Abstract
Background
miR-4293 rs12220909 polymorphism was reported associated with tumorigenesis, but the results are controversial. Thus, we planned to verify and obtain precise results.
Methods
Databases were searched and reviewed up to November, 2019. Case-control studies which concern about the association between cancer risks and miR-4293 polymorphisms were all enrolled. The odds ratios (ORs) and 95% confidence intervals (CIs) calculated by the Z test were used to assess the underlying links. We also prospected how the miR-4293 impacts biological process through its target genes.
Result
Finally, there are seven independent studies meet the enrolled criteria, along with 5147 cases and 6108 healthy controls. We revealed that there is a significant decrease effect of miR-4293 rs12220909 to cancer risks in heterozygote genetic model (BA vs. AA: OR = 0.857, p = .032), the similar results were also uncovered in PB control group, lung cancer and the studies conform to HWE. Results from GO items and KEGG pathway analysis illustrated that myeloid cell development, transcription factor complex, RNA polymerase II regulatory region DNA binding were regulated by miR-4293.
Conclusion
In summary, our meta-analysis chase down heterozygote rs12220909 polymorphism of miR-4293 is a protective factor to cancer initiation, especially for lung cancer.
Keywords:
Introduction
MicroRNAs (miRNAs) are a series of short length nucleotides in cells, which mostly affect the function of genes by suppressing them. In the past decades, miRNAs are the hotspots in cancer research and have been reported as the oncogene, as well as the biomarkers for cancer prognosis [Citation1–5]. Although miRNAs are smaller in size and number than messenger RNAs (mRNAs), they play a key role in regulating the expression and function of oncogenes, by binding to the 3′-UTR of mRNA [Citation6–8]. In most cell progress, such as proliferation, apoptosis or differentiation, miRNAs could alter the mechanisms by binding the translational region and inhibiting the translation of the target gene [Citation9,Citation10].
Polymorphisms located in the encoding gene of miRNA have recently been concerned with its possible role in the initialise or progression of cancer. Numerous studies have reported that polymorphism in miRNA encoded gene could affect the transcription of itself or its target gene, sharking the formatting of pro- or pre- miRNA, and changing the interaction association between miRNA and mRNA as well [Citation11–15]. miR-4293, a mature miRNA, is encoded by has-mir-4293, the 78 bases DNA sequence, located at the 10th chromosome. miR-4293 was first found in the embryonic stem cells and neural precursors by Ago2 assay [Citation16]. Recent studies have demonstrated the association between the aberrant miR-4293 polymorphism and cancer risk, Fan et al. [Citation17] reported that the rs12220909 polymorphism of miR-4293 links to a decreased risk to non-small cell lung cancer, however, Cai et al. [Citation18] illustrated a controversial result, they found that rs12220909 act as a promoter of cancer risk in female hepatocellular carcinoma (HCC) patients, but not affect the cancer risk in male patients. Whether rs12220909 polymorphism of miR-4293 is associated with cancer susceptibility, we planned to verify and obtain the precise result in the current study.
Materials and methods
Search strategy
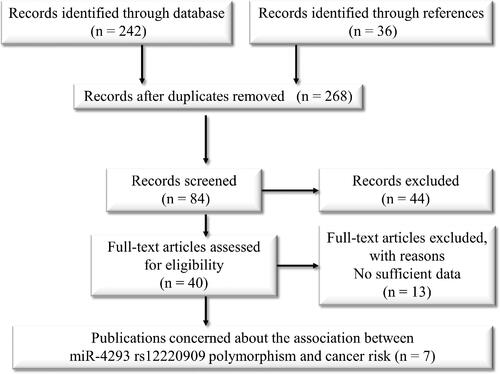
In order to enrol all the publications concerned about the cancer risk of miR-4293 rs12220909, PubMed, Medline, Google Scholar, as well as CNKI (China National Knowledge Infrastructure) database and Wanfang database from China were all searched and reviewed. The expiration date of literature searching up to November, 2019. The applied items including miR-4293, microRNA-4293 or rs12220909 to identify the specific polymorphism, as well as the usual items about cancer and polymorphisms. shows the selection process of all the enrolled publications.
