ABSTRACT
Babesiosis is a tick-transmitted intraerythrocytic zoonosis. In Korea, the first mortalities were reported in 2005 due to Babesia sp. detection in sheep; herein we report epidemiological and genetic characteristics of a second case of babesiosis. Microscopic analysis of patient blood revealed polymorphic merozoites. To detect Babesia spp., PCR was performed using Babesia specific primers for β-tubulin, 18S rDNA, COB, and COX3 gene fragments. 18S rDNA analysis for Babesia sp., showed 98% homology with ovine Babesia sp. and with Babesia infections in Korea in 2005. Moreover, phylogenetic analysis of 18S rDNA, COB, and COX3 revealed close associations with B. motasi. For identifying the infectious agent, Haemaphysalis longicornis (296) and Haemaphysalis flava (301) were collected around the previous residence of the babesiosis patient. Babesia genes were identified in three H. longicornis: one sample was identified as B. microti and two samples were 98% homologous to B. motasi. Our study is the first direct confirmation of the infectious agent for human babesiosis. This case most likely resulted from tick bites from ticks near the patient house of the babesiosis patient. H. longicornis has been implicated as a vector of B. microti and other Babesia sp. infections.
Introduction
Babesiosis, caused by intraerythrocytic protozoans of the Genus Babesia, is a zoonotic tick-borne disease, but also can be transmitted through blood transfusion [Citation1,Citation2]. Babesiosis is an emerging health concern and has been progressively diagnosed beyond known endemic areas [Citation3–5]. More than 100 Babesia species cause infections in many wild and domestic animals, with varying degrees of virulence among different species of Babesia in humans and animals. B. microti, B. divergens, and B. venatorum are the three most predominant species known to infect humans, while other species, B. ovis, B. major, B. bovis, B. bigemina, B. ovata, B. orientalis, B. motasi, and B. caballi are causal agents for animal infections [Citation6].
Four Genera, Rhipicephalus, Ixodes, Haemaphysalis, and Hyalomma within the Ixodidae have been reported as vectors of Babesia spp. Geographically, Ixodes scapularis in the USA, I. Ricinus in Europe, and I. persulcatus in Asia transmit Babesia parasites to natural hosts [Citation7–9]. B. ovis, B. motasi, and B. crassa are primarily transmitted via Haemaphysalis qinghaiensis and H. longicornis [Citation10]. In addition, B. bovis and B. bigemina are also transmitted primarily via Rhipicephalus microplus and R. annulatus. Recently, B. divergens, B. microti, and B. venatorum were detected in I. persulcatus and B. divergens in H. longicornis was detected in China.
Human cases of B. microti, B. duncani, and B. divergens-like infections have been reported in several regions in the USA [Citation1,Citation11–13]. Sporadic cases of infection by B microti-like or uncharacterized Babesia species were reported in Africa, South America, and Asia [Citation1,Citation14]. In East Asia, cases of human babesiosis have been reported in Japan [Citation15] and Taiwan [Citation16], caused by B. microti-like parasites; however, the patients were asymptomatic. In Korea, the first case of human babesiosis with a suspected B. ovis-like infection was reported in 2005; however, there were no epidemiological investigations conducted to determine the accepted vectors at that time [Citation17].
The purpose of this study was to: (i) identify the Babesia sp., reported as a novel large Babesia parasite infecting humans; (ii) investigate the distribution and diversity of Babesia sp. in ticks collected around the patient’s previous residence; and (iii) analyse the phylogenetic association of the Babesia sp. detected in the patient with that detected in associated ticks.
Materials and methods
Blood staining
A blood sample was obtained from the jugular vein in the patient that presented with dizziness and general weakness. Thin blood films were made on dry and clean slides, air-dried, and fixed with methyl alcohol for 5 min, followed by staining with 5% Giemsa solution [Citation18]. The slide was stained for 30 min and then dried for 30 min. Thereafter, the stained films were examined via light microscopy at ×100 magnification with an oil-immersion lens for blood parasites (Leica Microsystems, Heebrugg, Singapore). Percentage parasitemia was determined by counting the number of parasitized erythrocytes and dividing that by the total number of red blood cells (RBCs): % parasitemia = (infected RBCs/RBCs) × 100. A minimum of 500 RBCs were counted and RBCs infected with multiple parasites were counted as a single infected cell.
DNA isolation from blood
Genomic DNA was extracted from blood samples, using a DNeasy Blood and Tissue Kit (Qiagen, Hilden, Germany) in accordance with the manufacturer’s instructions. The extracted DNA was eluted in 100 µL of elution buffer. The quantity and quality of the isolated DNA were assessed with a spectrophotometer Nanodrop 2000c (Thermo Fisher Scientific, Massachusetts, USA) and stored at −20°C until use.
