ABSTRACT
Genotypic diversity and fluconazole susceptibility of 82 Cryptococcus neoformans and Cryptococcus gattii isolates from 60 renal transplant recipients in Brazil were characterized. Clinical characteristics of the patients and prognostic factors were analysed. Seventy-two (87.8%) isolates were C. neoformans and 10 (12.2%) were C. gattii. VNI was the most common molecular type (40 cases; 66.7%), followed by VNII (9 cases; 15%), VGII (6 cases; 10%), VNB (4 cases; 6.7%) and VNI/II (1 case; 1.7%). The isolates showed a high genetic diversity in the haplotype network and six new sequence types were described, most of them for VNB. There was a bias towards skin involvement in the non-VNI population (P = .012). VGII isolates exhibited higher fluconazole minimum inhibitory concentrations compared to C. neoformans isolates (P = 0.008). The 30-day mortality rate was 38.3%, and it was significantly associated with fungemia and absence of headache. Patients infected with VGII had a high mortality rate at 90 days (66.7%). A variety of molecular types produce disease in renal transplant recipients in Brazil and highlighted by VGII and VNB. We report the clinical appearance and impact of the molecular type, fluconazole susceptibility of the isolates, and clinical characteristics on patient outcome in this population.
Introduction
Cryptococcosis is a life-threatening invasive fungal disease caused by the encapsulated yeasts, Cryptococcus neoformans and Cryptococcus gattii [Citation1]. C. neoformans has a worldwide distribution affecting predominantly individuals with impaired cell-mediated immunity and C. gattii has a more limited environmental distribution and a higher percentage of disease within apparently normal hosts [Citation1]. In renal transplant recipients, cryptococcosis is recognized as the second most common invasive fungal infection, with incidence rates ranging from 0.3% to 5.8% and overall mortality rates as high as 20–50% [Citation2–11].
The nomenclature of C. neoformans/C. gattii species complexes is continuing to evolve under molecular evidences [Citation12,Citation13]. However, as a starting point, cryptococcosis is caused primarily by two species C. neoformans and C. gattii and currently these species can be further divided into ten molecular siblings known as VNI, VNII, VNB, VNIII, VNIV, VGI, VGII, VGIII and VGIV [Citation12] with a possible new molecular type designated VGV. The most widely utilized sequence-based genotyping method for the molecular identification of these complexes has been multilocus sequence typing (MLST). This method is robust and portable between laboratories [Citation14,Citation15].
Clinical comparative studies and understandings between different cryptococcal molecular types are still in their infancy and remain controversial whether or not these different molecular types represent specific characteristics in terms of clinical manifestations or attributable mortality rates [Citation13,Citation16–18]. Furthermore, most data related to strain distribution of C. neoformans and C. gattii species complexes in the transplant recipient relies on small series and case report [Citation5,Citation19–23].
The purpose of our study was to characterize the molecular types of C. neoformans and C. gattii isolated and to assess the clinical outcome of cryptococcosis and their molecular types in patients undergoing renal transplantation throughout Brazil. Interestingly, Brazil represents an environment with a diverse number of cryptococcal molecular types and likely has the most cryptococcal strain diversity of any country practising routine kidney transplantation [Citation24,Citation25].
Results
Clinical characteristics
We enrolled a total of 60 renal transplant recipients followed for a median period of 4 months (0 days to 11 years). One patient had received a liver transplant allograft one year before kidney transplantation and another patient had undergone simultaneous pancreas-kidney transplantation. The clinical characteristics are outlined in .
Table 1. Demographic and clinical characteristics of 60 renal transplant recipients infected by C. neoformans/C. gattii species complexes.
Molecular characterization and clinical associations
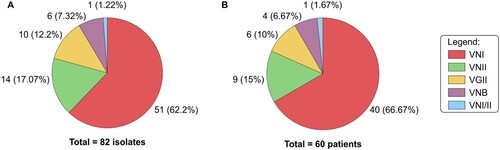
We collected 82 isolates of C. neoformans/C. gattii species complexes from 60 renal transplant recipients. Ten (12.2%) isolates were identified as C. gattii and 72 (87.8%) isolates as C. neoformans ((A)). Forty-seven isolates were from cerebrospinal fluid (CSF), 23 from blood, 6 from pulmonary secretions, 5 from skin biopsy and 1 from urine. The distribution of different molecular types among the patients is depicted in . The most common molecular type was VNI (51 isolates from 40 patients). For 20 episodes on which there were more than one isolate per patient, all but two exhibited similar molecular type within the same episode. In both patients infected by different molecular types we isolated VNI followed by VNII separated by 15 days in CSF or 44 days in blood. These two cases had their findings confirmed by three independent assays yielding the same results. In our entire cohort, only one isolate was diploid based on flow cytometry (VNI/VNII).