Eligible and exclusion criteria
Eligible criteria: First, Case-control studies which concern about the association between cancer risks and miR-4293 polymorphisms; Second, studies which provide the polymorphism allele frequency of cases and controls. Exclusion criteria: First, studies which not talk about miR-4293 polymorphism or not concern about cancer risk; Second, studies only explore the mechanisms in animal, cell line, or review articles; Third, studies which lack the allele frequency data; Fourth, duplicated studies will also be removed.
Extraction of data sources
We recorded several essential items of the enrolled studies and prepared for the next step analysis. The items include the last name of the first author, the year of the study published, the ethnicity of the individuals, the method of genotyping, the type of cancer, and the polymorphism distribution of cases and controls as well. All the data were recorded by two authors independently, any inconsistent record was re-confirmed by the third author.
Methods of Meta-analysis
We carried out the meta-analysis in pooled overall date, as well as in different stratified sub-groups. Crude odds ratios (ORs) and their 95% confidence intervals (CIs) calculated by Z-test were employed to estimate the potential interrelation between miR-4293 rs12220909 variant and cancer risks in five common hereditary models, including allele genetic model (B vs. A), homozygote genetic model (BB vs. AA), heterozygote genetic model (BA vs. AA), recessive contrast model (BB vs. BA + AA) and dominant contrast model (BB + BA vs. AA). The model used for Z-test was depended on the result of heterogeneity associated Q-test, P value of Q-test less than 0.1 indicated that there was meaningful heterogeneity among enrolled data, thus the random-effects model (Der Simonian and Laird method) was applied. Otherwise, fixed-effects model(the Mantel-Haenszel method) was used [Citation19]. Moreover, Hardy–Weinberg equilibrium (HWE) formula assessed by χ2 was evaluated in the control group of each study, p > .05 was considered to have reliable and representative controls [Citation20]. To assess the quality of each enrolled study, we applied Newcastle-Ottawa Scale (NOS) [Citation21].
Evaluation of the results
Begg’s funnel plot and Egger’s test were used to appraising any publication bias hide in the results [Citation22,Citation23]. On the other way, the underlying effect of every single study to overall results was evaluated by sensitivity analyses, with the method of deletion one independent study each time. All the statistic results shown in the current study were two-tailed, while p-values ≤0.05 were considered to be a remarkable difference.
Table 1. Characteristics of the enrolled studies on miR-4293 rs12220909 polymorphism and cancer.
In-silico analysis of miR-4293
In cellular, miRNAs could act as oncogene or tumour suppressor through the regulation of post-transcription to mRNAs, so we explored how miR-4293 impact cancer risk with bioinformatics analysis. miRMap, TargetScan7 and miRDB, three common online databases were employed to predict the downstream genes of miR-4293 [Citation24–26]. Metascape, a gene annotation and analysis resource, were used to draw the potential function enrichment of the merged downstream genes in the prior step, to illustrated the involved items of the target genes based on Gene Ontology (GO) term enrichment and Kyoto Encyclopaedia of Genes and Genomes (KEGG) pathway [Citation27].
Results
Study characteristics
Along with the general pre-set search items, as much as 268 articles were firstly taken into consideration from different databases. In the next two steps, we screened the relativity of each article by reading its abstract or whole manuscript. Finally, there are only seven articles that meet the inclusion criteria [Citation17,Citation18,Citation28–32]. Of the remaining seven case-control studies, there are about 5147 cancer patients and 6108 controls, and all the seven studies focussed on Asian population (). Two studies talked about the effect of rs12220909 to lung cancer, other studies explored the impact on hepatocellular carcinoma, breast cancer, nasopharyngeal carcinoma, thyroid cancer and oesophageal squamous cell carcinoma, respectively. Most of the sources are population-based, accompanied by the P value of HWE is higher than 0.05, these two results ensured the control group is representative and conformed to the law of genetic heredity. The Newcastle-Ottawa Scale (NOS) was performed to evaluate the quality of each enrolled study, results summarised in shown all the studies maintained high quality.
