 cells were inoculated into each well of 96-well plates, respectively and pre-incubated for 24 h. Next, the cells in each well were incubated with 10 μL of CCK-8 solution (APExBIO, Houston, TX, USA). After co-incubation for 1 h, the absorbance was recorded at 450 nm using Elx800 Reader (Bio-Tek, Winooski, VT, USA).
Apoptosis assay
The apoptotic cells were observed after stained in Annexin V-FITC/propidium iodide (PI) (Biosea, Beijing, China) under Flow Cytometer (BD Biosciences, San Jose, CA, USA). In short, ACHN and CAKI-1 cells were collected and exposed to trypsin (Pierce, Appleton, WI, USA) for digestion. After washed in pre-cold phosphate buffered saline (Sigma-Aldrich), the cells were collected by centrifugation. Then 5 μl of PI was added and co-incubated with the culture for 30 min. The apoptotic cells were analysed at 488 nm.
Western blot
The CAKI-1 and ACHN cells were lysed using RIPA (Beyotime, Shanghai, China) to extract the whole proteins. The extracted protein content was quantified using the BCATM Protein Assay Kit (Pierce). Then the protein extract was loaded onto sodium dodecyl sulphate polyacrylamide gel electrophoresis system for separation. Next, the separated proteins were transferred onto polyvinylidene difluoride (PVDF) membrane (Millipore, Bedford, MA, USA). The membranes carrying the proteins were sealed with 5% non-fat milk overnight at 4 °C. Next, the interest proteins were detected by incubating with primary antibodies overnight at 4 °C. Continually, the primary antibodies were hybridised by incubating with secondary antibody at room temperature for 1 h. Finally, protein signalling was captured with an ECL system (Amersham Pharmacia, Piscataway, NJ, USA).
Migration assay
To value the migratory activity of ACHN and CAKI-1 cells, a modified two-chamber carrying an 8 mm-pore was applied. Firstly, the cells suspended in the medium without serum were plated into the top chamber, and the complete medium (600 μL), FBS (with a final concentration of 10%) and epidermal growth factor (25 ng/mL) (Sigma-Aldrich) were supplemented into the lower compartment. Continually, the cells were incubated for 24 h, and then non-traversed cells on the upper surface of the filter were removed. Crystal violet was used to stain the traversed cells on the lower surface. Lastly, the cells were counted under a microscope (Olympus, Tokyo, Japan).
qRT-PCR
Total RNA in RCC tissues and cell was isolated by using Trizol reagent (Invitrogen). MultiScribe RT Kit (Applied Biosystems, Foster City, CA, USA) was used to perform reverse transcription reaction with random hexamers or oligo(dT) as primers. SYBR Green I Master Mix (Thermo Fisher Scientific, Waltham, MA, USA) was exploited for PCR on Applied Biosystems 7300 Sequence Detection system (Applied Biosystems). Relative expression levels of LINC01939 or miR-154 were normalised against those of GAPDH or U6 snRNA and calculated by 2-ΔΔCt method.
Statistical analysis
Obtained data was analysed using IBM SPSS 19.0 statistical software (IBM, Chicago, IL, USA), and the data were computed as the mean ± stand deviations (SD). Either one-way analysis of variance or student’s t test was applied for comparing the difference between two groups, and p-values less than .05 was accepted and suggested the statistical difference.
Results
LINC01939 was down-regulated in renal carcinoma tissues
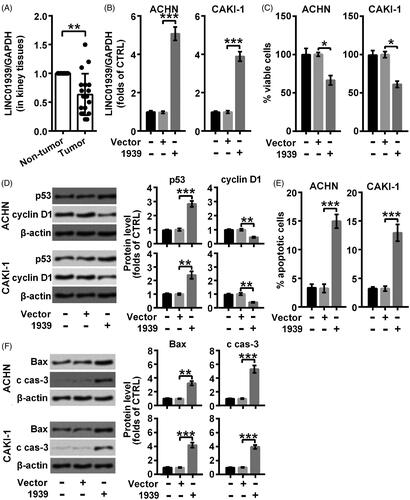
LINC01939 was examined in renal carcinoma tissues and the non-tumour tissues through qRT-PCR. As suggested in , the level of LINC01939 was visibly suppressed in renal carcinoma tissues compared with the para-carcinoma tissues (**p < .01). Consequently, we considered a possibility that the deficiency of LINC01939 might be associated with the process of renal carcinoma.