Figure 1. Molecular type distribution of 82 clinical isolates of C. neoformans/C. gattii species complexes (A) cultured from 60 renal transplants recipients (B).
A total of 81 isolates were analysed by MLST and compared to MLST database. Nine allele types have been identified for the URA5 locus (1 new), 10 for LAC1 (1 new), 9 SOD1 (1 new), 14 for IGS1 region (3 new), 10 for CAP59 (1 new), 11 for PLB1 and 8 for GPD1 (1 new). Based on the combined analysis of the 7 MLST loci for C. neoformans, 20 sequence types (ST) were found, in which 6 were new (ST581, ST587, ST588, ST589, ST590 and ST591). Of those new ST, four were VNB molecular type and two VNII (Supplementary Table 1).
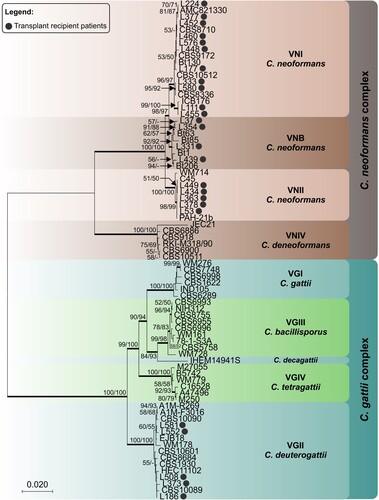
shows the phylogenetic tree of one representative sequence for each haplotype found for these transplant recipient isolates in relationship to reference strains. Our VGII isolates did not cluster with VGIIa, VGIIb or VGIIc.
Figure 2. Phylogenetic relationships as inferred from a maximum likelihood analysis of CAP59, LAC1, PLB1, SOD1, URA5, TEF1 and IGS1 sequences from 82 strains of C. neoformans and C. gattii from transplant patients and 63 reference strains, covering the main molecular types described. The numbers close to the branches represent indices of support (maximum likelihood/neighbor-joining) based on 1000 bootstrap replications. The branches with bootstrap support higher than 70% are indicated in bold.
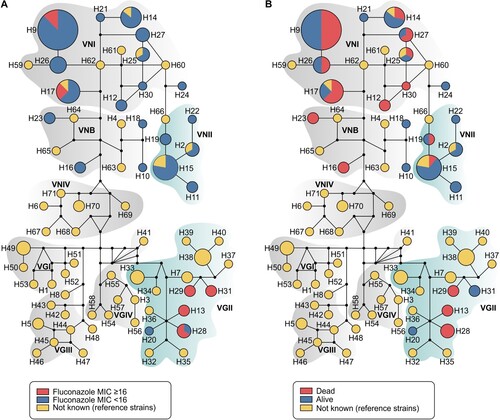
For haplotype analyses we employed a dataset of 144 sequences (7 loci) from C. neoformans/C. gattii species complexes, being 81 generated in this study and 63 recovered from GenBank (Supplementary Table 1). Haplotype and nucleotide diversities were high in our overall dataset (number of haplotypes = 71; Hd = 0.961; π = 0.07003) and also specifically within our 81 transplant recipient isolates (Hd = 0.890; π = 0.03379). Remarkably, 24 out of 71 different haplotypes found in the full dataset for C. neoformans/C. gattii species complexes originated from transplant recipients. Only four haplotypes (H14, H15, H17 and H25) were identical to reference strains, revealing the high diversity of our isolates (; Supplementary Table 1).
Figure 3. Median-joining haplotype network of 144 isolates of C. neoformans/C. gattii species complexes (81 isolates originated in this study in addition to 63 reference strains recovered from literature), covering all the concatenated loci CAP59, LAC1, PLB1, SOD1, URA5, TEF1 and IGS1 sequences. The isolates are coded, and their frequencies are represented by (A) fluconazole MIC ≥ than 16 mg l−1 from transplant recipients isolates or (B) 90-days mortality of transplant recipients. The size of the circumference is proportional to the haplotype frequency. The black dots (median vectors) represent unsampled or extinct haplotypes in the population. Further information about isolate source and GenBank accession number can be found in the Supplementary Table 1.
Regarding clinical correlations, patients infected by C. gattii were more likely to be a retransplant than those infected with C. neoformans (P = .046) (). Furthermore, patients infected with non-VNI molecular types were significantly more likely to have skin involvement than patients infected with VNI (40% vs. 10% patients, respectively; OR = 5.77; 95% CI = 1.47–22.56; P = .012).
Table 2. Comparisons of infections due to C. neoformans and C. gattii complex in 60 renal transplant recipients.