Tick collection and DNA extraction
Questing ticks were collected by flagging and a dry ice bait trap in Hoengseong-gun, Gangwon-do, Korea as described by H.S. Ginsberg and C.P. Ewing [Citation19]. The collection area was divided into each three parts, adjusting the differential collecting methods depending on short and long grasses for efficient tick sampling regardless of tick spp. The ticks with short grasses were collected by flagging and those with long grasses were collected by dry ice trap. A total of 597 ticks were collected in the survey area. Genomic DNA was isolated from individual ticks, using the DNeasy tissue kit (Qiagen) in accordance with the manufacturer’s instructions. Quantity and quality of the isolated DNA were assessed with a spectrophotometer Nanodrop 2000c (Thermo Fisher Scientific) and stored at −20°C until use.
Polymerase chain reaction (PCR) assays
Babesia spp. were detected via PCR for 18S rDNA, Cytochrome b (COB) and Cytochrome c oxidase subunit III (COX3) in accordance with a previously described method [Citation20–23] (). B. microti and B. divergens were detected via PCR, as described previously [Citation24–26]. Reactions were carried out in 20-μL reaction mixtures by using AccuPower PCR master mix (Bioneer, Daejeon, Korea) containing 1 μM each of the 1st forward and reverse primers, sterile water, and 500 ng of DNA template. Amplification products were electrophoresed using an auto-electrophoresis device (QIAxcel, Hilden, Germany) [Citation27]. The PCR products were then purified using an agarose gel DNA purification kit (Qiagen) [Citation27]. TA cloning was performed using the TOPO TA cloning kit with isolated PCR products for sequencing (Invitrogen, Carlsbad, CA, USA). These samples were sequenced using an ABI PRISM 3730xl Analyzer (Applied Biosystems, Foster City, CA, USA).
Table 1. Forward and reverse primers used for the detection of Babesia spp.
Sequence analysis
PCR products were purified and sequenced in both directions, using the specific primers. Nucleotide sequences were analysed using BLAST-N and aligned with ClustalW. The pairwise distance was analysed using the Kimura’s 2-parameter model. Phylogenetic analyses were conducted using the software MEGA 6 [Citation28]. The neighbor-joining method was employed to construct a phylogenetic tree. The reliability of the branches in the tree was evaluated via bootstrapping analysis with 1000 replicates [Citation29], and a bootstrap value more than 60% was considered significant.
Results
Clinical signs
A 70-year-old man was hospitalized with a history of dizziness and general weakness for 2 d. The patient received medication for diabetes mellitus, hypertension, and hyperlipidaemia, and underwent total pancreatectomy for intraductal papillary mucinous neoplasm 19 months ago. The patient resided in a suburban area in Hoengseong-gun, Gangwon-do, Korea. Initial analyses of vital signs were: a blood pressure 86/30 mmHg; pulse rate, 138 beats/min; respiratory rate, 20 breaths/min; body temperature, 39.5°C. Initial laboratory analysis: leukocyte count, 8840/μL with 93.5% neutrophils; haemoglobin, 14.5 g/dL; platelet count, 90,000/μL; blood urea nitrogen, 38.1 mg/dL; creatinine, 3.03 mg/dL; total bilirubin, 2.49 mg/dL; aspartate transaminase (AST), 336 IU/L; alanine transaminase (ALT), 86 IU/L; C-reactive protein, 8.2 mg/dL. Abdominopelvic computed tomography revealed no suspicious focal infection or local and distant metastasis. Piperacillin/tazobactam were administered as an initial empirical antimicrobial therapy. Blood analysis on the second day, 22 h after hospitalization changed as follows: leukocyte count, 6590/μL with 90.7% neutrophils; haemoglobin, 11.9 g/dL; platelet count, 21,000/μL; blood urea nitrogen, 33.0 mg/dL; creatinine, 2.21 mg/dL; total bilirubin, 7.98 mg/dL; AST, 1,010 IU/L; ALT, 191 IU. The patient’s consciousness was drowned and the oxygen saturation decreased to 82–83%. Chest X-ray revealed no abnormal findings. The patient was transferred to the intensive care unit and subjected to intubation and continuous renal replacement therapy; however, the patient died 36 h after hospitalization. No microorganisms were isolated from the blood culture.