Overall and subgroup analyses
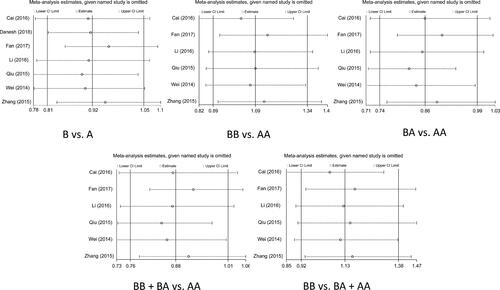
Analysis was handled to evaluated the feasible association between miR-4293 rs12220909 variant and cancer risk, the results are shown in . Overall, we exposed the decreased cancer risk of heterozygote genetic model (BA vs. AA: OR = 0.857, 95%CI = 0.745–0.987, p = .032), but no exceeding interrelation was sniffed out in other four genetic models (B vs. A: OR = 0.925, 95%CI = 0.811–1.054, p = .239; BB vs. AA: OR = 1.092, 95%CI = 0.887–1.343, p = .397; BB vs. BA + AA: OR = 0.879, 95%CI = 0.762–1.013, p = .076; BB + BA vs. AA: OR = 1.127, 95%CI = 0.918–1.383, p = .242) (). In the subgroup stratified by PB, we also chase down the similar result in heterozygote genetic model (BA vs. AA: OR = 0.835, 95%CI = 0.71–0.982, p = .029), as well as the subgroup analysis in lung cancer (BA vs. AA: OR = 0.778, 95%CI = 0.607–0.995, p < .001)and studies conformed to HWE (BA vs. AA: OR = 0.857, 95%CI = 0.745–0.987, p = .032).
Figure 2. Meta-analysis of the association between miR-4293 rs12220909 polymorphism and cancer risk.
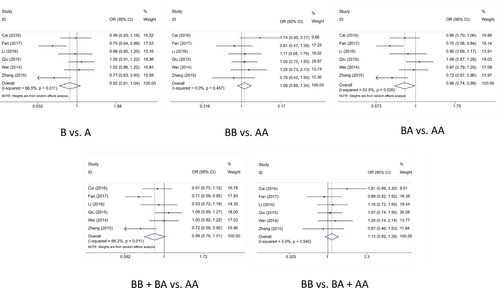
Table 2. Results of pooled analysis for miR-4293 rs12220909 polymorphism and cancer susceptibility.
Analyses of sensitivity and publication bias
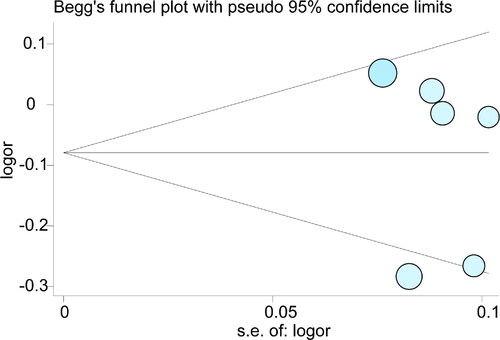
We completed the sensitivity analysis in the included literature by eliminating each study individually. The results show that a single article does not affect the stability of the overall results. (, Table S2). We also evaluated whether any inclusion studies have publication bias by the Begg’s funnel plot and the Egger’s test. As the results shown in Table S3, there is no P value less than 0.05 in both overall or subgroup analysis. Meanwhile, the distribution of studies in Begg’s plot also shown symmetrically result ().
In-silico analysis
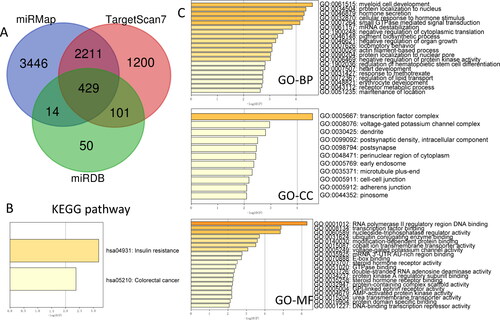
miRNAs always act as promoter or suppressor in cancer through its regulate function of downstream genes. So, we firstly predicted the downstream genes with online miRNA databases, including miRMap, TargetScan7 and miRDB, there are 429 target genes were predicted from all the three databases (). Then, we assessed the function of all the 429 genes with the GO and KEGG term enrichment. In the KEGG enrichment, we found that miRNA-4293 target genes are mostly involved in insulin resistance and colorectal cancer pathways (). GO-Biological Processes enrichment shows the function of these genes in myeloid cell development, protein localisation to nucleus, hormone secretion, cellular response to hormone stimulus and small GTPase mediated signal transduction (). GO-Cellular Components were mostly enrolled in the items of transcription factor complex (). Go-molecular function enrichment displays the involved function of RNA polymerase II regulatory region DNA binding, transcription factor binding, nucleoside-triphosphatase regulator activity, ubiquitin conjugation enzyme binding and modification-dependent protein binding ().