Figure 1. LINC01939 was lowly expressed in renal carcinoma tissues and LINC01939 overexpression exhibited a pro-apoptosis role in ACHN and CAKI-1 cells. (A). The expression of LINC01939 in renal carcinoma tissues and para-carcinoma tissues was examined through qRT-PCR. ACHN and CAKI-1 cells were transfected with the empty vector pcDNA3.1 (Vector) or pcDNA3.1-LINC01939 (1939) as indicated. (B). LINC01939 was analysed through qRT-PCR. (C). Viability of cells was valued through CCK8 assay. (D). Western blotting assay of proliferation related proteins p53 and cyclin D1 was carried out. (E). Apoptotic cells was examined through flow cytometry. (F). Western blots of apoptosis related proteins Bax and cleaved caspase-3 (c cas-3). Quantifications were expressed as means of three independent experiments ± SD. *p < .05, **p < .01, ***p < .001.
Overexpression of LINC01939 facilitated apoptosis process in renal carcinoma cell lines
Specific pc-LINC01939 was transduced into ACHN and CAKI-1 cells respectively to up-regulate the level of LINC01939 as shown in and the empty vector was used as a control (***p < .001). Then we found that cell viability was obviously suppressed in LINC01939-transfected cells (*p < .05) (). Further, results from Western blotting assay showed that p53 (**p < .01 or ***p < .001) was elevated and cyclin D1 (**p < .01) was suppressed in both CAKI-1 and ACHN cells transfected with LINC01939, suggesting that cell proliferation was suppressed by overexpression of LINC01939 (). In addition, we found that LINC01939 overexpression obviously induced apoptosis progress in both ACHN and CAKI-1 cells (***p < .001) (). At the same time, the expression of Bax (**p < .01 or ***p < .001) was also elevated by LINC01939, as well as the cleavage of caspase-3 (***p < .001) (). Thus, we concluded that overexpression of LINC01939 promoted the apoptosis of renal carcinoma cells.
Migration activity was weakened by LINC01939 overexpression in ACHN and CAKI-1 cells
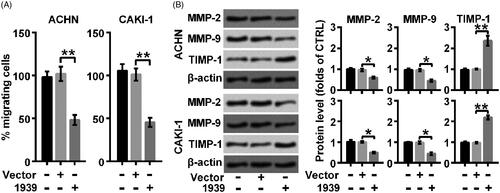
The effect of overexpression of LINC01939 on the migration of renal carcinoma cell lines was further examined through migration assay and Western blot, respectively. The statistical results indicated that the migration behaviour was largely suppressed by the overexpression of LINC01939 (**p < .01) (). Besides, LINC01939 overexpression caused the abundance of MMP-2 (*p < .05) and MMP-9 (*p < .05), while the production of TIMP-1 (**p < .01) was elevated by overexpressed LINC01939 (). Thus, we concluded that overexpression of LINC01939 suppressed the migration of renal carcinoma cell lines.
Figure 2. LINC01939 overexpression suppressed migration activity of ACHN and CAKI-1 cells. ACHN and CAKI-1 cells were transfected with the empty vector pcDNA3.1 (Vector) or pcDNA3.1-LINC01939 (1939) as indicated. (A). The migration behaviour was evaluated by wound healing assay. (B). Western blotting assay of migration related proteins matrix metallopeptidase (MMP)-2, MMP-9, and TIMP metallopeptidase inhibitor 1 (TIMP-1) was performed. Quantifications were expressed as means of three independent experiments ± SD. *p < .05 and **p < .01.