Microscopic findings
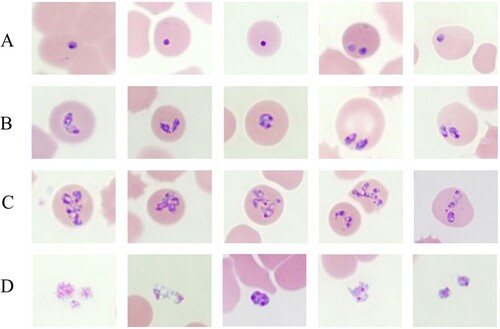
Upon light microscopic examination, variable intraerythrocytic parasites as ring forms, pear-shaped forms, paired pyriforms, pleomorphic ring forms, and multiple-infected parasites and clusters of extracellular rings were detected in Giemsa-stained blood smears. The percentage of parasitaemia was 1.8% (). Maltese cross forms comprising four masses in an erythrocyte that are often described as a characteristic of B. microti infection were not detected in most blood smears ().
Figure 1. Babesia spp in a thin blood smear stained with 5% Giemsa on May 30, 2017, from a patient in Hoengseong-gun, Gangwon-do, Korea, showing pleomorphic and pyriform rings and multiple-infected RBCs. Pigment not present in any of the parasites. (A) Ring-form parasites; (B) Paired-pyriform parasites; (C) Pleomorphic ring forms and multiply infected parasites; (D) Cluster of extracellular rings.
Molecular characteristics of Babesia sp. in human
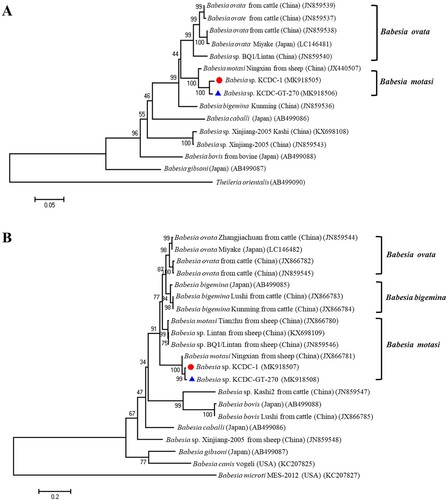
Genomic DNA was extracted from the patient’s blood sample and subjected to 18S rDNA analysis for Babesia species, Beta-tubulin for B. microti, and 18S rDNA for B. divergens. Only the 18S rDNA, Babesia sp. KCDC-1 (MK930513) was positive for Babesia species. Consequently, the samples were subjected to full-length (∼1.7 kb) amplification of 18S rDNA for Babesia spp. Samples with positive results upon PCR sequenced sequences then subjected to phylogenetic analysis with related Babesia species for 18S rRNA gene sequences in GenBank. The analysis demonstrated that the positive sequences were closely related to Babesia sp. KO1, ovine Babesia sp. Liaoning 2005, and ovine Babesia sp. Hebei-2005 (DQ346955, DQ159075, and DQ159074) (). To further evaluate the classification of each sample, the Babesia species was subjected to genomic DNA analysis using COB and COX3 gene fragments of Babesia species. Positive sequences from the analysis of both genes were identified and sequenced. The sequences were then phylogenetically analysed with COB, Babesia sp. KCDC-1 (MK918505) and COX3, Babesia sp. KCDC-1 (MK918507) sequences of related Babesia species in GenBank. This analysis demonstrated that the positive sequences of COB and COX3 were closely related with ovine Babesia motasi Ningxian (JX440507) ((A)) and ovine B. motasi Ningxian (JX866781) ((B)), respectively. As these sequences were clustered with B. motasi in the phylogenetic tree, the parasite detected from the patient’s blood belonged to ovine B. motasi.
