Abstract
MicroRNA-155 is over-expressed in many human cancers, but researches on its association with malignant oesophageal squamous cell carcinoma (ESCC) are limited. The aim of the present study was to evaluate the potential value of miR-155 as a biomarker for ESCC diagnosis and prognosis. In this study, we found that miR-155 was significantly increased in ESCC tissues compared with the paired adjacent tissues and healthy normal controls (p < .001), according to qRT-PCR, which suggested that miR-155 might act as an oncogene in ESCC. In addition, clinical features such as the depth of tumour invasion, tumour size, and TNM stage were all proved to impact the expression of miR-155 (p < .01). Then, ROC curve analysis, reaching an AUC of 0.870, and a sensitivity and specificity of 83.5% and 77.5%, respectively, revealed that miR-155 was a predictive factor for ESCC. As well, high expression of miR-155 was associated with poor overall survival of the patients (log-rank test, p = .004), according to Kaplan-Meier analysis. MiR-155 might be an independent predictor for overall survival in ESCC patients, manifested by Cox regression analysis (HR = 16.94, 95%CI = 3.33–86.12, p = .001). Taken together, miR-155 could be an independent diagnostic and prognostic biomarker for ESCC.
Introduction
Oesophageal cancer, an aggressive malignancy, is one of the leading causes of cancer-related mortality worldwide, showing a variable geographic distribution, while oesophageal squamous cell carcinoma (ESCC) is the most common histological type of this disease [Citation1,Citation2]. Despite remarkable advances in diagnostic and therapeutic techniques for the malignancy, like preoperative chemotherapeutic, radiotherapeutic and surgical treatment, the survival rate of ESCC patients has not been qualitatively improved [Citation3,Citation4], still witnessing poor prognoses. To improve the diagnosis and prognosis of ESCC patients, more predictive and useful tumour biomarkers need to be identified. Therefore, it is an important strategy to develop sensitive and specific screening technologies for patients with oesophageal cancer.
MiRNAs, about 18–25 nucleotides, are a large family of non-coding RNAs that are significant in regulating gene expressions at post-transcriptional level. They can play regulatory roles in cell development, metabolism, immunity, proliferation, differentiation, and apoptosis [Citation5,Citation6]. Moreover, miRNAs can improve mRNA degradation, prevent mRNA from being translated, and thus regulate gene expression by binding to complementary target mRNA [Citation7]. In addition, earlier findings demonstrated that miRNAs were involved in regulating tumorigenesis, tumour development and progression [Citation8], and clinical and quantitative studies confirmed that the expression levels of some miRNAs were associated with overall survival of patients with malignant tumours. Therefore, miRNAs may serve as potential diagnostic or prognostic biomarkers for cancers.
MicroRNA-155 (MiR-155) is a typical multifunctional miRNA involved in numerous biological processes including inflammation and immunity. Recent studies showed that miR-155 expression might be associated with the diagnosis or prognosis of colorectal cancer, gastric cancer, or non-small cell lung cancer [Citation9–11]. However, we still lack evidence for the prognostic role of miR-155 expression in ESCC. Thus, the aim of the study was to investigate clinical significance of miR-155 expression in ESCC, including its diagnostic and prognostic value in the malignancy.
Materials and methods
Patients and specimens
We collected tumour tissue samples from 96 ESCC patients who underwent surgery at the First Affiliated Hospital of Zhengzhou University, but received no preoperative treatments (chemotherapy or radiotherapy). In addition, paired adjacent normal tissues and 20 healthy specimens were used as controls. Clinicopathologic data of the patients were summarised in . Complete information from a follow-up for at least 5 years was available through telephone call and questionnaire letters to the patients or their relatives. The study protocol was approved by the Ethics Committee of the First Affiliated Hospital of Zhengzhou University. All patients agreed to the study procedure and signed consent forms. All specimens were anonymous and handled according to the ethical and legal standards.
Table 1. Relationship between miR-155 expression and clinicopathological characteristics.
Reverse transcription and qRT-PCR for miR-155
To detect miRNA level adopting real-time PCR, total RNA was purified from all ESCC samples and 20 normal control specimens using Trizol reagent (Invitrogen, Carlsbad, CA, USA). Total RNA samples only reaching an OD A260/A280 ratio close to 2.0 were adopted, which indicated that the RNA was pure. miR-155 and U6 specific cDNAs were synthesised with TaqMan Micro-RNA Reverse Transcription Kit, according to the manufacturer’s protocol. Then reverse transcription products were amplified and detected through quantitative real-time PCR (qRT-PCR) using TaqMan microRNA assay. MiR-155 expression levels were analysed as relative quantities, via normalised to endogenous control (U6). Primer sequences were as follows: miR-155 RT:5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTT-GAGACCCCTAT-3′; miR-155 F: 5′-ACACTCCAGCTGGGTTAATGCTAATCGTGAT-3′, R: 5′-TGGTGTCGTGGAGT CG-3′. U6: F: 5′-CTCGCTTCGGCAGCACA-3′, R: 5′-AACGCTTCACGAATTTGCGT-3′.
