The recently described Rickettsia felis is an emerging human pathogen causing flea-borne spotted fever [Citation1]. Although found in a wide range of arthropods including fleas, ticks, mites, and lice, the cat flea (Ctenocephalides felis) is currently believed to be the most likely primary vector of R. felis. There is growing evidence, however, that mosquitoes might be involved in R. felis transmission with the organism having been identified in a wide variety of mosquitoes in Africa and China [Citation1,Citation2]; mouse-model experiments have indicated transmission of R. felis by Anopheles gambiae [Citation3]; there is an association between malaria and flea-borne spotted fever cases in Africa [Citation2]. Although R. felis has been demonstrated in a variety of mammals and arthropods in the USA, there is only one study on its presence in mosquitoes [Citation4]. The organism was not identified by PCR in pools of Culex quinquefasciatus, Aedes albopictus, Culex pipiens complex, Anopheles punctipennis and Anopheles crucians from Georgia [Citation4].
To further investigate R. felis in mosquitoes in the USA, we studied 560 unfed adult mosquitoes trapped with CDC miniature light traps (John W. Hock, Gainesville, FL) throughout October 2019 on the campus of Auburn University College of Veterinary Medicine, Alabama. The mosquitoes were identified morphologically and with a PCR targeting the mitochondrial cytochrome c oxidase subunit [Citation5] before being pooled (n = 57; 4 to14 per pool) according to species, sex, trap number and collection site. After washing (once in PBS for one minute; once in 70% ethanol for ten minutes; four times, one minute each, in sterile PBS) to remove surface contaminants, DNA was extracted for three previously published and validated PCRs, a gltA-based FRET-PCR [Citation3], a nested-PCR targeting the gltA of Rickettsia [Citation3], and a R. felis species-specific BioB-based PCR [Citation4], which were performed to test for the presence of Rickettsia DNA in mosquitoes.
Nine percent (5/57) of the mosquito pools, including An. punctipennis (3/6), Aedes vexans (1/4) and Uranotaenia sapphirina (1/3), were positive by PCR, in each case with all three PCRs. One of the positive An. punctipennis, one of the positive Aedes vexans and the positive U. sapphirina pool contained only male mosquitoes. The remaining pools were negative: Culex erraticus (5), Culex nigripalpus (8), Culex coronator (2), Culex conspirator (9), Culex tarsalis (1), Cx. pipiens (13), Culex territans (1), Culex restuans (2), Cx. quinquefasciatus (2) and Ae. albopictus (1). The 120-bp nucleotide sequences of the five mosquito pools positive in R. felis-specific BioB-based PCRs were identical to one another, and to that of R. felis URRWXCal2 (CP000053.1). There was only a single base pair difference amongst the 446-bp nucleotide sequences of the positive gltA-based PCRs which were 99.7–100% identical to recognized R. felis strains in GenBank, and 84.0–95.7% identical to other Rickettsia spp. (). Most closely related organisms were Rickettsia lusitaniae, Rickettsia hoogstraalii and Candidatus Rickettsia senegalensis with which R. felis is known to cluster [Citation1].
Figure 1. Phylogenetic tree using a bootstrap analysis for the Rickettsia felis found in mosquitoes from USA. The 446-bp nucleotide sequences of the gltA PCR products were concatenated and aligned using CLUSTALW, and the phylogenetic inferences were obtained from a maximum likelihood analysis. The names of Rickettsia species and their GenBank accession numbers are provided. The numbers at the nodes are the bootstrap values obtained by repeating the analysis 100 times to generate a majority consensus tree. The sequences of R. felis identified in this study (in bold) were 99.7–100% identical to the recognized R. felis strains, and 84.0–95.7% identical to other Rickettsia spp. The bootstrap values < 80 were omitted in the phylogenetic tree.
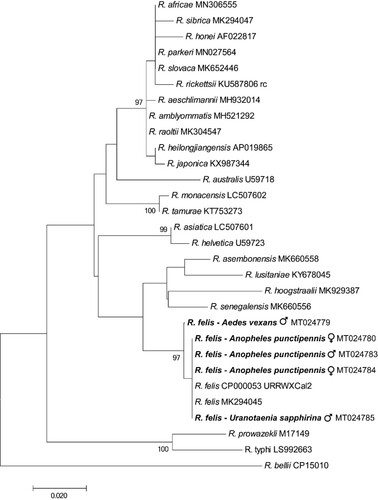
Our results show R. felis occurs in mosquitoes in the USA and adds An. punctipennis, Ae. vexans and U. sapphirina to the mosquitoes known to harbour the organism. Further more extensive studies are needed to determine the range of mosquitoes harbouring R. felis across the USA. It is noteworthy that one positive pool of An. punctipennis, one positive pool of Ae. vexans and one positive pool of U. sapphirina contained only male mosquitoes. As males do not take blood meals, it appears likely the infections were congenital and vertical transmission occurs in these mosquito species.