Figure 2. Phylogenetic relationships based on the 18S rRNA gene sequence of Babesia species in a human babesiosis sample and in ticks, Babesia sp. KCDC-GT-270, Babesia sp. KCDC-GR-272 and Babesia sp. KCDC-1, in accordance with the polymerase chain reaction-amplified sequence. The evolutionary history was inferred via the Neighbor-Joining method. The percentage of replicate trees wherein the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches. Evolutionary analyses were conducted using MEGA6 (![]()
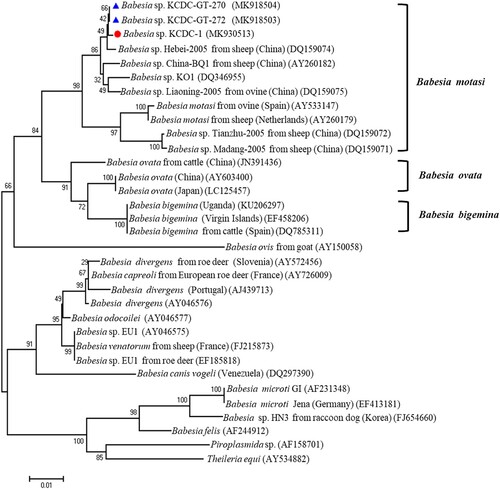
Figure 3. Phylogenetic relationships based on COB (A) and COX3 (B) sequence of Babesia species in a human babesiosis sample and ticks, Babesia sp. KCDC-GT-270 and Babesia sp. KCDC-1, in accordance with the polymerase chain reaction-amplified sequence. The evolutionary history was inferred via the Neighbor-Joining method. The percentage of replicate trees wherein the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches. Evolutionary analyses were conducted using MEGA6 (![]()
Identification and molecular detection of Babesia sp. in questing ticks
Tick collections were performed by dividing the area around the patient’s residence into six parts (1: front yard of patient’s residence, 2: patient’s residence hill III, 3: back yard of patient’s residence, a: Fields surrounding the patient’s residence, b: patient’s residence hill I, c: patient’s residence hill II) (). A total of 597 ticks were collected around the patient’s residence, including 296 H. longicornis (186 adult, 41 nymphs, and 68 larvae) and 301 Haemaphysalis flava (1 adult and 300 larvae) (). Among these, 94% of the ticks were collected in both the front yard of patient’s residence (442 ticks) and associated hill III (124 ticks). Based on the results of the amplification of Babesia genes in each tick, 2 (0.3%) were positive for 18S rDNA of Babesia species, 1 (0.2%) for COB and COX3, and 1 (0.2%) for β-tubulin gene of B. microti. While the nymph of H. longicornis yielded a positive result for only 18S rDNA, one female tick of H. longicornis yielded positive results for 18S rDNA, COB, and COX3 gene fragments. Also, one female tick of H. longicornis only yielded positive results for β-tubulin gene of B. microti ().
Figure 4. Location of the sampling site and collection methods surrounding the Babesia-positive patient’s residence in Hoengseong-gun, Gangwon-do, Korea.
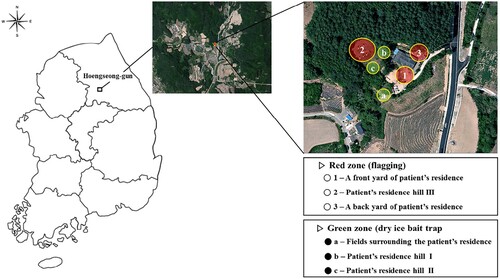
Table 2. Species, stage, gender, and number of collected ticks in Babesia-positive patient’s residence.
Table 3. Detection of Babesia DNA in the collected ticks via PCR.
Molecular characteristics of Babesia sp. in questing ticks
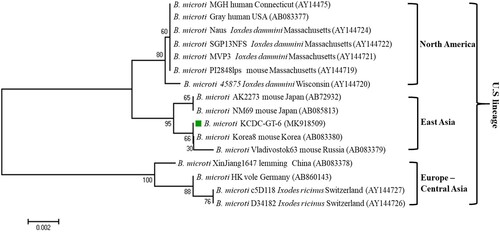
Sequencing of the PCR product allowed for the determination of Babesia species. The sequences are then phylogenetically analysed with related Babesia species for 18S rDNA, COB, COX3, and β-tubulin sequences in GenBank. Sequences of the 18S rRNA gene, Babesia sp. KCDC-GT-270 and KCDC-GT-272 (MK918504 and MK918503) in 2 H. longicornis were closely related with that of ovine Babesia sp. Hebei-2005 (DQ159074) (). Also, the sequence of COB, Babesia sp. KCDC-GT-270 (MK918506) and COX3, Babesia sp. KCDC-GT-270 (MK918508) in H. longicornis was very closely related with ovine Babesia sp. Ningxian (JX866781) ((A,B)). Since these sequences clustered with B. motasi, the Babesia sp. detected from H. longicornis was identified as ovine B. motasi. The sequence of the β-tubulin gene, B. microti KCDC-GT-6 (MK918509) in a female H. longicornis was very similar to that of the US-type B. microti (). Furthermore, the β-tubulin sequence in the positive samples in the present study was closely related to those reported previously in Korea and Russia in East Asia.