Statistical analysis
Statistical analyses were performed with SPSS 13.0 software (SPSS, Chicago, IL). Student’s t-test was used to analyse difference in miR-155 expression between ESCC tissues and the two groups of control specimens. Malignant samples were divided into high and low level groups, using their median miR-155 expression level as a cut-off value. Associations between miR-155 expression and clinicopathological characteristics were analysed via Chi-square test. Receiver operating characteristic (ROC) curve was used for diagnosis analysis. Survival curves were constituted using the Kaplan–Meier method with log-rank test. Univariate and multivariate analyses were performed with Cox regression analysis. All data were shown as mean ± SD, and p < .05 was considered as statistically significant level.
Results
The expression of miR-155 was increased in ESCC
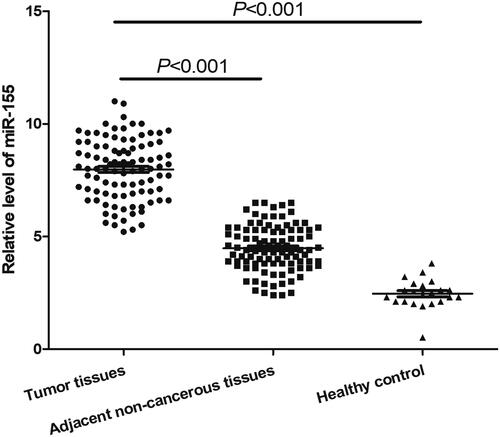
In this study, qRT-PCR was used to measure the expression levels of miR-155 in ESCC tissues, adjacent normal tissues and healthy controls (The expression level of every specimen was displayed in Supplementary_materials). The final results showed that after normalised to U6, the relative expression of miR-155 in ESCC specimens and matched adjacent normal tissues were 7.98 ± 1.35 and 4.47 ± 1.04, respectively, such figure in 20 normal control specimens was 2.55 ± 0.51 (). The differences in miR-155 level between the ESCC samples and two groups of controls were both statistically significant (p < .001), indicating that miR-155 expression was increased in ESCC. The results demonstrated that miR-155 might play an oncogenic role in ESCC.
Relationship between miR-155 and clinicopathological characteristics of ESCC patients
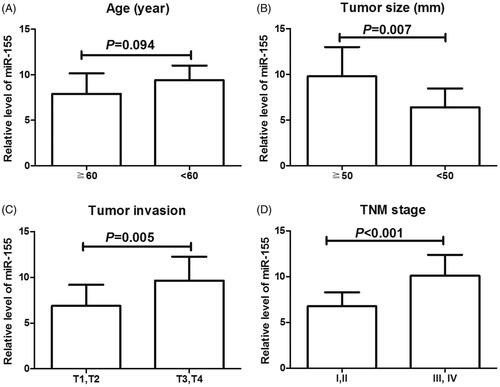
To explore correlation between miR-155 expression and pathological characteristics in ESCC, we manually divided ESCC specimens into two groups: high group with a relative expression level of miR-155 higher than 7.98 (mean value for the expression) and low group with miR-155 level lower than 7.98. Accordingly, 51 cases were classified into high expression group while 45 in low expression group. Then we investigated the association of miR-155 expression with clinicopathological data of the patients. Results showed that miR-155 level was significantly associated with tumour size (p = .001), depth of tumour invasion (p = .002) and TNM stage (p = .000). However, there was no association between miR-155 expression and age, sex, lymph node metastasis or distant metastasis (all, p > .05, ).
Table 2. Multivariate analysis of prognostic factors in ESCC.
In addition, we also compared the expression levels of miR-155 among ESCC patients based on their age, tumour size, tumour invasion and TNM stages. As displayed in , patients with large tumour size, deep tumour invasion and advanced TNM stages exhibited significantly high levels of miR-155 (p < .05 for all), but there was no obvious association between age and miR-155 expression (p = .094).