LINC01939 overexpression-caused down-regulation of miR-154 functioned as a mediator to induce apoptosis
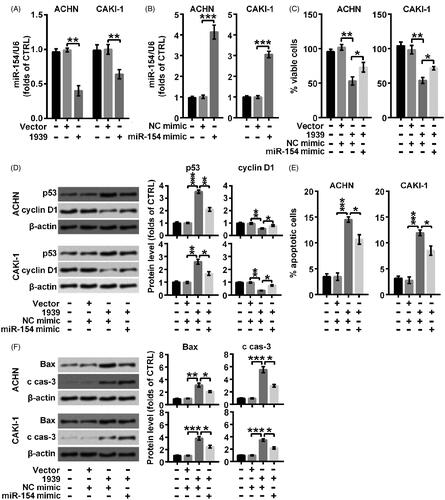
Results from indicated that miR-154 was obviously suppressed (**p < .01) by overexpressed LINC01939, suggesting that there might be a targeting relationship between LINC01939 and miR-154. In order to explore the connection between LINC01939 and miR-154, specific miR-154 mimic was used in our following experiments to elevate the accumulation of miR-154. As shown in , miR-154 mimic apparently elevated the level of miR-154 in ACHN and CAKI-1 cells (***p < .001). Then we observed that the viability inhibited by overexpressed LINC01939 was elevated by the combination of miR-154 mimic (*p < .05) (). Besides, the inhibiting effect of LINC01939 on cell proliferation was also abolished by the adding of miR-154 mimic through suppressing the level of p53 (*p < .05 or **p < .01) and elevating the level of cyclin D1 (*p < .05) (). In addition, elevated cell apoptosis rate by LINC01939 was then suppressed by overexpressed miR-154 (*p < .05) (). At the same time, elevated expression of Bax and cleaved caspase-3 induced by LINC01939 was both suppressed by the combination of miR-154 mimic in CAKI-1 (*p < .05) and ACHN (*p < .05) cells (). Thus, we concluded that overexpression of LINC01939 accelerated the apoptosis process by repressing the abundance of miR-154.
Figure 3. LINC01939 promoted apoptosis process via controlling the abundance of miR-154. (A) ACHN and CAKI-1 cells were transfected with the empty vector pcDNA3.1 (Vector) or pcDNA3.1-LINC01939 (1939) as indicated. miR-154 was examined through qRT-PCR. (B) ACHN and CAKI-1 cells were transfected with NC (negative control) mimic or miR-154 mimic as indicated. The expression of miR-154 was examined through qRT-PCR. ACHN and CAKI-1 cells were transfected with pcDNA3.1-LINC01939 and miR-154 mimic. (C) Viability was valued through CCK8 assay. (D) Western blotting assay of proliferation related proteins p53 and cyclin D1 was carried out. (E) Apoptotic cells were examined through flow cytometry. (F) Western blotting assay of apoptosis related proteins Bax and cleaved caspase-3 (c cas-3). Quantifications were expressed as means of three independent experiments ± SD. *p < .05, **p < .01, ***p < .001.
LINC01939 overexpression impeded migration behaviour by repressing the enrichment of miR-154
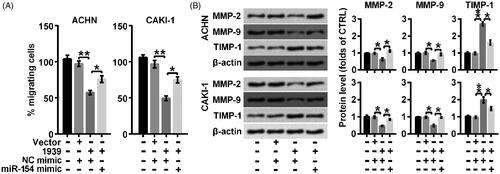
This synergetic modulation was also verified in cell migration. As shown in , overexpressed LINC01939 suppressed cell migration rate but the inhibiting effect was largely weakened by the combination of miR-154 mimic (*p < .05). At the same time, suppressed level of MMP-2 and MMP-9 was elevated and elevated expression of TIMP-1 was suppressed by the adding of miR-154 mimic compared with the effect of LINC01939 separately (both *p < .05) (). Thus, we concluded that overexpression of LINC01939 suppressed cell migration by down-regulating miR-154.