Figure 5. Phylogenetic relationships based on the β-tubulin gene sequence of Babesia microti in a tick, B. microti KCDC-GT-6, in accordance with the polymerase chain reaction-amplified sequence. The evolutionary history was inferred via the Neighbor-Joining method. The percentage of replicate trees wherein the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches. Evolutionary analyses were conducted using MEGA6 (![]()
Discussion
The severity of Babesia infections in humans is rapidly becoming apparent, irrespective of whether the disease is transmitted via a tick bite or secondarily transmitted via transfusion of infected blood [Citation30–32]. Previously, seven different Babesia spp., B. microti, B. divergens, B. bovis, B. canis, B. duncani, B. venatorium, and a novel Babesia sp. similar to ovine babesias were reported to cause human babesiosis [Citation5,Citation17,Citation26,Citation32,Citation33]. Regarding the geographical distribution of human babesiosis, B. microti is the primary species responsible for disease in the USA [Citation34] while B. divergens is the causative agent in Europe [Citation35,Citation36]. In Asia, human cases of a B. microti infections have been reported in Taiwan, Japan, China, and Mongolia [Citation16,Citation37,Citation38]. In Korea, the first case of human babesiosis (KO1) was reported in 2007 and KO1 was highly related to Chinese ovine Babesia sp. [Citation17]. Unfortunately, there was no epidemiological surveys conducted involving patients to determine the causative infects. Human babesiosis (KCDC-1) in 2017 was the second case identified in Korea and the sequence of Babesia sp. was very closely related to that of KO1 and Liaoning, China. These large Babesia are clearly distinct from other agents of human babesiosis based on their shape and phylogeny. These results suggest that the causative agent in their case of babesiosis is a novel large Babesia parasite infecting humans and may be highly fatal.
Human babesiosis infections and virulence depend on a number of factors, such as parasite species, patient, age, and host immune competency [Citation39]. In general, old age and reduced cellular immunity are associated with a higher risk of symptomatic infection and more severe illness. In particular, most cases of severe babesiosis were reported among individuals lacking a spleen [Citation14,Citation40]. When Babesia sp. sporozoites first infect humans, they immediately target the host RBCs. Further, infected RBCs remain circulating in peripheral blood and frequently penetrate the host’s spleen [Citation41–43]. The spleen with its lympho-reticular filter function is essential in resisting primary infections of Babesia sp. by eliminating infected cells from circulation. The spleen and cellular immune response play an essential role in resisting both primary and challenge infections of Babesia species [Citation26]. However, the spleen is not critical for the development of immunity and its primary function is to eliminate infected cells from circulation and to stimulate phagocytosis of infected cells [Citation36,Citation44]. In this study, the patient had a splenectomy and red blood cells were considered to be infected with Babesia sp. and were not eliminated, thus resulting in the fatality, which is highly suggestive that splenectomy is one of the critical risk factors for Babesia sp. symptomatic infections and potential fatality.
Inside RBCs, Babesia sp. begins a cycle of maturation and growth. In the initial stages of their life cycle, Babesia spp. are morphologically indistinguishable from Plasmodium sp., both appearing as ring-like parasites. Duplication occurs via budding, where one ring divides into two rings. That is commonly referred to as the “figure eight” form. Budding may recur, giving rise to the tetrad form known as a Maltese Cross [Citation42]. Both these morphological forms are unique to Babesia spp. and form the basis of a microscopy-based conclusive diagnosis. Based on morphology, Babesia spp. are divided into the small babesias including B. gibsoni, B. microti, and B. divergens and large babesias including B. bovis, B. caballi, and B. canis. While small Babesia spp. are 1.0–2.5-µm-long and regularly appear in the ring form and Maltese cross form, large Babesia spp. are 2.5–5-µm-long and regularly appears as a unique paired pyriform [Citation26,Citation45]. The form of parasites detected in the patient’s blood showed pleomorphic and paired pyriform rings. Hence, the Babesia sp. KCDC-1 was classified as large Babesia spp. on the basis of appearance and size.
Regarding the molecular characteristics of the parasite obtained from the patient’s blood, we performed nested PCR for 18S rDNA and the β-tubulin gene of B. microti, which has been regarded as the most probable cause of human babesiosis and has been detected in wild mice and other mammals in Korea [Citation46,Citation47]. When we sequenced full-length sequence of 18S rDNA of Babesia sp. [Citation37,Citation38], the sequence was identical to that of Babesia sp. KO1 (DQ346955) in Korea. Phylogenetic analysis, Babesia sp. KCDC-1 in our study formed a clade including several ovine Babesia spp. reported previously in China [Citation48]. Based on the analysis of 18S rDNA amplification from the collected ticks, two sequences of Babesia spp. KCDC-GT-270 and KCDCGT-272 were identical to that of Babesia sp. KO1 (DQ346955) and Babesia sp. Hebei-2005 (DQ159074). Previous studies confirmed that the occurrence of additional viable sites in COB and COX3 allowed for an improved interspecies differentiation of Babesia species [Citation22,Citation23]. The phylogenetic analysis for both COB and COX3 indicates that Babesia sp. KCDC-1 and KCDC-GT-270 were highly identical and formed a clade including ovine B. motasi Ningxian in China (JX440507) based on COB and ovine B. motasi Ningxian in China (JX866781) based on COX3. This indicated that the identified Babesia parasites might be B. motasi, and this is the first study to detect B. motasi in human babesiosis and H. longicornis in Korea.