Opossums are the probable main mammalian reservoirs of R. felis in endemic areas in the USA [Citation4] while dogs have been implicated elsewhere in the world [Citation6]. None of the mosquitoes we found positive for R. felis have a reported tendency to feed on these species. An. punctipennis occurs widely in the eastern USA and feeds mostly on mammals, especially deer and sheep, but also on birds and people [Citation7]. The cosmopolitan Ae. vexans feeds on people and other mammals, especially deer, cattle, horses, rabbits, sheep, and dogs [Citation8]. U. sapphirina is found in the eastern, central and southern US, and is the only mosquito known to feed on invertebrates, mainly earthworms and leeches [Citation9]. Our finding that this species was infected with R. felis is of note as leeches have previously been suggested to be vectors of the organism [Citation10].
There is still much to be understood about the vector and reservoir role of the wide range of arthropods that harbour R. felis. The growing reports of R. felis occurring in mosquito species around the world and the known role of mosquitoes in transmitting a wide range of very important human and animal pathogens indicate an urgent need for further studies to determine the role mosquitoes might play in the epidemiology of R. felis infections in people. Further, since R. felis might play a role in parthenogenesis in arthropods [Citation1], its role in the biology of mosquitoes would thus also appear to warrant detailed investigation.
Acknowledgments
This research was funded in part a USDA-ARS program (58-6040-9-017), and by Alabama Agricultural Experimental Station and the USDA National Institute of Food and Agriculture, Hatch project (ALA052-1-17026).
Additional information
Funding
References
- Angelakis E, Mediannikov O, Parola P, et al. Rickettsia felis: the complex journey of an emergent human pathogen. Trends Parasitol. 2016;32:554–564. doi: 10.1016/j.pt.2016.04.009
- Mediannikov O, Abat C, Sokhna C, et al. Parallel decline of malaria and Rickettsia felis infections in Senegal. Am J Trop Med Hyg. 2018;99:360–361. doi: 10.4269/ajtmh.17-0194
- Zhang J, Lu G, Li J, et al. Molecular detection of Rickettsia felis and Rickettsia bellii in mosquitoes. Vector Borne Zoonotic Dis. 2019;19:802–809. doi: 10.1089/vbz.2019.2456
- Anderson ML, Rustin RC, Eremeeva ME. Pilot survey of mosquitoes (Diptera: Culicidae) from southeastern Georgia, USA for Wolbachia and Rickettsia felis (Rickettsiales: Rickettsiaceae). J Vector Borne Dis. 2019;56:92–97. doi: 10.4103/0972-9062.263714
- Folmer O, Black M, Hoeh W, et al. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol. 1994;3:294–299.
- Moonga LC, Hayashida K, Nakao R, et al. Molecular detection of Rickettsia felis in dogs, rodents and cat fleas in Zambia. Parasit Vectors. 2019;12:168. doi: 10.1186/s13071-019-3435-6
- Molaei G, Farajollahi A, Armstrong PM, et al. Identification of bloodmeals in Anopheles quadrimaculatus and Anopheles punctipennis from eastern equine encephalitis virus foci in northeastern USA. Med Vet Entomol. 2009;23:350–356. doi: 10.1111/j.1365-2915.2009.00838.x
- Greenberg JA, Lujan DA, DiMenna MA, et al. Identification of blood meal sources in Aedes vexans and Culex quinquefasciatus in Bernalillo County, New Mexico. J Insect Sci. 2013;13:75. doi: 10.1673/031.013.7501
- Reeves LE, Holderman CJ, Blosser EM, et al. Identification of Uranotaenia sapphirina as a specialist of annelids broadens known mosquito host use patterns. Commun Biol. 2018;1:92. doi: 10.1038/s42003-018-0096-5
- Slesak G, Inthalath S, Dittrich S, et al. Leeches as further potential vectors for rickettsial infections. Proc Natl Acad Sci USA. 2015;112:E6593–4. doi: 10.1073/pnas.1515229112
