ABSTRACT
Adenoviruses (AdVs) are major contributors to clinical illnesses. Novel human and animal AdVs continue to be identified and characterized. Comparative analyses using bioinformatic methods and Omics-based technologies allow insights into how these human pathogens have emerged and their potential for host cross-species transmission. Systematic review of literature published across ProQuest, Pubmed, and Web of Science databases for evidence of adenoviral zoonotic potential identified 589 citations. After removing duplicates, 327 citations were screened for relevance; of which, 74 articles received full-text reviews. Among these, 24 were included here, of which 16 demonstrated evidence of zoonotic transmission of AdVs. These documented instances of AdV crossing host species barriers between humans and non-human primate, bat, feline, swine, canine, ovine, and caprine. Eight studies sought to but did not find evidence of zoonosis. The findings demonstrate substantial evidence suggesting AdVs have previously and will continue crossing host species barriers. These have human health consequences both in terms of novel pathogen emergence and epidemic outbreaks, and of appropriate and safe use of non-human adenoviruses for therapeutics. As routine human clinical diagnostics may miss a novel cross-species adenovirus infection in humans, next generation sequencing or panspecies molecular diagnostics may be necessary to detect such incursions.
Introduction
Human adenoviruses (HAdVs) were among the first human viral pathogens to be isolated and characterized with their near-simultaneous discoveries by Rowe, et al. in 1953 [Citation1] and Hilleman, et al. in 1954 [Citation2]. Subsequently, HAdVs have increasingly been recognized as a major contributor to clinical illnesses including pneumonia, upper and lower respiratory tract diseases, pharyngitis, bronchiolitis, meningitis, rhinorrhea, hemorrhagic cystitis, hepatitis, conjunctivitis, keratoconjuntivitis, and obesity with certain clinical diseases associated with specific adenoviral species and genotypes. With the genomics era, high-resolution sequence data have led to a better and more accurate view beyond the limited epsilon and gamma antigens used originally for serotyping [Citation3,Citation4]. These nonenveloped double-stranded DNA viruses are transmitted via aerosol particles or droplets, the fecal-oral route, the hand-ocular route, and fomites. HAdV infection may also occur upon reactivation from latency following organ transplantation and hematopoietic stem cell transplantation [Citation5].While illnesses associated with HAdV infection are typically asymptomatic or mild, infants, children, the elderly, and immunocompromised patients are at increased risk for severe disease and death [Citation6]. Furthermore, there have been instances of severe and highly contagious disease associated with several genotypes, including HAdV-4, HAdV-7, and HAdV-14, which have caused acute febrile respiratory disease among military trainees [Citation7–9]. HAdV-14 and other genotypes, including HAdV-3, HAdV-5, HAdV-7, HAdV-11, and HAdV-21, have also been associated with acute respiratory illness in healthy civilian populations [Citation10–13]. It should be noted that while one serotype of HAdV-11, genome typed as “HAdV-11a”, is associated with respiratory disease, all other HAdV-11 genome types are associated with renal disease; with whole genome data, the incongruity of “HAdV-11a”, i.e. a renal pathogen associated with respiratory disease, has been resolved, with the strain renamed genotype HAdV-55. Genomic and bioinformatic analyses indicate it is a “Trojan Horse” virus with a recombinant genome that serotypes as a renal pathogen, HAdV-11, but genotypes as HAdV-14, a respiratory pathogen [Citation14,Citation15] as it contains much of the genome chassis of parental HAdV-14 [Citation16]. HAdV-55 is a potent, circulating respiratory pathogen reported in several large epidemic outbreaks [Citation17,Citation18], illustrating the prominent role of genome recombination in the genesis of emergent adenoviral pathogens. Another example is HAdV-53, a recombinant exhibiting a “non-pathogen” HAdV-22 signature but is associated with highly contagious epidemic keratoconjunctivitis [Citation19]. Later in this report, a zoonotic human respiratory pathogen, HAdV-4, will be discussed in detail within the context of recombination providing a host adaptation to allow optimal viral replication in the new host [Citation20].
