ABSTRACT
Tigecycline is one of the last-resort antibiotics to treat severe infections. Recently, tigecycline resistance has sporadically emerged with an increasing trend, and Tet(X) family represents a new resistance mechanism of tigecycline. In this study, a novel chromosome-encoded tigecycline resistance gene, tet(X14), was identified in a tigecycline-resistant and colistin-resistant Empedobacter stercoris strain ES183 recovered from a pig fecal sample in China. Tet(X14) shows 67.14-96.39% sequence identity to the other variants [Tet(X) to Tet(X13)]. Overexpression of Tet(X14) in Escherichia coli confers 16-fold increase in tigecycline MIC (from 0.125 to 2 mg/L), which is lower than that of Tet(X3), Tet(X4) and Tet(X6). Structural modelling predicted that Tet(X14) shared a high homology with the other 12 variants with RMSD value from 0.003 to 0.055, and Tet(X14) can interact with tetracyclines by a similar pattern as the other Tet(X)s. tet(X14) and two copies of tet(X2) were identified on a genome island with abnormal GC content carried by the chromosome of ES183, and no mobile genetic elements were found surrounding, suggesting that tet(X14) might be heterologously obtained by ES183 via recombination. Blasting in Genbank revealed that Tet(X14) was exclusively detected on the chromosome of Riemerella anatipestifer, mainly encoded on antimicrobial resistance islands. E. stercoris and R. anatipestifer belong to the family Flavobacteriaceae, suggesting that the members of Flavobacteriaceae maybe the major reservoir of tet(X14). Our study reports a novel chromosome-encoded tigecycline resistance gene tet(X14). The expanded members of Tet(X) family warrants the potential large-scale dissemination and the necessity of continuous surveillance for tet(X)-mediated tigecycline resistance.
Introduction
Antimicrobial resistance (AMR) represents a major global public health challenge in the twenty-first century [Citation1]. The clinical infections caused by AMR bacteria, especially carbapenem-resistant Enterobacteriaceae (CRE) and Acinetobacter spp. (CRA), largely limit the effective prevention and treatment strategies resulting in a high mortality [Citation2,Citation3]. Tigecycline, the minocycline derivative 9-tert-butyl-glycylamido-minocycline, is the third generation of tetracycline family antibiotic which negates most tetracyclines resistance mechanisms due to ribosomal protection and drug efflux [Citation4,Citation5]. This expanded spectrum antibiotics approved by US FDA in 2005 can be used to treat multidrug-resistant gram-positive and gram-negative pathogens [Citation6]. Currently, tigecycline is one of last-resort antibiotics frequently used as a major treatment regimen for the infections caused by CRE and CRA.
Tigecycline resistance has emerged in the clinical setting since then and the resistance is frequently caused by the overexpression of non-specific active efflux pumps or mutations within the drug-binding site in the ribosome [Citation7,Citation8]. Additionally, tigecycline resistance can be mediated by a flavin-dependent monooxygenase gene tet(X) and its variants in a small proportion of tigecycline-resistant Enterobacteriaceae and Acinetobacter isolates through the degradation of tigecycline [Citation5]. The tigecycline breakpoint for Escherichia coli and Citrobacter koseri has been set down from 2 mg/L in version 8–0.5 mg/L in version 9 and version 10 by European Committee on Antimicrobial Susceptibility Testing (EUCAST) [Citation9,Citation10].
The tet(X) gene was firstly identified in Tn4351 and Tn4400 carried by the chromosome of anaerobe Bacteroides fragilis [Citation11]. Subsequently, a few chromosome-encoded and plasmid-mediated novel tet(X) variants have been found. Chromosome-encoded tet(X), tet(X1), tet(X2), tet(X3) and tet(X6) have been identified in Bacteroides fragilis, B. thetaiotaomicron, Pseudomonas aeruginosa, Myroides phaeus, Acinetobacter spp. and Proteus spp., mainly isolated from chickens and pigs [Citation11–15]. Plasmid-mediated tet(X3), tet(X3.2), tet(X4), tet(X5) and tet(X6) have been detected in A. baumannii, Empedobacter brevis, E. falsenii, A. indicus, A. schindleri, A. lwoffiiand and A. towneri isolated from chickens, pigs, cattle, shrimp, avian and human [Citation16–22]. Most recently, another 7 variants including tet(X7) to tet(X13) have been detected in 244 gut-derived metagenomic libraries in America [Citation23]. Of concern, Tet(X4) and Tet(X6) have recently been found to co-exist with mcr-1 in E. coli [Citation24,Citation25]. The convergence of the last-store antibiotic resistance warns the emergence of superbug in the near future.