Discussion
miR-4293 is encoded by the 77 bases length sequence located on the 10th chromosome, firstly transcript to the pre-mir-4293, and finally altered to the mature form. The rs12220909 G > C polymorphism is located on the seed region of miR-4239, the C allele alteration of rs12220909 could decrease the free energy of miR-4239, which could make the miRNA more stable and increase the mature miRNA expression [Citation33]. Moreover, the rs12220909 mutation of miR-4293 would lose 1,735 target genes and only gain 199 target genes after the G replaced by the C allele. Li et al. [Citation31] evaluated the effect of rs12220909 in miR-4293 to cancer risks, however, they found no remarkable difference in the polymorphisms to cancer. Meanwhile, Fan et al. [Citation17] illustrated that rs12220909 of miR-4293 might play a significant role in the initial development of lung cancer, and act as a tumour suppressor. As to other types of cancer, Qiu et al. [Citation28] reported that rs12220909 would not affect the cancer risk of nasopharyngeal carcinoma, and Danesh et al. [Citation32] obtained a similar conclusion in breast cancer. Oppositely, Zhang et al. [Citation30] reported that miR-4293 rs12220909 significantly decreased the risk of oesophageal squamous cell carcinoma.
Because of these contradictory results, we managed the current study to eliminate the confusion. As the results show above, we concluded that the heterozygote allele might associate with decreased cancer risks in overall cancers, especially for lung cancer. And the results from bioinformatics analysis show that miRNA-4293 might be involved in the insulin resistance, colorectal cancer, myeloid cell development, hormone secretion, transcript factor complex, as well as RNA polymerase II regulatory region DNA binding. These fields should be paid more attention in the future.
This is the first meta-analysis of the relationship between miR-4293 rs12220909 polymorphism and cancer risk, we have obtained some useful results and found that polymorphisms are associated with reduced tumour risk, there are some limitations should be concerned. First, in the overall analysis, we enrolled all the seven studies based on different cancers, of which might affect the heterogeneity of the results, so we evaluated the potential heterogeneity by Q-test, and chosen the appropriate random- or fixed-effects model to calculate the overall results. Second, in the subgroup analysis, there are fewer samples for each group to be included in the analysis, which may lead to unstable results. Second, the study does not consider the effects of factors other than genetic factors on tumorigenesis, such as living environment, living habits, or other disease history.
To sum up, the current study successfully figured out the reduced cancer risks associated with the miR-4293 rs12220909 polymorphism, especially for lung cancer. These results require more extensive samples to confirm.
Supplementary_table1-3.doc
Download MS Word (79.5 KB)Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Liu T, Zhang Q, Zhang J, et al. EVmiRNA: a database of miRNA profiling in extracellular vesicles. Nucleic Acids Res. 2019;47(D1):D89–D93.
- Lokeshwar SD, Talukder A, Yates TJ, et al. Molecular characterization of renal cell carcinoma: a potential three-microRNA prognostic signature. Cancer Epidemiol Biomarkers Prev. 2018;27(4):464–472.
- Heublein S, Albertsmeier M, Pfeifer D, et al. Association of differential miRNA expression with hepatic vs. peritoneal metastatic spread in colorectal cancer. BMC Cancer. 2018;18(1):201.
- Ratert N, Meyer HA, Jung M, et al. miRNA profiling identifies candidate mirnas for bladder cancer diagnosis and clinical outcome. J Mol Diagn. 2013;15(5):695–705.
- White NM, Khella HW, Grigull J, et al. miRNA profiling in metastatic renal cell carcinoma reveals a tumour-suppressor effect for miR-215. Br J Cancer. 2011;105(11):1741–1749.
- Erhard F, Haas J, Lieber D, et al. Widespread context dependency of microRNA-mediated regulation. Genome Res. 2014;24(6):906–919.
- O’Brien J, Hayder H, Zayed Y, et al. Overview of microRNA biogenesis, mechanisms of actions, and circulation. Front Endocrinol (Lausanne). 2018;9:402.
- Quevillon Huberdeau M, Simard MJ. A guide to microRNA-mediated gene silencing. Febs J. 2019;286(4):642–652.