Fluconazole minimum inhibitory concentration (MIC) distributions for C. neoformans and C. gattii isolates varied as depicted in . Modal MICs were 2–16 mg l−1, with the higher mode for VGII (16 mg l−1) and lowest mode for VNII (2 mg l−1). For example, 13 out of 82 (15.85%) isolates exhibited fluconazole MIC values that were ≥16 mg l−1, in which 8 (61.54%) were represented by molecular type VGII ((A)). By multivariate analysis, fluconazole MIC values were higher in patients infected by C. gattii when compared to C. neoformans (OR = 12.62; 95% CI = 1.97–80.99; P = .008) ().
Table 3. Fluconazole MIC distribution for 82 isolates of C. neoformans/C. gattii species complexes tested.
Mortality and prognostic factors
In our study, 23 out of 60 patients (38.3%) died within 30 days after diagnosis of cryptococcosis, and 4 more (total of 45%) died within 90 days. Finally, the overall mortality rate in our study group was 61.7% (37/60), over the entire follow-up period. Based on the univariate analysis, the following factors were significantly associated with high mortality at 30 days: induction immunosuppressive therapy, deceased donor, receipt of a calcineurin-inhibitor agent, pulmonary infection, fungemia, somnolence and absence of headache at admission, CSF antigen titre >1:512 at diagnosis, positive pulmonary culture, discontinuation of tacrolimus after infection, graft loss within 30 days and patients infected with isolates showing MIC ≥ 16 mg l−1. After multivariate regression analysis, the factors independently associated with 30-day mortality were fungemia (OR = 3.79; p = .044) and absence of headache (OR = 0.13; p = .001) (). Most of our patients infected by VGII died within 90 days after cryptococcosis onset. In contrast, there was high survival rate of patients infected by VNII haplotype ((B)). Furthermore, the probability of survival after 12 weeks of cryptococcosis was significantly lower in patients infected with isolates exhibiting fluconazole MICs ≥ 16 mg l−1compared to those with MICs < 16 mg l−1 (P < .001, log rank test) ().
Figure 4. Kaplan–Meier analysis of 12 weeks survival of 60 renal transplant recipient infected by C. neoformans/C. gattii species complexes according to fluconazole MIC ≥ 16 mg l−1 (n = 8) or MIC < 16 mg l−1 (n = 52).
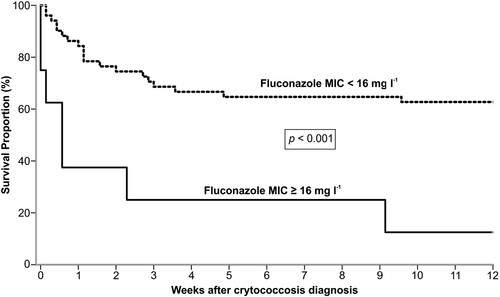
Table 4. Variables associated with 30-day mortality after cryptococcosis in 60 renal transplant recipients.
Discussion
Our retrospective analysis describes 60 cases of cryptococcosis among renal transplant recipients documented during a period of 26 years. The patients presented with high rates of fungemia (38.3%), common disseminated infection (63.3%), and substantial 90-day mortality (45%). These data are comparable to some prior studies evaluating cryptococcosis in renal transplant recipients [Citation3,Citation5–8,Citation26,Citation27]. However, lower rates of mortality have been recently reported among all types of solid transplant recipients in high resource-available countries [Citation2,Citation9–11,Citation26–28]. Furthermore, in our cohort of transplant recipients infected by C. gattii, there was an impressive 66.7% mortality rate within 90 days. Similarly, other investigators have found high rates of dissemination (63.6%) and a high 90-day mortality (36%) for solid organ transplant (SOT) recipients infected by C. gattii [Citation19]. These observations emphasize the potential need for clinicians to know the identification of the species of Cryptococcus in an individual transplant recipient infection for prognostic determination and possibly management alterations.
We observed clinically that fungemia and absence of headache were significantly correlated with decreased survival in our kidney transplant recipients. These characteristics are likely to identify a patient with a high burden of yeasts and/or late diagnosis. These clinical characteristics have been found to be poor prognostic features in other patient populations [Citation29,Citation30].
In Brazil, most cryptococcal molecular types in infections are represented. Indeed, MLST and whole-genome based population analyses suggest that Brazil could be a global centre for diversity of C. neoformans/C. gattii species complexes and even a location for species origin [Citation21,Citation25,Citation31–33]. However, studies addressing the distribution of specific molecular types of Cryptococcus infecting transplant recipients remain scarce worldwide. In the setting of SOT, one study in China revealed 9 cases of VNI and a recent study described 10 strains of molecular type VGII and one of VGI from the Pacific Northwest of USA outbreak. In this last study, 18% of the strains in transplant recipients were C. gattii [Citation5,Citation19]. We found in our cohort of renal transplant recipients that C. gattii represented 12% of isolates (VGII). While VNI was the most common molecular type infecting our patients (66.7%), some genotypic diversity with C. neoformans was also found as we detected 14 VNII and 6 VNB isolates. The molecular predominance of VGII may represent either a specific tropism for transplant recipients and/or increased resistance to tacrolimus/cyclosporine or more likely higher exposure to VGII in the environment over the other VG molecular types. A higher frequency of C. gattii disease observed in retransplantation recipients supports the hypothesis that cryptococcal disease of immunocompromised hosts with C. gattii may be more a primary disease while C. neoformans disease more frequently represents secondary or reactivation disease [Citation4].