The B. microti Group is a varied group of globally distributed parasites of various lineages, such as the USA, Kobe, Hobetsu, and Munich [Citation49,Citation50]. The major lineage of the B. microti Group, the US lineage, includes B. microti sensu stricto, a causal agent of human babesiosis in the northeastern and upper midwestern US, where most human cases worldwide have been reported [Citation9]. While pathogenicity may vary among parasite populations, the B. microti US lineage parasites are infectious to humans with or without a splenectomy. Recently, the US lineage parasites have been established to genetically vary in their β-tubulin gene sequences [Citation51]. Moreover, parasites of the US lineage are phylogenetically clustered into 3 different sub-lineages, North America, Europe-Central Asia, and East Asia, each of which reflects the geographic origin of the parasites [Citation9]. Previous reports from Korea have shown that B. microti detected in small mammals and wild animals closely aligned with the US strain [Citation46,Citation47,Citation51]. Similarly, in this study, the B. microti detected in H. longicornis were closely related with the US linage and clustered into East Asia with Korea8 from a mouse in Korea and Vladivostok63 from mice in Russia. This study is the first to detect B. microti, of the US lineage in H. longicornis in Korea.
Babesia parasites are transmitted by the bite of ticks that have distinct geographical distributions [Citation52]. Ixodes spp. ticks are common in the USA and Europe [Citation53,Citation54]. The primary vector of B. microti is I. scapularis. Other vectors of various strains of B. micorti include I. spinipalpis, I. angustus, I. muris, and I. ricinus. Furthermore, the vector of B. divergens and B. venatorum are I. ricinus and I. persulcatu, respectively [Citation14]. In Asia, tick-borne pathogens were detected in both Haemaphysalis and Ixodes ticks [Citation16,Citation55–57]. In particular, H. longicornis is the primary tick species in China and essentially serves as a vector for several pathogens that cause anaplasmosis, babesiosis, and rickettsiosis [Citation58]. In Korea, H. longicornis is the most commonly tick species infected with Babesia, collected from grass and vegetation [Citation59,Citation60]. The present results indicate that most of the collected ticks around the patient’s residence were H. longicornis and H. flava. Correspondingly, we confirmed that two of the H. longicornis were infected with B. motasi and the one was infected with B. microti. Recent reports indicate that H. longicornis is the potential carrier for B. microti to the vertebrate host and is assumed to serve as a tick vector of B. motasi-like isolates [Citation10]. The present results suggest that H. longicornis ticks may serve as the predominant vector of B. motasi and B. microti in Gangwon-do, Korea.
Acknowledgements
SHH, JRR, SHC and SEL conceived the study and designed the experiments. SHH performed the experiment. SYK and BGS performed tick collection and species identification. CRC, THU, CNK and YGK preformed the acquisition and interpretation of the clinical data. SHH and SEL analysed the data and wrote the manuscript.
Disclosure statement
No potential conflict of interest was reported by the authors.
Correction Statement
This article has been republished with minor changes. These changes do not impact the academic content of the article.
Additional information
Funding
References
- Vannier E, Krause PJ. Human babesiosis. N Engl J Med. 2012;366:2397–2407.
- Herwaldt BL, Linden JV, Bosserman E, et al. Transfusion-associated babesiosis in the United States: a description of cases. Ann Intern Med. 2011;155:509–519.
- Jones KE, Patel NG, Levy MA, et al. Global trends in emerging infectious diseases. Nature. 2008;451:990–993.
- Centers for Disease Control and Prevention. Babesiosis surveillance – 18 states, 2011. Morb Mortal Wkly Rep. 2012;61:505–509.
- Schnittger L, Rodriguez AE, Florin-Christensen M, et al. Babesia: a world emerging. Infect Genet Evol. 2012;12:1788–1809.
- Fang LQ, Liu K, Li X-L, et al. Emerging tick-borne infections in mainland China: an increasing public health threat. Lancet Infect Dis. 2015;15:1467–1479.
- Brasseur P, Gorenflot A. Human babesial infections in Europe. Rocz Akad Med Bialymst. 1996;41:117–122.
- Sun Y, Liu G, Yang L, et al. Babesia microti-like rodent parasites isolated from Ixodes persulcatus (Acari: Ixodidae) in Heilongjiang Province, China. Vet Parasitol. 2008;156:333–339.
- Zamoto-Niikura A, Morikawa S, Hanaki KI, et al. Ixodes persulcatus ticks as vectors for the Babesia microti U.S. lineage in Japan. Appl Environ Microbiol. 2016;82:6624–6632.
- Niu Q, Liu Z, Yang J, et al. Genetic diversity and molecular characterization of Babesia motasi-like in small ruminants and ixodid ticks from China. Infect Genet Evol. 2016;41:8–15.