Within the genus Mastadenovirus, there were 51 serotypes that are now integrated into the 103 human AdV genotypes described in high resolution using genomics [Citation21]. These are parsed into seven species (A-G) that were originally distinguished by biological, clinical, and restriction enzyme digestion properties and have been reconfirmed using Omics data. In addition to humans, AdVs within the genus Mastadenovirus infect a wide range of mammalian hosts including bats, bovines, canines, deer, dolphins, equines, murines, non-human primates (NHPs), ovines, swine, sea lions, skunks, squirrels, and tree shrews. Further, there are AdVs in four other genera within the Adenoviridae family that infect avian, reptilian, and fish species- essentially covering all vertebrate species studied to date. As with humans, AdV infections in animals can cause diseases that range from asymptomatic to fatal.
In addition to increased attention to HAdVs due to infection-associated morbidity, the biochemical and clinical characteristics of the virus, including its stability and ability to induce innate and adaptive immune responses in mammals, have made AdVs particularly attractive vectors for vaccines and gene therapy. The use of non-human simian AdVs (SAdVs) [Citation22] to circumvent prior exposure associated immunity in humans is of importance. A recent review of HAdV seroprevalence studies reports that by 2018 there were hundreds of vaccine, gene therapy, and cancer trials using HAdV-based vectors [Citation23].
Given that humans and animals are known hosts to AdVs, it is reasonable to consider the possibility of zoonotic or cross-species transmission of AdVs. Hence, we sought to review literature surrounding the zoonotic potential of adenoviruses. For the purposes of this systematic review, “zoonoses” was considered using a One Health definition, as a “two-way street, with humans infecting animals as well as the other way round” [Citation24]. In instances where we sought to specify the directionality of transmission, we used the term “anthropozonoosis” to describe a pathogen moving from animals to humans, and the term “zooanthroponosis” to describe a pathogen moving from humans to animals. Directionality was assigned empirically based on the species of the host identified in the study and the natural host of the virus in question given the available serologic or virologic data.
Methods
On November 8, 2018, we conducted a search of literature for evidence that AdV can be transmitted between humans and other animals through ProQuest, Pubmed, and Web of Science. This search followed the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines. To be captured in the search, the terms “adenovirus” (or genus members including “mastadenovirus”, “atadenovirus”, “aviadenovirus”, “ichtadenovirus”, and “siadenovirus”) along with a term implying transmission of virus between humans and other animals in either direction (e.g. “cross-species”, “interspecies”, “zoonoses”, “anthropozoonoses”, etc) must have been included in the text. The complete search string used was, “((adenovirus* OR mastadenovirus OR atadenovirus OR aviadenovirus OR ichtadenovirus OR siadenovirus) AND (cross-species OR interspecies OR zoono* OR anthropono* OR anthropozoono* OR zooanthropono*)).” Citations were exported from the three databases into Endnote, where duplicates were removed. All abstracts and titles were screened, and relevant articles available in English or French were selected for a full review if they mentioned adenovirus and language indicating an investigation of evidence of cross-species virus transmission. At each step, articles were screened separately by J.K.F. and L.K.B. and then reviewed and discussed until the authors were in agreement with respect to interpretation.
Results
Search results
The initial search generated 589 citations with 221 through ProQuest, 228 through Pubmed, and 140 through Web of Science (). After removing duplicates, the titles and abstracts of the remaining 327 articles were screened for relevance to receive a full review. Following the title and abstract review, 74 articles received a full-text review. Among these, 24 articles were selected for inclusion in the review (). Sixteen articles (67%) successfully demonstrated evidence of zoonoses and eight articles (33%) sought but failed to demonstrate evidence of zoonoses.
Figure 1. Diagram of article screening process for a systematic review of evidence of adenovirus transmission between humans and other animals. The search captured 327 articles from 3 databases, 24 of which were included in the final review.