Surprisingly we found VNB isolates of C. neoformans in this Brazilian cohort, a molecular type that was considered to be geographically restricted to Africa [Citation34]. However, recently, almost a dozen clinical and environmental isolates from Italy, Portugal, China and Brazil have been identified as VNB strains [Citation20,Citation35–Citation39]. In fact, a recent study comparing Brazilian VNB isolates from transplant recipients with African isolates showed a high diversity within these isolates, except for one isolate from Brazil that nested deeply within the African clade on the phylogeny tree [Citation31]. This isolate was recovered from a mulatto patient living in São Paulo and might have corresponded to a recent migration event. However, VNB isolates as a whole may support the fact that their geographical niche was separated during the Pangea period in which continents split [Citation40].
The Next Generation MLST (NGMLST) methodology and primers that we used were not able to properly sequence the GPD1 locus for VGII molecular type as previously reported [Citation14]. Another issue for our genotyping analysis was our inability to precisely separate our VGII isolates into three distinct clonal lineages (VGIIa, VGIIb and VGIIc). MLST and whole-population genome studies comparing the VGII outbreak strains with VGII isolates from other regions showed that isolates, especially from south of Brazil, have not clustered with any of the three specific lineages [Citation25,Citation33,Citation41].
The clinical impact of different cryptococcal molecular types on patient outcome is still unknown. Controversial results have been generated in the correlation between molecular types and virulence based on experimental studies [Citation13,Citation16,Citation18] and clinical data [Citation17,Citation20,Citation42,Citation43]. We failed to demonstrate substantial differences in clinical presentation or outcome by molecular types, except that non-VNI molecular strains were significantly more likely to show skin involvement than patients infected by VNI strains. There were also no significant differences in outcomes between the C. neoformans and C. gattii infections with the important caveat that there were fewer C. gattii strains and all were VGII. Consequently, the statistical power to detect differences in our study is limited. However, these initial observations support the hypothesis that virulence is not consistently associated with a single major molecular type or subtype but may be more related to the distinct properties of individual isolates.
Fluconazole susceptibility profile is a potentially important clinical issue but its relevance in terms of precisely determining prognosis is still controversial, especially in the setting of the transplant population [Citation5,Citation19,Citation44,Citation45]. In general, direct correlation between species type and antifungal susceptibility has varied [Citation18,Citation21,Citation46–48]. Our VGII isolates showed high fluconazole MICs. Furthermore, in our study, infection by C. neoformans/C. gattii species complex isolates exhibiting high fluconazole MICs did correlate with a worse patient survival rate. However, the precise clinical correlation of in vitro antifungal susceptibility testing of Cryptococcus and break points remains to be further defined [Citation49–53].
In conclusion, cryptococcosis in the setting of SOT in less-resourced countries should be considered a life-threatening fungal disease with substantial mortality rates. The high mortality is probably due to late diagnosis, suboptimal antifungal therapy, including lack of 5FC, as well as the virulence properties of these fungi in SOT. In fact, this transplant population in Brazil represents a relatively uniform host with high rates of mortality for cryptococcosis that may allow for the detection and characterization of particularly virulent strains. With this diversity of strains and a similar high-risk host population for mortality, further examination of these transplant cohorts in Brazil and their fungal strains may give us signals into how these yeasts specifically cause aggressive disease.
Materials and methods
Isolates
We evaluated all C. neoformans and C. gattii strains isolated from patients who underwent renal transplantation and subsequently developed cryptococcosis from 1987 to 2013 at São Paulo Hospital or the Kidney Hospital in São Paulo. The isolates were stored in cryopreservative medium at −70°C with Yeast Peptone Dextrose (YPD) and 20% glycerol at the Special Mycology Laboratory – Federal University of São Paulo.
Clinical data and definitions
The following variables were collected from the medical records using a standard clinical report form: demographic data, complications prior to and after transplantation, immunosuppressive regimens, antifungal therapy, and laboratory data. Disseminated cryptococcosis was defined as the involvement of at least two noncontiguous organ systems or the presence of fungemia [Citation6]. Primary therapy was defined as the first systemic antifungal regimen administered for at least two consecutive days [Citation54]. The study was approved by the local ethic committee (UNIFESP Number: 318847, 2013).