- Liu B, Shyr Y, Cai J, et al. Interplay between miRNAs and host genes and their role in cancer. Brief Funct Genomics. 2019;18(4):255–266.
- Thomas J, Ohtsuka M, Pichler M, et al. MicroRNAs: clinical relevance in colorectal cancer. Int J Mol Sci. 2015;16(12):28063–28076.
- Hrovatin K, Kunej T. Classification of miRNA-related sequence variations. Epigenomics. 2018;10(4):463–481.
- Ibrahim W, Sakr BR, Obaya E, et al. MicroRNA-146a expression and microRNA-146a rs2910164 polymorphism in Behcet’s disease patients. Clin Rheumatol. 2019;38(2):397–402.
- Sibin MK, Harshitha SM, Narasingarao KV, et al. Effect of rs11614913 polymorphism on mature miR196a2 expression and its target gene HOXC8 expression in human glioma. J Mol Neurosci. 2017;61(2):144–151.
- Quan HY, Yuan T, Hao JF. A microRNA-125a variant, which affects its mature processing, increases the risk of radiation-induced pneumonitis in patients with non-small-cell lung cancer. Mol Med Rep. 2018;18(4):4079–4086.
- Li H, Wang XL, Wu YQ, et al. Correlation of the predisposition of Chinese children to cerebral palsy with nucleotide variation in pri-miR-124 that alters the non-canonical apoptosis pathway. Acta Pharmacol Sin. 2018;39(9):1453–1462.
- Goff LA, Davila J, Swerdel MR, et al. Ago2 immunoprecipitation identifies predicted microRNAs in human embryonic stem cells and neural precursors. PLoS One. 2009;4(9):e7192.
- Fan L, Chen L, Ni X, et al. Genetic variant of miR-4293 rs12220909 is associated with susceptibility to non-small cell lung cancer in a Chinese Han population. PLoS One. 2017;12(4):e0175666.
- Cai. Min WZ, Jun Z, Fengtao C, et al. Genetic polymorphisms of microRNA-4293 and hepatocellular carcinoma susceptibility in female. Liver (Chinese). 2016;21(5):347–350.
- DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7(3):177.
- Lau J, Ioannidis JP, Schmid CH. Quantitative synthesis in systematic reviews. Ann Intern Med. 1997;127(9):820–826.
- Wells M, Chande N, Adams P, et al. Meta-analysis: vasoactive medications for the management of acute variceal bleeds. Aliment Pharmacol Ther. 2012;35(11):1267–1278.
- Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50(4):1088–1101.
- Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21(11):1539–1558.
- Wong N, Wang X. miRDB: an online resource for microRNA target prediction and functional annotations. Nucleic Acids Res. 2015;43:D146–D152.
- Agarwal V, Bell GW, Nam JW, et al. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4:e05005.
- Vejnar CE, Blum M, Zdobnov EM. miRmap web: comprehensive microRNA target prediction online. Nucleic Acids Res. 2013;41:W165–W168.
- Zhou Y, Zhou B, Pache L, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10(1):1523.
- Qiu F, Yang L, Zhang L, et al. Polymorphism in mature microRNA-608 sequence is associated with an increased risk of nasopharyngeal carcinoma. Gene. 2015;565(2):180–186.
- Wei WJ, Wang YL, Li DS, et al. Association study of single nucleotide polymorphisms in mature microRNAs and the risk of thyroid tumor in a Chinese population. Endocrine. 2015;49(2):436–444.
- Zhang P, Wang J, Lu T, et al. miR-449b rs10061133 and miR-4293 rs12220909 polymorphisms are associated with decreased esophageal squamous cell carcinoma in a Chinese population. Tumour Biol. 2015;36(11):8789–8795.
- Li D, Zhu G, Di H, et al. Associations between genetic variants located in mature microRNAs and risk of lung cancer. Oncotarget. 2106;7(27):41715–41724.
- Danesh H, Hashemi M, Bizhani F, et al. Association study of miR-100, miR-124-1, miR-218-2, miR-301b, miR-605, and miR-4293 polymorphisms and the risk of breast cancer in a sample of Iranian population. Gene. 2018;647:73–78.
- Gong J, Tong Y, Zhang HM, et al. Genome-wide identification of SNPs in microRNA genes and the SNP effects on microRNA target binding and biogenesis. Hum Mutat. 2012;33(1):254–263.