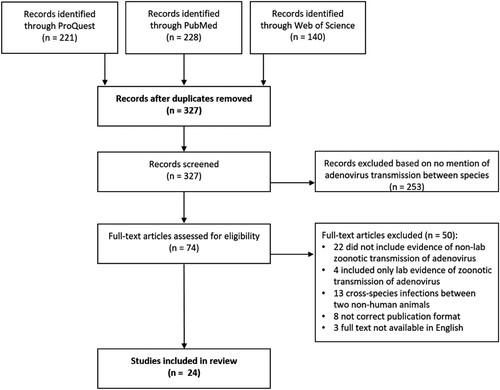
Table 1. Publications considered important in evaluating the zoonotic potential of adenoviruses
Strength of evidence
Of the 24 studies, 18 (75%) provided phylogenetic evidence, four (17%) provided serological evidence, one provided both phylogenetic and serological data, and one relied on molecular evidence supporting or refuting the zoonotic potential of the viruses. Of the studies presenting phylogenetic evidence of zoonoses, three demonstrated interspecies recombination of HAdVs and simian AdVs (SAdVs) [Citation20,Citation25,Citation35] and one found evidence of an intraspecies recombination between HAdV-B detected in wild gorillas and chimpanzees [Citation36]. Ten (10) studies used genomic sequencing to identify human or non-human AdV species in biological samples.
The study by Chen, et al. provided both phylogenetic evidence using whole-genome sequencing and serological evidence of human infections with a titi monkey adenovirus (TMAdV) producing arguably the strongest evidence of a anthropozoonotic transmission of adenoviruses [Citation31]. In this study, a researcher at a California primate centre who had been in closest contact with a monkey infected with TMAdV developed an acute respiratory illness and had a convalescent serum sample that was seropositive for TMAdV [antibody (Ab) titre 1:32]. Subsequently, two members of this researcher’s family developed influenza-like-illnesses one to two weeks after the researcher initially fell ill; and a year later, one of those family members was also found positive for neutralizing antibodies to TMAdV (Ab titre 1:8). The TMAdV was a novel adenovirus, distinct from all species A-G HAdVs and with only 54% to 56.3% identity with the closest SAdV relatives, SAdV-3, -18, and -21.
Another report supporting anthropozoonotic transmission of adenoviruses to humans from primates noted a 1997 outbreak of acute respiratory illness in baboons at a primate research facility in Texas. In this outbreak, six novel baboons adenoviruses (BaAdVs) comprising a proposed novel “simian adenovirus C (SAdV-C)” species were isolated from asymptomatic and sick baboons, including two fatalities. Although no humans showed symptoms, antibodies to these new BaAdVs were found in staff personnel at the facility, as well as in other baboons [Citation34].
DNA sequencing data show genomic “footprints” of cross-species transmission as two nearly identical SAdVs, isolated independently from a chimpanzee at the New Iberia Research Center (SAdV-B35.1; University of Louisiana; Lafayette, LA) and a bonobo in a zoological setting (SAdV-B35.2; San Diego Zoo; San Diego, CA) by Roy, et al. [Citation29], contain recombinant genomes with contributions from SAdV-21 (chimpanzee), HAdV-21(human), SAdV-27.1 (chimpanzee), and HAdV-16 (human) [Citation35]. Interestingly, an emergent human pathogen, HAdV-76, with a nearly identical genome [Citation40] to both SAdV-35.1 and -35.2 was associated with a respiratory disease fatality in Texas in 1967 [Citation49].
Evidence of anthropozoonoses
We identified six studies (25%) demonstrating evidence of anthropozoonoses, five (21%) of which present transmission of AdVs from NHPs to humans. In one of these studies, Xiang et al. presented serological evidence of anthropozoonotic transmission of AdV to NHPs from five countries across three continents [Citation28]. The study found humans in Côte D`Ivoire, Cameroon, and Nigera with neutralizing antibodies for several chimpanzee-derived AdV-C; however, none of the 50 humans working in U.S. zoos in close contact with primates had detectable antibodies.