Figure 4. LINC01939 overexpression suppressed migration via up-regulating miR-154. ACHN and CAKI-1 cells were transfected with NC (negative control) mimic or miR-154 mimic as indicated. (A) The migration behaviour was evaluated by wound healing assay. (B) Western blotting assay of migration related proteins matrix metallopeptidase (MMP)-2, MMP-9, and TIMP metallopeptidase inhibitor 1 (TIMP-1) was performed. Quantifications were expressed as means of three independent experiments ± SD. *p < .05, **p < .01.
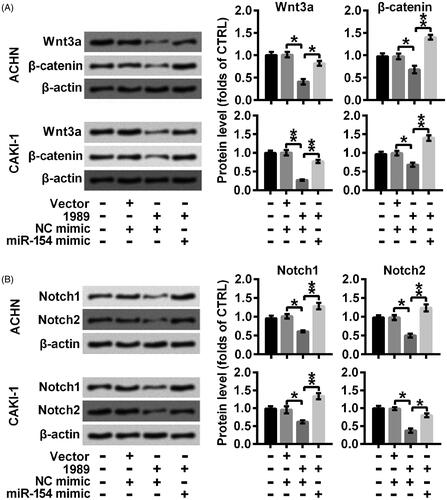
Overexpression of LINC01939 retarded the activation of Wnt/β-catenin and Notch by decreasing miR-154
Related signal pathways were further investigated through Western blotting assay. As shown in ), transducers of Wnt/β-catenin (Wnt3a and β-catenin) were downregulated in the cells overexpressing LINC01939, and were then elevated by the combination of miR-154 mimic (*p < .05 or **p < .01), indicating that overexpressed LINC01939 inactivated Wnt/β-catenin via down-regulating miR-154. Similarly, the inhibiting effect of overexpressed LINC01939 on the Notch signal pathway was abolished by miR-154 mimic through elevating the expression of Notch1 and Notch2 (*p < .05 or **p < .01). Thus, we concluded that overexpression of LINC01939 inactivated Wnt/β-catenin and Notch via repressing miR-154.
Figure 5. LINC01939 overexpression-caused blockade of Wnt3a/β-catenin and Notch might be mediated by the down-regulation of miR-154. ACHN and CAKI-1 cells were transfected with NC (negative control) mimic or miR-154 mimic as indicated. Western blotting assay of signalling mediators of (A) Wnt3a/β-catenin (Wnt3a and β-catenin) and (B) Notch (Notch1 and Notch2) pathways was performed. Quantifications were expressed as means of three independent experiments ± SD. *p < .05, **p < .01.
Discussion
RCC originates from the renal parenchymal urinary tubular epithelial system, and it is widely accepted as a common malignant tumour in urinary system, accounting for nearly 2%-5% of adult malignant tumours worldwide [Citation16]. With the advancement of imaging technology, many small RCC less than 3 cm were found, but many patients had already been in the advanced stage of the disease when they were found. Therefore, it is urgent to improve the diagnosis of RCC and to find new therapeutic targets in RCC treatment.
Recently, lncRNAs have become a research hotspot, and studies verified that lncRNAs play a more and more important role in tumorigenesis and development. Studies have revealed that lncRNAs play an essential role in differentiation, proliferation processes, and metastasis processes, and its abnormal expression causes the functional changes which might be a causal association with malignant tumours and other diseases [Citation17]. Also, up to now, numerous lncRNAs have been found to participate in the tumorigenic and tumour-suppressive regulatory network in RCC. As an example, in metastatic ccRCC tissues, lncRNA termed metastatic renal cell carcinoma-associated transcript 1 (MRCCAT1) is ectopic up-regulated, which might cause the metastatic capacity of ccRCC via mediating associated signalling transduction systems [Citation18]. Besides, lncRNA UCA1 is overexpressed in ccRCC and the overexpression of UCA1 has been regarded as a poor prognostic biomarker in ccRCC diagnosis [Citation19]. In our present study, we found LINC01939 was obviously down-regulated in RCC tissues. Further study found that overexpressed LINC01939 through exogenous transfection effectively repressed proliferation, accelerated apoptosis and blocked migration process of RCC cell lines. Consequently, we concluded that LINC01939 could be regarded as a novel diagnostic and therapeutic interest lncRNA for RCC.