Diagnostic value of miR-155 in ESCC
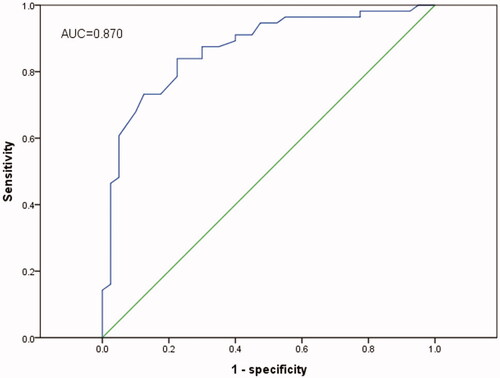
ROC curve analysis was performed to distinguish ESCC patients from healthy controls with an area under the curve (AUC) of 0.870 (95%CI = 0.797–0.943, p < .001, ) accompanied by a sensitivity of 83.5% and a specificity of 77.5% at the cut-off of 7.75. The results showed that miR-155 was an useful marker in diagnosing ESCC.
Correlation between miR-155 expression and prognosis in ESCC patients
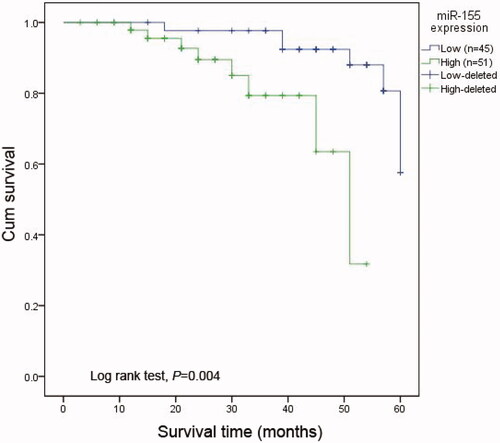
Studies revealed that miR-155 expression in ESCC was significantly associated with tumour progression. Thus, we evaluated its prognostic value in ESCC patients. Kaplan-Meier analysis with the log-rank test was utilised to evaluate difference in overall survival between high and low expression groups. Compared to those with low miR-155 expression, patients with high expression had significantly worse overall survival rates (log-rank test, p < .001, ). In addition, Cox regression analysis was performed to evaluate the prognostic value of miR-155 and clinicopathological features. The results showed that patients with high levels of miR-155 expression had a 5.87-fold higher risk of death (95%CI = 1.606–21.445, p < .001) , while clinicopathological characteristics of TNM stage(p = .004), tumour size (p = .017) and depth of tumour invasion (p = .007) were also associated with overall survival among ESCC patients. However, sex, age, lymph node metastasis and distant metastasis had no prognostic value regarding to overall survival.
Figure 4. Kaplan-Meier analysis for the overall survival of patients with ESCC according to the expression of miR-155. Patients with high miR-155 expression had shorter overall survival than those with low expression. Log rank test proved that the difference was significant (p < .01).
Then, multivariate analysis was used to evaluate whether miR-155 could serve as an independent prognostic marker for patients with ESCC. MiR-155 expression was found to be independently associated with overall survival (HR = 16.94, 95%CI = 3.33–86.12; p = .001), indicating that high miR-155 expression was correlated with decreased overall survival in ESCC patients.
Discussion
As a main histological type of oesophageal carcinoma, ESCC shows horrendous invasive capacity in gastrointestinal system, with a five-year survival rate less than 20% despite remarkable advances in its treatments [Citation12,Citation13]. In addition, delay in diagnosis is also a key reason for its poor survival outcomes. Therefore, it is essential to discover effective biomarkers that could predict the malignancy progression and improve its prognosis.
In previous studies, miR-155 has been found to facilitate tumour invasion and migration in many tumours, acting as a mediator of EMT [Citation14]. Evidences from abundant studies have indicted that the up-regulation of miR-155 played oncogenic function in solid tumours [Citation15], such as breast cancer [Citation16], Burkitt lymphoma [Citation17], pancreatic ductal adenocarcinoma [Citation18,Citation19], hepatocellular carcinoma [Citation20], primary mediastinal, diffuse large-cell lymphoma [Citation21], lung cancer [Citation22], thyroid carcinoma [Citation23], and cervical cancer [Citation24]. Furthermore, miR-155 was also over-expressed in haematopoietic malignancies inducing early B cell polyclonal proliferation, followed by high-grade lymphoma-pre-B leukaemia [Citation25]. Relevant researches showed that the oncogenic properties of miR-155 were related to its anti-apoptotic function through blocking caspase-3 activity or suppressing proapoptotic genes such as TP53BP1 [Citation26,Citation27]. Moreover, miR-155 reduced PTEN and PDCD4 or SHIP1 with the up-regulation of phosphorylated AKT in NK-cell lymphoma/leukaemia, and promoted cell proliferation by down-regulating the suppressor of cytokine signalling 1 gene [Citation28,Citation29]. All these evidences revealed miR-155 might function as an oncogenic miRNA in human cancers and could be a valuable diagnostic or prognostic biomarker in malignant diseases. MiR-155 expression could serve as a prognostic marker in lung cancer [Citation30], breast cancer, pancreatic ductal adenocarcinomas, hepatocellular carcinomas, and adult T-cell leukaemia [Citation16,Citation19,Citation20].