- Persing DH, Herwaldt BL, Glaser C, et al. Infection with a babesia-like organism in northern California. N Engl J Med. 1995;332:298–303.
- Beattie JF, Michelson ML, Holman PJ. Acute babesiosis caused by Babesia divergens in a resident of Kentucky. N Engl J Med. 2002;347:697–698.
- Sonenshine DE, Michael RR. Biology of ticks. 2nd ed. New York (NY): Oxford University Press; 2014.
- Hunfeld KP, Hildebrandt A, Gray JS. Babesiosis: recent insights into an ancient disease. Int J Parasitol. 2008;38:1219–1237.
- Saito-Ito A, Tsuji M, Wei Q, et al. Transfusion-acquired, autochthonous human babesiosis in Japan: isolation of Babesia microti-like parasites with hu-RBC-SCID mice. J Clin Microbiol. 2000;38:4511–4516.
- Shih CM, Liu LP, Chung WC, et al. Human babesiosis in Taiwan: asymptomatic infection with a Babesia microti-like organism in a Taiwanese woman. J Clin Microbiol. 1997;35:450–454.
- Kim JY, Cho S-H, Joo H-N, et al. First case of human babesiosis in Korea: detection and characterization of a novel type of Babesia sp. (KO1) similar to ovine babesia. J Clin Microbiol. 2007;45:2084–2087.
- Saal JR. Giemsa stain for the diagnosis of bovine babesiosis. I. Staining properties of commercial samples and their component dyes. J Protozool. 1964;11:573–582.
- Ginsberg HS, Ewing CP. Comparison of flagging, walking, trapping, and collecting from hosts as sampling methods for northern deer ticks, Ixodes dammini, and lone-star ticks, Amblyomma americanum. Exp Appl Acarol. 1989;7:313–322.
- Zintl A, Finnerty EJ, Murphy TM, et al. Babesias of red deer (Cervus elaphus) in Ireland. Vet Res. 2011;42:7.
- Medlin L, Elwood HJ, Stickel S, et al. The characterization of enzymatically amplified eukaryotic 16S-like rRNA-coding regions. Gene. 1988;71:491–499.
- Tian Z, Luo J, Zheng J, et al. Phylogenetic analysis of Babesia species in China based on Cytochrome b (COB) gene. Infect Genet Evol. 2013;13:36–40.
- Tian Z, Liu G, Yin H, et al. Cytochrome c oxidase subunit III (COX3) gene, an informative marker for phylogenetic analysis and differentiation of Babesia species in China. Infect Genet Evol. 2013;18:13–17.
- Zamoto A, Tsuji M, Kawabuchi T, et al. US-type Babesia microti isolated from small wild mammals in Eastern Hokkaido, Japan. J Vet Med Sci. 2004;66:919–926.
- Tavassoli M, Tabatabaei M, Mohammadi M, et al. PCR-based detection of Babesia spp. infection in collected ticks from cattle in West and North-West of Iran. J Arthropod Borne Dis. 2013;7:132–138.
- Homer MJ, Aguilar-Delfin I, Telford SR 3rd, et al. Babesiosis. Clin Microbiol Rev. 2000;13:451–469.
- Talameh J, Misher A, Hoskins J. A capillary electrophoresis method for genotyping the 9-bp exon 1 insertion/deletion in BDKRB2. Pharmacogenomics. 2012;13:353–358.
- Kumar S, Nei M, Dudley J, et al. MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform. 2008;9:299–306.
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791.
- Leiby DA. Transfusion-transmitted Babesia spp.: bull’s-eye on Babesia microti. Clin Microbiol Rev. 2011;24:14–28.
- Leiby DA. Transfusion-associated babesiosis: shouldn’t we be ticked off? Ann Intern Med. 2011;155:556–557.
- Gubernot DM, Nakhasi HL, Mied PA, et al. Transfusion-transmitted babesiosis in the United States: summary of a workshop. Transfusion. 2009;49:2759–2771.
- Calvo de Mora A, Garcia Castellano JM, Herrera C, et al. Human babesiosis: report of a case with fatal outcome. Med Clin. 1985;85:515–516.
- Vannier E, Krause PJ. Update on babesiosis. Interdiscip Perspect Infect Dis. 2009;2009:984568.
- Centeno-Lima S, do Rosario V, Parreira R, et al. A fatal case of human babesiosis in Portugal: molecular and phylogenetic analysis. Trop Med Int Health. 2003;8:760–764.
- de Vos AJ, Dalgliesh RJ, Callow LL. Babesia. In: Soulsby EJL, editor. Immune responses in parasitic infections. Boca Raton, FL: CRC Press; 1987; Vol. III, Protozoa; p. 183–222.