miRNAs not only regulate the development and differentiation of cells, but also are potential molecular markers of tumours. They have good application prospects in early diagnosis, treatment, prognosis and chemotherapy resistance of tumours. MiRNAs act as a cancer-promoting or anti-cancer factor through epigenetic regulation and regulation of the expression of processing genes and proteins [Citation20,Citation21]. For example, Wang et al. proved miR-154 is down-regulated in glioblastoma while miR-154 overexpression suppresses the growth of glioblastoma cells, which could act as a beneficial therapeutic target [Citation22]. However, Lin et al. revealed that miR-154 is overexpressed in RCC specimens and cells, which exaggerates the growth of RCC cells [Citation13]. Therefore, the same miRNA may play opposite roles in different tumours through different regulatory mechanisms.
Many lncRNAs have structures similar to the mRNA targeted by miRNAs, so miRNAs can negatively regulate lncRNA through mechanisms similar to those acting on mRNA. The regulatory mechanisms between lncRNA and microRNAs include the following types: lncRNA competes with 3′-UTR of target RNA of microRNAs to indirectly inhibit the negative regulation of target genes by miRNAs [Citation23]; lncRNA act as competitive endogenous RNA and plays the role of molecular sponge of miRNAs to inhibit the expression of miRNAs [Citation24]. In addition, lncRNA can indirectly regulate the expression of miRNAs through other proteins to regulate the progression of tumours. In our study, we found that LINC01939 overexpression suppressed miR-154 expression. Further study showed that miR-154 mimic abolished inhibitory role of LINC01939 in proliferation and migration, and the promoting effect of LINC01939 on apoptosis. Thus, we concluded that overexpressed LINC01939 suppressed proliferation and migration and induced apoptosis dependent on the down-regulation of miR-154.
Wnt/β-catenin and Notch signal pathways play important roles in kinds of cancers. Jin et al. reported that miR-135b stimulated the recurrence and metastasis of osteosarcoma via activating Wnt/β-Catenin and Notch by multiple negative regulators [Citation25]. In our present study, both of Wnt/β-catenin and Notch signal pathways was inactivated by overexpressed LINC01939. However, the inhibiting role of LINC01939 overexpression on Wnt/β-catenin and Notch was abolished by the combination of miR-154 mimic. Thus, we conclude that overexpressed LINC01939 inactivated Wnt/β-catenin and Notch by suppressing miR-154.
Taken together, we found that LINC01939 was down-regulated in RCC tissues. Overexpression of LINC01939 suppressed proliferation and migration and induced apoptosis by down-regulating miR-154. Further study also revealed that Wnt/β-catenin and Notch were inactivated by overexpression of LINC01939 via suppressing miR-154. Our present study for the first time revealed the regulating effect and associated regulation mechanism of LINC01939 in RCC, providing novel diagnostic and therapeutic targets for RCC.
Ethics approval and consent to participate
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. The research was encouraged by the Medical Ethics Committee of the First Affiliated Hospital of Soochow University.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Additional information
Funding
References
- Siegel R, Ma J, Zou Z, et al. Cancer statistics, 2014. CA Cancer J Clin. 2014;64(1):9–29.
- Youssef YM, White NM, Grigull J, et al. Accurate molecular classification of kidney cancer subtypes using microRNA signature. Eur Urol. 2011;59(5):721–730.
- Gremel G, Djureinovic D, Niinivirta M, et al. A systematic search strategy identifies cubilin as independent prognostic marker for renal cell carcinoma. BMC Cancer. 2017;17(1):9.
- Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23(13):1494–1504.
- Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet. 2014;15(1):7–21.
- Hong Q, Li O, Zheng W, et al. LncRNA HOTAIR regulates HIF-1alpha/AXL signaling through inhibition of miR-217 in renal cell carcinoma. Cell Death Dis. 2017;8(5):e2772.
- Lu Y, Liu WG, Lu JH, et al. LncRNA UCA1 promotes renal cell carcinoma proliferation through epigenetically repressing p21 expression and negatively regulating miR-495. Tumour Biol. 2017;39(5):1010428317701632.
- Chen S, Ma P, Li B, et al. LncRNA CCAT1 inhibits cell apoptosis of renal cell carcinoma through up-regulation of Livin protein. Mol Cell Biochem. 2017;434(1–2):135–142.
- Chen CL, Ke Q, Luo M, et al. Loss of LINC01939 expression predicts progression and poor survival in gastric cancer. Pathology, Research and Practice. 2018;214(10):1539–1543.
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233.
- Leucci E, Patella F, Waage J, et al. microRNA-9 targets the long non-coding RNA MALAT1 for degradation in the nucleus. Sci Rep. 2013;3(1):2535.
- Szabo Z, Szegedi K, Gombos K, et al. Expression of miRNA-21 and miRNA-221 in clear cell renal cell carcinoma (ccRCC) and their possible role in the development of ccRCC. Urol Oncol. 2016;34(12):533.e21–533.e27.
- Lin C, Li Z, Chen P, et al. Oncogene miR-154-5p regulates cellular function and acts as a molecular marker with poor prognosis in renal cell carcinoma. Life Sciences. 2018;209:481–489.
- Chi JR, Yu ZH, Liu BW, et al. SNHG5 promotes breast cancer proliferation by sponging the miR-154-5p/PCNA Axis. Mol Ther Nucleic Acids. 2019;17:138–149.
- Xu M, Chen X, Lin K, et al. The long noncoding RNA SNHG1 regulates colorectal cancer cell growth through interactions with EZH2 and miR-154-5p. Mol Cancer. 2018;17(1):141.
- Cheng SK, Chuah KL. Metastatic renal cell carcinoma to the pancreas: a review. Arch Pathol Lab Med. 2016;140(6):598–602.
- Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Mol Cancer. 2011;10(1):38.
- Li JK, Chen C, Liu JY, et al. Long noncoding RNA MRCCAT1 promotes metastasis of clear cell renal cell carcinoma via inhibiting NPR3 and activating p38-MAPK signaling. Mol Cancer. 2017;16(1):111.
- Wang Y, Gao W, Xu J, et al. The long noncoding RNA urothelial carcinoma-associated 1 overexpression as a poor prognostic biomarker in clear cell renal cell carcinoma. Tumour Biol. 2017;39(5):1010428317698377.
- Erdmann K, Kaulke K, Thomae C, et al. Elevated expression of prostate cancer-associated genes is linked to down-regulation of microRNAs. BMC Cancer. 2014;14(1):82.
- Mulrane L, Klinger R, McGee SF, et al. microRNAs: a new class of breast cancer biomarkers. Expert Rev Mol Diagn. 2014;14(3):347–363.
- Wang X, Sun S, Tong X, et al. MiRNA-154-5p inhibits cell proliferation and metastasis by targeting PIWIL1 in glioblastoma. Brain Res. 2017;1676:69–76.
- Faghihi MA, Modarresi F, Khalil AM, et al. Expression of a noncoding RNA is elevated in Alzheimer’s disease and drives rapid feed-forward regulation of beta-secretase. Nat Med. 2008;14(7):723–730.
- Salmena L, Poliseno L, Tay Y, et al. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146(3):353–358.
- Jin H, Luo S, Wang Y, et al. miR-135b stimulates osteosarcoma recurrence and lung metastasis via notch and Wnt/beta-catenin signaling. Mol Ther Nucleic Acids. 2017;8:111–122.