Significantly, one of the first human viral pathogens isolated and characterized in 1953 as cytopathogenic respiratory illness agent RI-67 [Citation2], later renamed as HAdV-4, has been shown to contain a genome with near identity to several chimpanzee adenoviruses [Citation20,Citation26], which suggests anthropozoonosis. Prior to the Omics era, it was speculated that this unique and odd human adenovirus, HAd4, was more closely related to the few chimpanzee adenoviruses characterized and was grouped in its own taxonomic clade, as species E [Citation50]. It was thought to be the “archetype”, having limited amino acid and nucleic acid sequence similarities to human adenoviruses of species B and C [Citation50,Citation51]. Additional and more whole genome DNA sequences from the dataset including all human adenovirus genotypes and more simian adenoviruses [Citation20,Citation26] suggest a chimpanzee origin for this still-significant human pathogen [Citation52]. A long-standing curiosity of HAdV-4 was its apparent host range limitation to the U.S. military basic trainee population [Citation52]. A study by Dehghan et al. found that two recent HAdV-4 isolates from U.S. military trainees shared genome near identities with five simian AdV genomes isolated from chimpanzees [Citation20]. Furthermore, unlike the AdV-4 prototype and vaccine field strains [Citation26,Citation53], these recent circulating isolates contain a modified Inverted Terminal Repeat (ITR) sequence [Citation20], apparently recombined from a parental human adenovirus, that provides all three viral replication factor binding sites that have been reported as required for optimal replication in human cells [Citation54–58]. HAdV-4 outbreaks have been recently reported globally and in civilian populations [Citation59,Citation60]; DNA sequencing reveals the recombinant ITR [Citation59].
Another study by Phan et al. reported RT–PCR detection of feline AdV in a one-year old child hospitalized for acute gastroenteritis [Citation27]. Through sequencing, the specimen was identified as HAdV with 100% amino acid sequence identity in seven hypervariable regions of the hexon gene and 97% in fibre genes (BLAST and CLUSTAL X).
Evidence of zooanthroponoses
The review found eight studies (33%) demonstrating evidence of zooanthroponoses, including six studies (25%) that provided evidence of transmission of AdVs from humans to NHPs. Across those six studies, the most common HAdV species detected in NHPs were HAdV-B, -C, -E, and -F, though the global study of fecal, blood and tissue samples from wild and captive NHPs conducted by Wevers et al. found all HAdVs species A-G [Citation30].
Baker et al. isolated a novel AdV from bat urine in Ghana and sequenced the hexon gene [Citation33]. This Eidolon helvum or fruit bat AdV-1 hexon forms a phylogenetic clade separate from previously identified bat AdVs, and is distinct in that it clusters with the HAdV clade. The high bootstrap value of 93 in their phylogenetic analysis indicates strong support that will likely be preserved with additional sampling. It was noted by Baker et al. that the hexon shared 77% and 90% amino acid similarities with the HAdV counterpart across 58 and 63 amino acids, respectively, of the 900 amino acid hexon protein [Citation33].
Additionally, Pauly et al. detected HAdV in fecal samples collected from pigs, dogs, sheep, and goats in Côte d’Ivoire [Citation38]. Even more intriguing is the isolation of a HAdV-1-like virus from cats, following a large sample screening of domestic cats using a HAdV-1 hexon antigen and ELISA in the mid-1990s by Ongradi et al. [Citation61]. Sequence determination of the feline hexon and fibre genes showed near-identity to the human counterparts [Citation62]. This virus was reported to replicate in both human and monkey cells in vitro [Citation61], which was updated recently (2019) to include a screen of various permissive and non-permissive human and other animal cells, along with additional immunochemical analyses [Citation62].
Studies Lacking evidence of zoonoses
The review captured eight studies which did not find evidence of the zoonotic potential of AdVs. Of those studies, four specifically sought to find evidence of zoonanthroponoses and three specifically sought to find evidence of anthropozoonoses. Five studies provided phylogenetic evidence characterizing fecal samples from bats, rodents or NHPs, determining that the AdVs detected were genetically distinct from HAdVs. One negative study, Wang et al., was premised on the absence of molecular presence of AdV [Citation47]. The Nkogue et al. study of western lowland gorillas and humans in Gabon concluded detected HAdV-C in both hosts. However, characterization of the HAdV-C species detected in the human and gorilla samples were genetically distinct, leading Nkogue and colleagues to interpret this as a lack of evidence for zoonoses [Citation45].