- Wei Q, Tsuji M, Zamoto A, et al. Human babesiosis in Japan: isolation of Babesia microti-like parasites from an asymptomatic transfusion donor and from a rodent from an area where babesiosis is endemic. J Clin Microbiol. 2001;39:2178–2183.
- Yao LN, Wei R, Zeng CY, et al. Pathogen identification and clinical diagnosis for one case infected with Babesia. Zhongguo Ji Sheng Chong Xue Yu Ji Sheng Chong Bing Za Zhi. 2012;30:118–121.
- Ord RL, Lobo CA. Human babesiosis: pathogens, prevalence, diagnosis and treatment. Curr Clin Microbiol Rep. 2015;2:173–181.
- Gorenflot A, Moubri K, Precigout E, et al. Human babesiosis. Ann Trop Med Parasitol. 1998;92:489–501.
- Cursino-Santos JR, Halverson G, Rodriguez M, et al. Identification of binding domains on red blood cell glycophorins for Babesia divergens. Transfusion. 2014;54:982–989.
- Lobo CA, Rodriguez M, Cursino-Santos JR. Babesia and red cell invasion. Curr Opin Hematol. 2012;19:170–175.
- Lobo CA. Babesia divergens and Plasmodium falciparum use common receptors, glycophorins A and B, to invade the human red blood cell. Infect Immun. 2005;73:649–651.
- Khan N, editor. Emerging protozoan pathogens. London: Taylor and Francis; 2008; p. 303–349.
- Ramgopal Laha MD, Sen A. Morphology, epidemiology, and phylogeny of Babesia: an overview. Trop Parasitol. 2015;5:94–100.
- Hong SH, Kim H-J, Jeong Y-I, et al. Serological and molecular detection of Toxoplasma gondii and Babesia microti in the blood of rescued wild animals in Gangwon-do (Province), Korea. Korean J Parasitol. 2017;55:207–212.
- Hong SH, Lee S-E, Jeong Y-I, et al. Prevalence and molecular characterizations of Toxoplasma gondii and Babesia microti from small mammals captured in Gyeonggi and Gangwon Provinces, Republic of Korea. Vet Parasitol. 2014;205:512–517.
- Liu AH, Yin H, Guan GQ, et al. At least two genetically distinct large Babesia species infective to sheep and goats in China. Vet Parasitol. 2007;147:246–251.
- Goethert HK, Telford SR. What is Babesia microti? Parasitology. 2003;127:301–309.
- Tsuji M, Wei Q, Zamoto A, et al. Human babesiosis in Japan: epizootiologic survey of rodent reservoir and isolation of new type of Babesia microti-like parasite. J Clin Microbiol. 2001;39:4316–4322.
- Zamoto A, Tsuji M, Wei Q, et al. Epizootiologic survey for Babesia microti among small wild mammals in northeastern Eurasia and a geographic diversity in the beta-tubulin gene sequences. J Vet Med Sci. 2004;66:785–792.
- Spielman A, Wilson ML, Levine JF, et al. Ecology of Ixodes dammini-borne human babesiosis and Lyme disease. Annu Rev Entomol. 1985;30:439–460.
- Spielman A. Human babesiosis on Nantucket Island: transmission by nymphal Ixodes ticks. Am J Trop Med Hyg. 1976;25:784–787.
- Walter G. Transmission and course of parasitemia of Babesia microti (Hannover I strain) in the bank vole (Clethrionomys glareolus) and field vole (Microtus agrestis). Acta Trop. 1984;41:259–264.
- Uilenberg G. International collaborative research: significance of tick-borne hemoparasitic diseases to world animal health. Vet Parasitol. 1995;57:19–41.
- Fujisaki K. Development of acquired resistance precipitating antibody in rabbits experimentally infested with females of Haemaphysalis longicornis (Ixodoidea: Ixodidae). Natl Inst Anim Health Q (Tokyo). 1978;18:27–38.
- Ica A, Vatansever Z, Yildirim A, et al. Detection of Theileria and Babesia species in ticks collected from cattle. Vet Parasitol. 2007;148:156–160.
- Yongshuai P, Qi M, Jian F, et al. Molecular identification of tick-borne pathogens in tick Haemaphysalis longicornis from sheep in Henan, China. Turk J Vet Anim Sci. 2017;41:51–55.
- Kim CM, Yi Y-H, Yu D-H, et al. Tick-borne rickettsial pathogens in ticks and small mammals in Korea. Appl Environ Microbiol. 2006;72:5766–5776.
- Kang SW, Doan HTT, Choe SE, et al. Molecular investigation of tick-borne pathogens in ticks from grazing cattle in Korea. Parasitol Int. 2013;62:276–282.