Previous studies revealed that miR-155 was dramatically increased in ESCC tissues and could act as an oncogene through targeting TP53INP1 in the malignancy [Citation31]. Meanwhile, a significant association between circulating miR-155 level and oesophageal cancer development also revealed that miR-155 might be a potential biomarker for the diagnosis of the disease [Citation32]. However, whether miR-155 could be a diagnostic and/or prognostic biomarker for ESCC has not been reported so far. With the aim of evaluating biological function of miR-155, we detected the expression of miR-155 in ESCC cases, matched adjacent tissues and normal controls adopting qRT-PCR. The results revealed that miR-155 expression was significantly increased, and its high expression reduced survival time of patients with ESCC. Moreover, ROC analysis with AUC (0.870) revealed that miR-155 expression was a diagnostic indicator for ESCC. In addition, qRT-PCR is a sensitive, faster and affordable technology for gene detection [Citation33]. Thus, miR-155 detection based on qRT-PCR technology might be a practical and economical approach for early screening ESCC. These evidences revealed that miR-155 could be an independent diagnostic and prognostic factor for ESCC. However, the sample size was relatively small in our research that might influence the accuracy of our results. Second, the results obtained in our study were not verified in other populations. Lastly, the diagnosis specificity of miR-155 was relatively low. Thus, applying miR-155 in early detection of ESCC might cause diagnostic errors, so miR-155 might be employed as an auxiliary tool for such operation. Studies covering larger samples in different populations are necessary to recognise the clinical significance of miR-155 in ESCC more clearer.
In addition, the expression level of miR-155 was associated with tumour size, tumour grade, tumour stage and lymph node metastasis in patients with malignant tumours [Citation16,Citation34]. Therefore, we investigated the association of miR-155 expression with the clinicopathological characters of ESCC patients. The results revealed that the expression of miR-155 was positively correlated with some clinical features such as depth of tumour invasion, tumour size, and TNM stage. However, there was no association between miR-155 expression and age, sex, lymph node metastasis or distant metastasis.
In conclusion, a significant relationship exists between increased expression of miR-155 and the development of ESCC. MiR-155 might serve as a new biomarker in the diagnosis and prognosis of ESCC.
Supplemental Material
Download MS Excel (54.5 KB)Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary materials
The original data were loaded as Supplementary materials.
References
- Enzinger PC, Mayer RJ. Esophageal cancer. N Engl J Med. 2003;349(23):2241–2252.
- Pennathur A, Gibson MK, Jobe BA, et al. Oesophageal carcinoma. Lancet. 2013;381(9864):400–412.
- van Hagen P, Hulshof MC, van Lanschot JJ, et al. Preoperative chemoradiotherapy for esophageal or junctional cancer. N Engl J Med. 2012;366(22):2074–2084.
- Matsuda T, Ajiki W, Marugame T, et al. Population-based survival of cancer patients diagnosed between 1993 and 1999 in Japan: a chronological and international comparative study. Jpn J Clin Oncol. 2011;41(1):40–51.
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297.
- Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350–355.
- Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15(8):509–524.
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843–854.
- Onyeagucha BC, Mercado-Pimentel ME, Hutchison J, et al. S100P/RAGE signaling regulates microRNA-155 expression via AP-1 activation in colon cancer. Exp Cell Res. 2013;319(13):2081–2090.
- Li CL, Nie H, Wang M, et al. microRNA-155 is downregulated in gastric cancer cells and involved in cell metastasis. Oncol Rep. 2012;27(6):1960–1966.
- Yang M, Shen H, Qiu C, et al. High expression of miR-21 and miR-155 predicts recurrence and unfavourable survival in non-small cell lung cancer. Eur J Cancer. 2013;49(3):604–615.
- Jatoi A, Soori G, Foster NR, et al. Phase II study of preoperative pemetrexed, carboplatin, and radiation followed by surgery for locally advanced esophageal cancer and gastroesophageal junction tumors. J Thorac Oncol. 2010;5(12):1994–1998.