Genotyping
A single colony of each isolate was isolated, grown in YPD broth and frozen at −70°C with glycerol. To isolate genomic DNA, the isolates were streaked from these frozen stocks onto fresh YPD agar, grown for 2–4 days. Next, several colonies of each sample were used to extract and purify the genomic DNA, using the MasterPure Yeast DNA Purification Kit (Epicentre Biotechnologies, Madison, WI, USA). We analysed the sequences from eight loci, including seven standard gene regions (CAP59, LAC1, PLB1, SOD1, URA5, TEF1, GPD1) and the intergenic spacer region 1 (IGS1) of the nuclear ribosomal RNA gene. The primer sequences and the preparation of the sequencing libraries were based on the NGMLST method previously described and the raw sequencing data was processed using MLSTEZ [Citation14]. The allele type of each generated consensus sequence for every locus was determined using the MLST database (http://mlst.mycologylab.org).
The phylogenetic analysis was performed using MEGA 7.0 [Citation55]. The evolutionary relationships, with 1000 bootstrap replicates of the concatenated nucleotide sequences, were inferred using maximum likelihood and the neighbor-joining methods [Citation56]. Major molecular types were confirmed according to phylogenetic clustering with reference strains. Evolutionary relationships at the intraspecific level were evaluated using haplotype networks in order to visualize differences and diversity among isolates. Haplotype and nucleotide diversities were estimated using DNAsp v5.0 [Citation57]. Median-joining networks for the dataset were obtained and visualized using the software Network 5.0 (Fluxus Technology). Flow cytometry was used to check ploidy of the hybrid isolate [Citation58].
Antifungal susceptibility testing
The determination of MICs for fluconazole was performed using the CLSI broth microdilution assay, according to the M27-A3 document [Citation59]. Candida parapsilosis ATCC 22019 and Candida krusei ATCC 6258 were included as quality controls. Assays were performed in RPMI with endpoints read after 72 h at 35°C. The MIC was defined as the lowest concentration that produced 50% growth inhibition compared with the drug-free growth control. The interpretation of MIC values was based on epidemiological cutoff values [Citation60].
Statistical analysis
Continuous data were presented as either the mean ± SD or the median and range, and categorical data were presented as proportions. Univariate analyses were performed to compare categorical variables using either the Chi-square or Fisher's exact test as appropriate and continuous variables by Student's t-test. Variables whose univariate test result had a P-value <.1 were considered candidates for the multivariate model. Binary logistic models were generated using forward stepwise selection for factors associated with death and comparisons between molecular types and species. All analyses were performed using SPSS software for Windows, version 22 (SPSS, Chicago, IL). A value of P ≤ .05 was considered statistically significant.
Acknowledgements
The authors are grateful to the clinical staff of the hospitals involved, the Perfect Laboratory and all adjacent facilities at Duke University School of Medicine and the Special Mycology Laboratory, Division of Infectious Disease – Federal University of São Paulo. V.P., A.L.C. and J.R.P. were responsible for study design, data analysis and manuscript writing. V.P., J.L.P. and D.L.T. carried out experiments. V.P. performed statistical analysis. V.P., Y.C. and A.M.R. performed bioinformatics analysis and manuscript writing. All the authors reviewed, approved and contributed to the final version of the manuscript.
Data availability
The sequences of all newly identified allele types have been submitted to the MLST database (http://mlst.mycologylab.org) and GenBank (https://www.ncbi.nlm.nih.gov/genbank/).
Disclosure statement
V.P.: Educational grants from Pfizer, Gilead Sciences-United Medical (Brazil), Merck Sharp and Dohme, Astellas, TEVA and Sanofi. Y.C., A.M.R., J.L.T., D.L.T, J.O.M.: report no conflicts of interest. A.L.C.: Educational grants from Pfizer, Gilead Sciences-United Medical (Brazil), Merck Sharp and Dohme, and a research grant from Astellas and Pfizer; J.R.P.: Grants, consultant fees, and honorariums from Astellas, Pfizer, Merck, TEVA, F-2G, Matinas, Viamet, Amplyx, Vical and Cidara.
Additional information
Funding
References
- Maziarz EK, Perfect JR. Cryptococcosis. Infect Dis Clin North Am. 2016;30:179–206. doi: 10.1016/j.idc.2015.10.006
- Pappas PG, Alexander BD, Andes DR, et al. Invasive fungal infections among organ transplant recipients: results of the transplant-associated infection surveillance network (TRANSNET). Clin Infect Dis. 2010;50:1101–1111. doi: 10.1086/651262
- Ponzio V, Camargo LF, Medina-Pestana J, et al. Outcomes of cryptococcosis in renal transplant recipients in a less-resourced health care system. Transplant Infect Dis. 2018;20:e12910. doi: 10.1111/tid.12910
- BaddleyJW, Forrest GN, AST Infectious Diseases Community of Practice. Cryptococcosis in solid organ transplantation. Am J Transplant. 2013;13(Suppl 4):242–249.