Discussion
Unlike our previously published systematic review of the zoonotic potential of enteroviruses (EVs) [Citation63], in which nearly half of the publications included in the final review were clustered in Central Africa, the studies included in this review varied geographically. Of the 16 publications demonstrating phylogentic and/or serological evidence of zoonotic adenovirus transmission, four studies were conducted globally with samples spanning three or more continents, four were conducted across several countries in West and Central Africa, six studies were conducted in the United States, one study was conducted in Japan, and one in Thailand.
Of the negative studies, five were conducted in the United States, and one each in China, Gabon, and Tanzania. While this regional distribution may suggest the zoonotic potential of adenoviruses is not geographically isolated or clustered, it is important to note that the two U.S.-based studies demonstrating zooanthroponosis were based in primate facilities [Citation31,Citation34]. Additionally, the serological study by Xiang et al. examining sera from humans and chimpanzees in Cameroon, Côte D`Ivoire, Nigeria, Thailand, and the United States found neutralizing antibodies in human serum to chimpanzee-derived antibodies were more prevalent in humans living in sub-Saharan Africa than humans living in Thailand and the United States [Citation28].
The majority of the studies identified as providing evidence for zoonotic transmission of AdVs reported species crossing between humans and non-human primates. In addition to these 13 reports, a recent study that was published after the search date for this review by Dehghan et al. unearthed evidence of a unique “ping-pong” pattern of AdV transmission between humans, chimpanzees, and bonobos when conducting genomics analyses of archived AdV sequences [Citation40]. Of note, an emergent recombinant adenovirus typed by the authors, HAdV-B76, was associated with a human fatality in 1965.
Additional relevant studies were published following the initial review in November 2018, including the previously cited Dehghan et al. study [Citation40], Ongrádi et al. study of feline AdV [Citation62], the Zhang et al. study of HAdV-E4 circulating in Hong Kong [Citation59].
This systematic review had several limitations. While a number of studies with minimal or no evidence of zoonotic adenovirus transmission were identified, these negative studies may have been more likely to be missed in this search. Additionally, our search strategy excluded studies with evidence of cross-species infections among nonhuman animals. Indeed, of the 50 articles excluded following the full-text review, 13 were excluded from the final analysis as they strictly investigated cross-species infections between two non-human animals.
Conclusions
Given the evidence in this review, it is apparent that AdVs have crossed, and will continue to cross host species. This has been especially true for the species barrier between humans and NHPs (and likely between different NHP species), but it may also occur between humans and other animal species. Humans with intense exposure to NHPs, such as though occupational exposure at primate facilities, may be at higher risk of becoming infected with a zoonotic adenovirus. Routine clinical diagnostics are not likely to detect a novel animal adenovirus infection in humans. Hence, clinicians must have a strong degree of suspicion and apply next-generation sequencing and/or panspecies diagnostics (not yet commercially available) in detecting such a novel strain. Wellehan et al [Citation64] have reported a conventional panspecies PCR approach, which coupled with limited sequencing, may be so employed.
Disclosure Statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Rowe WP, Huebner RJ, Gilmore LK, et al. Isolation of a cytopathogenic agent from human adenoids undergoing spontaneous degeneration in tissue culture. Proc Soc Exp Biol Med. 1953;84(3):570–573.
- Hilleman MR, Werner JH. Recovery of new agent from patients with acute respiratory illness. Proc Soc Exp Biol Med. 1954;85(1):183–188.
- Seto D, Chodosh J, Brister JR, et al. Using the whole-genome sequence to characterize and name human adenoviruses. J Virol. 2011;85(11):5701–5702.
- Seto D, Jones MS, Dyer DW, et al. Characterizing, typing, and naming human adenovirus type 55 in the era of whole genome data. J Clin Virol. 2013;58(4):741–742.
- Garnett CT, Jones MS, Dyer DW, et al. Latent species C adenoviruses in human tonsil tissues. J Virol. 2009;83(6):2417–2428.