- Dipetrillo T, Suntharalingam M, Ng T, et al. Neoadjuvant paclitaxel poliglumex, cisplatin, and radiation for esophageal cancer: a phase 2 trial. Am J Clin Oncol. 2012;35(1):64–67.
- Kong W, Yang H, He L, et al. MicroRNA-155 is regulated by the transforming growth factor beta/Smad pathway and contributes to epithelial cell plasticity by targeting RhoA. Mol Cell Biol. 2008;28(22):6773–6784.
- Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6(4):259–269.
- Chen J, Wang BC, Tang JH. Clinical significance of microRNA-155 expression in human breast cancer. J Surg Oncol. 2012;106(3):260–266.
- Metzler M, Wilda M, Busch K, et al. High expression of precursor microRNA-155/BIC RNA in children with Burkitt lymphoma. Genes Chromosomes Cancer. 2004;39(2):167–169.
- Szafranska AE, Davison TS, John J, et al. MicroRNA expression alterations are linked to tumorigenesis and non-neoplastic processes in pancreatic ductal adenocarcinoma. Oncogene. 2007;26(30):4442–4452.
- Greither T, Grochola LF, Udelnow A, et al. Elevated expression of microRNAs 155, 203, 210 and 222 in pancreatic tumors is associated with poorer survival. Int J Cancer. 2010;126(1):73–80.
- Han ZB, Chen HY, Fan JW, et al. Up-regulation of microRNA-155 promotes cancer cell invasion and predicts poor survival of hepatocellular carcinoma following liver transplantation. J Cancer Res Clin Oncol. 2012;138(1):153–161.
- Kluiver J, Poppema S, de Jong D, et al. BIC and miR-155 are highly expressed in Hodgkin, primary mediastinal and diffuse large B cell lymphomas. J Pathol. 2005;207(2):243–249.
- Donnem T, Eklo K, Berg T, et al. Prognostic impact of MiR-155 in non-small cell lung cancer evaluated by in situ hybridization. J Transl Med. 2011;9:6.
- Nikiforova MN, Tseng GC, Steward D, et al. MicroRNA expression profiling of thyroid tumors: biological significance and diagnostic utility. J Clin Endocrinol Metab. 2008;93(5):1600–1608.
- Wang X, Tang S, Le SY, et al. Aberrant expression of oncogenic and tumor-suppressive microRNAs in cervical cancer is required for cancer cell growth. PloS One. 2008;3(7):e2557
- Costinean S, Zanesi N, Pekarsky Y, et al. Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in E(mu)-miR155 transgenic mice. Proc Natl Acad Sci USA. 2006;103(18):7024–7029.
- Gironella M, Seux M, Xie MJ, Cano C, et al. Tumor protein 53-induced nuclear protein 1 expression is repressed by miR-155, and its restoration inhibits pancreatic tumor development. Proc Natl Acad Sci USA. 2007;104(41):16170–16175.
- Ovcharenko D, Kelnar K, Johnson C, et al. Genome-scale microRNA and small interfering RNA screens identify small RNA modulators of TRAIL-induced apoptosis pathway. Cancer Res. 2007;67(22):10782–10788.
- Jiang S, Zhang HW, Lu MH, et al. MicroRNA-155 functions as an OncomiR in breast cancer by targeting the suppressor of cytokine signaling 1 gene. Cancer Res. 2010;70(8):3119–3127.
- Yamanaka Y, Tagawa H, Takahashi N, et al. Aberrant overexpression of microRNAs activate AKT signaling via down-regulation of tumor suppressors in natural killer-cell lymphoma/leukemia. Blood. 2009;114(15):3265–3275.
- Yanaihara N, Caplen N, Bowman E, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9(3):189–198.
- Zhang J, Cheng C, Yuan X, et al. microRNA-155 acts as an oncogene by targeting the tumor protein 53-induced nuclear protein 1 in esophageal squamous cell carcinoma. Int J Clin Exp Pathol. 2014;7(2):602–610.
- Liu R, Liao J, Yang M, et al. Circulating miR-155 expression in plasma: a potential biomarker for early diagnosis of esophageal cancer in humans. J Toxicol Environ Health Part A. 2012;75(18):1154–1162.
- Gadkar V, Filion M. New developments in quantitative real-time polymerase chain reaction technology. Curr Issues Mol Biol. 2014;16:1–6.
- Shinmei S, Sakamoto N, Goto K, et al. MicroRNA-155 is a predictive marker for survival in patients with clear cell renal cell carcinoma. Int J Urol. 2013;20(5):468–477.