- Yang YL, Chen M, Gu J-l, et al. Cryptococcosis in kidney transplant recipients in a Chinese university hospital and a review of published cases. Int J Infect Dis. 2014;26:154–161. doi: 10.1016/j.ijid.2014.05.028
- Vilchez R, Shapiro R, McCurry K, et al. Longitudinal study of cryptococcosis in adult solid-organ transplant recipients. Transpl Int. 2003;16:336–340. doi: 10.1111/j.1432-2277.2003.tb00309.x
- Guimaraes LF, Halpern M, de Lemos AS, et al. Invasive fungal disease in renal transplant recipients at a Brazilian center: local epidemiology matters. Transplantation Proc. 2016;48:2306–2309. doi: 10.1016/j.transproceed.2016.06.019
- Marques S, Carmo R, Ferreira I, et al. Cryptococcosis in renal transplant recipients: a single-center experience. Transplantation Proc. 2016;48:2289–2293. doi: 10.1016/j.transproceed.2016.06.006
- Osawa R, Alexander BD, Forrest GN, et al. Geographic differences in disease expression of cryptococcosis in solid organ transplant recipients in the United States. Ann Transplant. 2010;15:77–83.
- Davis JA, Horn DL, Marr KA, et al. Central nervous system involvement in cryptococcal infection in individuals after solid organ transplantation or with AIDS. Transplant Infect Dis. 2009;11:432–437. doi: 10.1111/j.1399-3062.2009.00424.x
- Gassiep I, McDougall D, Douglas J, et al. Cryptococcal infections in solid organ transplant recipients over a 15-year period at a state transplant center. Transpl Infect Dis. 2017;19:e12639. doi: 10.1111/tid.12639
- Kwon-Chung KJ, Bennett JE, Wickes BL, et al. The case for adopting the ‘species complex’ nomenclature for the etiologic agents of cryptococcosis. mSphere. 2017;2:pii: e00357-16. doi: 10.1128/mSphere.00357-16
- Hagen F, Khayhan K, Theelen B, et al. Recognition of seven species in the Cryptococcus gattii/Cryptococcus neoformans species complex. Fungal Genet Biol. 2015;78:16–48. doi: 10.1016/j.fgb.2015.02.009
- Chen Y, Frazzitta AE, Litvintseva AP, et al. Next generation multilocus sequence typing (NGMLST) and the analytical software program MLSTEZ enable efficient, cost-effective, high-throughput, multilocus sequencing typing. Fungal Genet Biol. 2015;75:64–71. doi: 10.1016/j.fgb.2015.01.005
- Munoz M, Camargo M, Ramirez JD. Estimating the intra-taxa diversity, population genetic structure, and evolutionary pathways of Cryptococcus neoformans and Cryptococcus gattii. Front Genet. 2018;9:148. doi: 10.3389/fgene.2018.00148
- Firacative C, Duan S, Meyer W, et al. Galleria mellonella model identifies highly virulent strains among all major molecular types of Cryptococcus gattii. PloS One. 2014;9:e105076. doi: 10.1371/journal.pone.0105076
- Beale MA, Sabiiti W, Robertson EJ, et al. Genotypic diversity is associated with clinical outcome and phenotype in cryptococcal meningitis across Southern Africa. PLoS Negl Trop Dis. 2015;9:e0003847. doi: 10.1371/journal.pntd.0003847
- Thompson GR, Albert N, Hodge G, et al. Phenotypic differences of cryptococcus molecular types and their implications for virulence in a drosophila model of infection. Infect Immun. 2014;82:3058–3065. doi: 10.1128/IAI.01805-14
- Forrest GN, Bhalla P, DeBess EE, et al. Cryptococcus gattii infection in solid organ transplant recipients: description of Oregon outbreak cases. Transplant Infect Dis. 2015;17:467–476. doi: 10.1111/tid.12370
- Andrade-Silva LE, Ferreira-Paim K, Ferreira TB, et al. Genotypic analysis of clinical and environmental Cryptococcus neoformans isolates from Brazil reveals the presence of VNB isolates and a correlation with biological factors. PloS One. 2018;13:e0193237. doi: 10.1371/journal.pone.0193237
- Herkert PF, Meis JF, Lucca de Oliveira Salvador G, et al. Molecular characterization and antifungal susceptibility testing of Cryptococcus neoformans sensu stricto from southern Brazil. J Med Microbiol. 2018;67:560–569. doi: 10.1099/jmm.0.000698
- Cicora F, Petroni J, Formosa P, et al. A rare case of Cryptococcus gattii pneumonia in a renal transplant patient. Transplant Infect Dis. 2015;17:463–466. doi: 10.1111/tid.12371
- Chen SC, Slavin MA, Heath CH, et al. Clinical manifestations of Cryptococcus gattii infection: determinants of neurological sequelae and death. Clin Infect Dis. 2012;55:789–798. doi: 10.1093/cid/cis529