- Lion T. Adenovirus infections in immunocompetent and immunocompromised patients. Clin Microbiol Rev. 2014;27(3):441–462.
- Dudding BA, Wagner SC, Zeller JA, et al. Fatal pneumonia associated with adenovirus type 7 in three military trainees. N Eng J Med. 1972;286(24):1289–1292.
- Barraza EM, Ludwig SL, Gaydos JC, et al. Reemergence of adenovirus type 4 acute respiratory disease in military trainees: report of an outbreak during a lapse in vaccination. J Infec Dis. 1999;179(6):1531–1533.
- Centers for Disease Control and Prevention. Acute respiratory disease associated with adenovirus serotype 14 - four states, 2006-2007. Morb Mortal Wkly Rep. 2007;56(45):1181–1184.
- Centers for Disease Control and Prevention. Civilian outbreak of adenovirus acute respiratory disease–south Dakota, 1997. Morb Mortal Wkly Rep. 1998;47(27):567.
- Gray GC, McCarthy T, Lebeck MG, et al. Genotype prevalence and risk factors for severe clinical adenovirus infection, United States 2004–2006. Clin Infec Dis. 2007;45(9):1120–1131.
- Zhao S, Wan C, Ke C, et al. Re-emergent human adenovirus genome type 7d caused an acute respiratory disease outbreak in Southern China after a twenty-one year absence. Sci Rep. 2014;4:7365.
- Killerby ME, Rozwadowski F, Lu X, et al. Respiratory illness associated with emergent human adenovirus genome type 7d, New Jersey, 2016-2017. Open Forum Infect Dis. 2019;6(2):ofz017.
- Seto J, Walsh MP, Mahadevan P, et al. Genomic and bioinformatics analyses of HAdV-14p, reference strain of a re-emerging respiratory pathogen and analysis of B1/B2. Virus Res. 2009;143(1):94–105.
- Carr MJ, Kajon AE, Lu X, et al. Deaths associated with human adenovirus-14p1 infections, Europe, 2009-2010. Emerg Infec Dis. 2011;17(8):1402–1408.
- Walsh MP, Seto J, Jones MS, et al. Computational analysis identifies human adenovirus type 55 as a re-emergent acute respiratory disease pathogen: FIG. 1. J Clin Microbiol. 2010;48(3):991–993.
- Lu QB, Tong Y-G, Wo Y, et al. Epidemiology of human adenovirus and molecular characterization of human adenovirus 55 in China, 2009-2012. Influenza Other Resp. 2014;8(3):302–308.
- Yang Z, Zhu Z, Tang L, et al. Genomic analyses of recombinant adenovirus type 11a in China. J Clin Microbiol. 2009;47(10):3082–3090.
- Walsh MP, Chintakuntlawar A, Robinson CM, et al. Evidence of molecular evolution driven by recombination events influencing tropism in a novel human adenovirus that causes epidemic keratoconjunctivitis. PLoS ONE. 2009;4(6):e5635.
- Dehghan S, Seto J, Liu EB, et al. Computational analysis of four human adenovirus type 4 genomes reveals molecular evolution through two interspecies recombination events. Virology. 2013;443(2):197–207.
- Human Adenovirus Working Group [Internet]. [cited 2019 Jul 30]. Available from: http://hadvwg.gmu.edu/.
- Roy S, Gao G, Lu Y, et al. Characterization of a family of chimpanzee adenoviruses and development of molecular clones for gene transfer vectors. Hum Gene Ther. 2004;15(5):519–530.
- Mennechet FJD, Paris O, Ouoba AR, et al. A review of 65 years of human adenovirus seroprevalence. Expert Rev Vaccines. 2019;18(6):597–613.
- Evans B, Leighton F. A history of One Health. Rev Sci Tech. 2014;33(2):413–420.
- Madisch I, Harste G, Pommer H, et al. Phylogenetic analysis of the main neutralization and hemagglutination determinants of all human adenovirus prototypes as a basis for molecular classification and taxonomy. J Virol. 2005;79(24):15265–15276.