- Ferreira-Paim K, Andrade-Silva L, Fonseca FM, et al. MLST-based population genetic analysis in a global context reveals clonality amongst Cryptococcus neoformans var. grubii VNI isolates from HIV patients in Southeastern Brazil. PLoS Negl Trop Dis. 2017;11:e0005223. doi: 10.1371/journal.pntd.0005223
- Souto AC, Bonfietti LX, Ferreira-Paim K, et al. Population genetic analysis reveals a high genetic diversity in the Brazilian Cryptococcus gattii VGII population and shifts the global origin from the Amazon rainforest to the semi-arid desert in the Northeast of Brazil. PLoS Negl Trop Dis. 2016;10:e0004885. doi: 10.1371/journal.pntd.0004885
- Sun HY, Wagener MM, Singh N. Cryptococcosis in solid-organ, hematopoietic stem cell, and tissue transplant recipients: evidence-based evolving trends. Clin Infect Dis. 2009;48:1566–1576. doi: 10.1086/598936
- Singh N, Alexander BD, Lortholary O, et al. Cryptococcus neoformans in organ transplant recipients: impact of calcineurin-inhibitor agents on mortality. J Infect Dis. 2007;195:756–764. doi: 10.1086/511438
- George IA, Spec A, Powderly WG, et al. Comparative epidemiology and outcomes of human immunodeficiency virus (HIV), Non-HIV non-transplant, and solid organ transplant associated cryptococcosis: a population-based study. Clin Infect Dis. 2018;66:608–611. doi: 10.1093/cid/cix867
- Dismukes WE, Cloud G, Gallis HA, et al. Treatment of cryptococcal meningitis with combination amphotericin B and flucytosine for four as compared with six weeks. N Engl J Med. 1987;317:334–341. doi: 10.1056/NEJM198708063170602
- Brizendine KD, Baddley JW, Pappas PG. Predictors of mortality and differences in clinical features among patients with cryptococcosis according to immune status. PloS One. 2013;8:e60431. doi: 10.1371/journal.pone.0060431
- Rhodes J, Desjardins CA, Sykes SM, et al. Tracing genetic exchange and biogeography of Cryptococcus neoformans var. grubii at the global population level. Genetics. 2017;207:327–346. doi: 10.1534/genetics.117.203836
- Hagen F, Ceresini PC, Polacheck I, et al. Ancient dispersal of the human fungal pathogen Cryptococcus gattii from the Amazon rainforest. PloS One. 2013;8:e71148. doi: 10.1371/journal.pone.0071148
- Engelthaler DM, Hicks ND, Gillece JD, et al. Cryptococcus gattii in North American Pacific Northwest: whole-population genome analysis provides insights into species evolution and dispersal. MBio. 2014;5:e01464-14. doi: 10.1128/mBio.01464-14
- Litvintseva AP, Thakur R, Vilgalys R, et al. Multilocus sequence typing reveals three genetic subpopulations of Cryptococcus neoformans var. grubii (serotype A), including a unique population in Botswana. Genetics. 2005;172:2223–2238. doi: 10.1534/genetics.105.046672
- Barreto de Oliveira MT, Boekhout T, Theelen B, et al. Cryptococcus neoformans shows a remarkable genotypic diversity in Brazil. J Clin Microbiol. 2004;42:1356–1359. doi: 10.1128/JCM.42.3.1356-1359.2004
- Bovers M, Hagen F, Kuramae EE, et al. Unique hybrids between the fungal pathogens Cryptococcus neoformans and Cryptococcus gattii. FEMS Yeast Res. 2006;6:599–607. doi: 10.1111/j.1567-1364.2006.00082.x
- Ngamskulrungroj P, Gilgado F, Faganello J, et al. Genetic diversity of the cryptococcus species complex suggests that Cryptococcus gattii deserves to have varieties. PloS One. 2009;4:e5862. doi: 10.1371/journal.pone.0005862
- Dou HT, Xu YC, Wang HZ, et al. Molecular epidemiology of Cryptococcus neoformans and Cryptococcus gattii in China between 2007 and 2013 using multilocus sequence typing and the DiversiLab system. Eur J Clin Microbiol Infect Dis. 2015;34:753–762. doi: 10.1007/s10096-014-2289-2
- Cogliati M, Prigitano A, Esposto MC, et al. Epidemiological trends of cryptococcosis in Italy: molecular typing and susceptibility pattern of Cryptococcus neoformans isolates collected during a 20-year period. Med Mycol. 2018;56(8):963–971.
- Casadevall A, Freij JB, Hann-Soden C, et al. Continental drift and speciation of the Cryptococcus neoformans and Cryptococcus gattii species complexes. mSphere. 2017;2:pii: e00103-17.
- Herkert PF, Hagen F, de Oliveira Salvador GL, et al. Molecular characterisation and antifungal susceptibility of clinical Cryptococcus deuterogattii (AFLP6/VGII) isolates from Southern Brazil. Eur J Clin Microbiol Infect Dis. 2016;35:1803–1810. doi: 10.1007/s10096-016-2731-8
- Wiesner DL, Moskalenko O, Corcoran JM, et al. Cryptococcal genotype influences immunologic response and human clinical outcome after meningitis. MBio. 2012;3: pii: e00196-12. doi: 10.1128/mBio.00196-12
- Day JN, Qihui S, Thanh LT, et al. Comparative genomics of Cryptococcus neoformans var. grubii associated with meningitis in HIV infected and uninfected patients in Vietnam. PLoS Negl Trop Dis. 2017;11:e0005628. doi: 10.1371/journal.pntd.0005628
- Tarai B, Tarai B, Kher V, et al. Early onset primary pulmonary cryptococcosis in a renal transplant patient. Indian J Med Microbiol. 2010;28:250–252. doi: 10.4103/0255-0857.66489
- Trpkovic A, Pekmezovic M, Barac A, et al. In vitro antifungal activities of amphotericin B, 5-fluorocytosine, fluconazole and itraconazole against Cryptococcus neoformans isolated from cerebrospinal fluid and blood from patients in Serbia. J Mycol Med. 2012;22:243–248. doi: 10.1016/j.mycmed.2012.06.002
- Trilles L, Meyer W, Wanke B, et al. Correlation of antifungal susceptibility and molecular type within the Cryptococcus neoformans/C. gattii species complex. Med Mycol. 2012;50:328–332. doi: 10.3109/13693786.2011.602126
- Andrade-Silva L, Ferreira-Paim K, Mora DJ, et al. Susceptibility profile of clinical and environmental isolates of Cryptococcus neoformans and Cryptococcus gattii in Uberaba, Minas Gerais, Brazil. Med Mycol. 2013;51:635–640. doi: 10.3109/13693786.2012.761737
- Lockhart SR, Iqbal N, Bolden CB, et al. Epidemiologic cutoff values for triazole drugs in Cryptococcus gattii: correlation of molecular type and in vitro susceptibility. Diagn Microbiol Infect Dis. 2012;73:144–148. doi: 10.1016/j.diagmicrobio.2012.02.018
- Dannaoui E, Abdul M, Arpin M, et al. Results obtained with various antifungal susceptibility testing methods do not predict early clinical outcome in patients with cryptococcosis. Antimicrob Agents Chemother. 2006;50:2464–2470. doi: 10.1128/AAC.01520-05
- Van Wyk M, Govender NP, Mitchell TG, et al. Multilocus sequence typing of serially collected isolates of cryptococcus from HIV-infected patients in South Africa. J Clin Microbiol. 2014;52:1921–1931. doi: 10.1128/JCM.03177-13
- Rossi SA, Trevijano-Contador N, Scorzoni L, et al. Impact of resistance to fluconazole on virulence and morphological aspects of Cryptococcus neoformans and Cryptococcus gattii isolates. Front Microbiol. 2016;7:153.
- Aller AI, Martin-Mazuelos E, Lozano F, et al. Correlation of fluconazole MICs with clinical outcome in cryptococcal infection. Antimicrob Agents Chemother. 2000;44:1544–1548. doi: 10.1128/AAC.44.6.1544-1548.2000
- Bicanic T, Harrison T, Niepieklo A, et al. Symptomatic relapse of HIV-associated cryptococcal meningitis after initial fluconazole monotherapy: the role of fluconazole resistance and immune reconstitution. Clin Infect Dis. 2006;43:1069–1073. doi: 10.1086/507895
- Dromer F, Mathoulin-Pelissier S, Launay O, et al. Determinants of disease presentation and outcome during cryptococcosis: The CryptoA/D study. PLoS Med. 2007;4:e21. doi: 10.1371/journal.pmed.0040021
- Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054
- Tamura K, Stecher G, Peterson D, et al. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197
- Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187
- Tanaka R, Taguchi H, Takeo K, et al. Determination of ploidy in Cryptococcus neoformans by flow cytometry. J Med Vet Mycol. 1996;34:299–301. doi: 10.1080/02681219680000521
- Clinical and Laboratory Standards Institute (CLSI). Reference method for broth dilution antifungal susceptibility testing of yeasts. Approved standard – 3rd ed. CLSI document M27-A3. Wayne (PA): Clinical and Laboratory Standards Institute; 2008. ISBN 1-56238-666-2.
- Espinel-Ingroff A, Aller AI, Canton E, et al. Cryptococcus neoformans-Cryptococcus gattii species complex: an international study of wild-type susceptibility endpoint distributions and epidemiological cutoff values for fluconazole, itraconazole, posaconazole, and voriconazole. Antimicrob Agents Chemother. 2012;56:5898–5906. doi: 10.1128/AAC.01115-12