- Purkayastha A, Ditty SE, Su J, et al. Genomic and bioinformatics analysis of HAdV-4, a human adenovirus causing acute respiratory disease: Implications for gene therapy and vaccine vector development. J Virol. 2005;79(4):2559–2572.
- Phan TG, Shimizu H, Nishimura S, et al. Human adenovirus type 1 related to feline adenovirus: evidence of interspecies transmission. Clin Lab. 2006;52(9-10):515–518.
- Xiang ZQ, Li Y, Cun A, et al. Chimpanzee adenovirus antibodies in humans, sub-Saharan Africa. Emerg Infec Dis. 2006;12(10):1596–1599.
- Roy S, Vandenberghe LH, Kryazhimskiy S, et al. Isolation and characterization of adenoviruses persistently shed from the gastrointestinal tract of non-human primates. PLoS Pathog. 2009;5(7):e1000503.
- Wevers D, Metzger S, Babweteera F, et al. Novel adenoviruses in wild primates: a high level of genetic diversity and evidence of zoonotic transmissions. J Virol. 2011;85(20):10774–10784.
- Chen EC, Yagi S, Kelly KR, et al. Cross-species transmission of a novel adenovirus associated with a fulminant pneumonia outbreak in a new world monkey colony. PLoS Pathog. 2011;7(7):e1002155.
- Roy S, Sandhu A, Medina A, et al. Adenoviruses in fecal samples from asymptomatic rhesus macaques, United States. Emerg Infec Dis. 2012;18(7):1081–1088.
- Baker KS, Leggett RM, Bexfield NH, et al. Metagenomic study of the viruses of African straw-coloured fruit bats: detection of a chiropteran poxvirus and isolation of a novel adenovirus. Virol. 2013;441(2):95–106.
- Chiu CY, Yagi S, Lu X, et al. A novel adenovirus species associated with an acute respiratory outbreak in a baboon colony and evidence of coincident human infection. Mbio. 2013;4(2):e00084–13.
- Dehghan S, Seto J, Jones MS, et al. Simian adenovirus type 35 has a recombinant genome comprising human and simian adenovirus sequences, which predicts its potential emergence as a human respiratory pathogen. Virol. 2013;447(1-2):265–273.
- Hoppe E, Pauly M, Gillespie TR, et al. Multiple cross-species transmission events of human adenoviruses (HAdV) during hominine evolution. Mol Biol Evol. 2015;32(8):2072–2084.
- Hoppe E, Robbins M, Boji Mungu-Akonkwa D-d, et al. Phylogenomic evidence for recombination of adenoviruses in wild gorillas. J Gen Virol. 2015;96(10):3090–3098.
- Pauly M, Akoua-Koffi C, Buchwald N, et al. Adenovirus in rural Côte D`Ivoire: high diversity and cross-species detection. EcoHealth. 2015;12(3):441–452.
- Sukmak M, Wajjwalku W, Ostner J, et al. A first report of non-invasive adenovirus detection in wild Assamese macaques in Thailand. Primates. 2017;58(2):307–313.
- Dehghan S, Seto J, Liu EB, et al. A zoonotic adenoviral human pathogen emerged through genomic recombination amongst human and nonhuman simian hosts. J Virol. 2019;93(18):e00564-19.
- Miller LT, Yates VJ. Reactions of human sera to avian adenoviruses and Newcastle disease virus. Avian Dis. 1971;15(4):781–788.
- Kayali G, Ortiz EJ, Chorazy ML, et al. Lack of evidence of avian adenovirus infection among Turkey workers. J Agromedicine. 2009;14(3):299–305.
- Li L, Victoria JG, Wang C, et al. Bat guano virome: predominance of dietary viruses from insects and plants plus novel mammalian viruses. J Virol. 2010;84(14):6955–6965.
- Phan TG, Kapusinszky B, Wang C, et al. The fecal viral flora of wild rodents. Plos Pathog. 2011;7(9):e1002218.
- Nkogue CN, Horie M, Fujita S, et al. Molecular epidemiological study of adenovirus infecting western lowland gorillas and humans in and around Moukalaba-Doudou National Park (Gabon). Virus Genes. 2016;52(5):671–678.
- Zheng XY, Qiu M, Chen H-f, et al. Molecular detection and phylogenetic characterization of bat and human adenoviruses in Southern China. Vector Borne Zoonotic Dis. 2016;16(6):423–427.
- Wang X, Anderson BD, Pulscher LA, et al. Epidemiological study of people living in rural North Carolina for novel respiratory viruses. Zoonoses Public Health. 2018;65(1):e265–e269.
- Dadáková E, Brožová K, Piel AK, et al. Adenovirus infection in savanna chimpanzees (Pan troglodytes schweinfurthii) in the Issa Valley, Tanzania. Arch Virol. 2018;163(1):191–196.
- Crandell RA, Dowdle WR, Holcomb TM, et al. A fatal illness associated with two viruses: An intermediate adenovirus type(21-16) and influenza A2. J Pediatr. 1968;72(4):467–473.
- Wadell G, Hammarskjöld M-L, Winberg G, et al. Genetic variability of adenoviruses. Ann N Y Acad Sci. 1980;354:16–42.
- Gruber WC, Russell DJ, Tibbetts C. Fiber gene and genomic origin of human adenovirus type 4. Virology. 1993;196(2):603–611.
- Graham FL, Prevec L. Adenovirus-based expression vectors and recombinant vaccines. Biotechnol. 1992;20:363–390.
- Purkayastha A, Su J, McGraw J, et al. Genomic and bioinformatics analyses of HAdV-4vac and HAdV-7vac, two human adenovirus (HAdV) strains that constituted original prophylaxis against HAdV-related acute respiratory disease, a reemerging epidemic disease. J Clin Microbiol. 2005;43(7):3083–3094.
- Mul YM, Verrijzer CP, van der Vliet PC. Transcription factors NFI and NFIII/oct-1 function independently, employing different mechanisms to enhance adenovirus DNA replication. J Virol. 1990;64(11):5510–5518.
- Stillman BW, Topp WC, Engler JA. Conserved sequences at the origin of adenovirus DNA replication. J Virol. 1982;44(2):530–537.
- Rosenfeld PJ, Kelly TJ. Purification of nuclear factor I by DNA recognition site affinity chromatography. J Biol Chem. 1986;261(3):1398–1408.
- Rosenfeld PJ, O'Neill EA, Wides RJ, et al. Sequence-specific interactions between cellular DNA-binding proteins and the adenovirus origin of DNA replication. Mol Cell Biol. 1987;7(2):875–886.
- Nagata K, Guggenheimer RA, Enomoto T, et al. Adenovirus DNA replication in vitro: identification of a host factor that stimulates synthesis of the preterminal protein-dCMP complex. Proc Natl Acad Sci USA. 1982;79(21):6438–6442.
- Zhang J, Kang J, Dehghan S, et al. A survey of recent adenoviral respiratory pathogens in Hong Kong reveals emergent and recombinant human adenovirus type 4 (HAdV-e4) circulating in civilian populations. Viruses. 2019;11(2):129.
- Li J, Lu X, Sun Y, et al. A swimming pool-associated outbreak of pharyngoconjunctival fever caused by human adenovirus type 4 in Beijing, China. Int J Infec Dis. 2018;75:89–91.
- Ongradi J. Identification of a feline adenovirus isolate that replicates in monkey and human cells in vitro. Am J Vet Res. 1999;60(12):1463.
- Ongradi J, Chatlynne LG, Tarcsai KR, et al. Adenovirus isolated from a cat is related to human adenovirus 1. Front Microbiol. 2019;10:1430.
- Fieldhouse JK, Wang X, Mallinson KA, et al. A systematic review of evidence that enteroviruses may be zoonotic. Emerg Microbes Infec. 2018;7(1):164.
- Wellehan JF, Johnson AJ, Harrach B, et al. Detection and analysis of six lizard adenoviruses by consensus primer PCR provides further evidence of a reptilian origin for the atadenoviruses. J Virol. 2004;78(23):13366–13369.