The rapid emergence of new resistance mechanisms and phenotypes has worsened the current status of AMR controls, and has elevated the public health significance of this issue. Consequently, the identification of novel tet(X) variants is important for us to fully understand the landscape of tigecycline resistance mechanism to control its further dissemination. In this study, we reported a novel chromosome-encoded tet(X) variant, designated tet(X14), in a livestock-associated E. stercoris strain.
Materials and methods
Bacterial strains
Stool samples were collected from 6 livestock farms in China in 2019. Two hundred and ninety-two strains, including 215 Acinetobacter spp. strains and 77 strains of other species, were recovered from stool samples by plating on CHROMagarTM Acinetobacter medium (CHROMagar, Paris, France). PCR screening of tet(X) variants in the collection was performed as previously described [Citation26].
Antimicrobial susceptibility testing (AST)
AST was performed by using broth microdilution method according to CLSI guidelines (29th edition) [Citation27]. The breakpoints of antibiotics tested here were interpreted according to the recommended points for Enterobacteriaceas by EUCAST version 10.0 [Citation10]. E. coli strain ATCC25922 was used for the quality control.
Whole genome sequencing (WGS) and bioinformatic analysis
Total genomic DNA of the tigecycline-resistant isolate was extracted by Puregene Yeast/Bact Kit B (Qiagen, Maryland, US), and was sequenced by using Hiseq 4000 system (Illumina, San Diego, US) and PromethION platform (Nanopore, Oxford, UK). Hybrid assembly was performed by using Unicycler version 0.4.8 [Citation28]. Antibiotic resistance genes were identified by ResFinder 3.2 [Citation29] and CARD (https://card.mcmaster.ca/) with identity >80% and coverage >60%. Plasmid replicon typing was performed using PlasmidFinder v2.1 (https://cge.cbs.dtu.dk/services/PlasmidFinder/) with at least 95% identity and 60% coverage. Synteny analysis was performed using Easyfig [Citation30]. Fragments >5 kb that were absent in at least one genome were detected by BLAST and were defined as genomic islands (GEIs) in this study as previously described [Citation31]. Phylogenetic analysis with amino-acid sequences of Tet(X)s was performed by using the maximum likelihood method with default parameters by using Mega X Version 10.0.5 [Citation32]. The amino acid sequences of Tet(X)s were submitted to ESPript 3 server [Citation33] to perform the alignment and predict the secondary structure elements.
Functional cloning of tet(X14)
The fragment from 219 bp upstream to 53 bp downstream of tet(X14) including the predicted promoter of tet(X14) was amplified using primers pUC19-tet(X14)-F (5’-cgctgcagCAAAAGAGCGGGTTAAGTGG-3’) and p-tet(X14)-R (5’-cgtctagaTACTTCACCGGCTCTATTGC-3’). The amplicon was ligated into pUC19, and the recombinant plasmid was transformed into E. coli DH5α competent cells by heat shock. Transformants were selected on LB agar plates containing 100 mg/L ampicillin. In parallel, tet(X3), tet(X4) and tet(X6) were cloned into pUC19 as positive controls.
Structural modelling of Tet(X14)
The amino acid sequences of Tet(X) variants were submitted to SwissModel [Citation34] to construct 3D structures and 4A6N (PDB entry code) was employed as the template [Citation35]. The overlays of these structures and protein-molecule docking were generated by using AutoDock Vina [Citation36]. Totally hydrogenated Tet(X14), tigecycline and tetracycline were used to perform flexible ligand docking in AutoDock Vina with default parameters. The conformation of ligand which is the most similar with its in 4A6N was chose to construct the recipient-ligand complex to predict the binding sites between Tet(X14) and tigecycline or tetracycline.
In silico screening of tet(X14) in GenBank
We screened the sequences of tet(X14) in GenBank (https://blast.ncbi.nlm.nih.gov/Blast.cgi, accessed by 10 Jun 2020). Matches with >99.74% identity and >97% coverage were retrieved from GenBank. The retrieved sequences with the reference of each tet(X) variant were submitted to phylogenetic analysis to confirm the variant types.
Nucleotide sequence accession numbers
The complete sequences of the chromosome and plasmids (pES183-1, pES183-2 and pES183-3) of strain ES183 have been submitted to GenBank under the accession numbers CP053698-CP053701.
Results
A novel tigecycline resistance gene, tet(X14), was identified in E. stercoris
A tigecycline-resistant strain ES183 recovered from a pig fecal sample obtained in 2019 was positive for tet(X) screening. The strain was identified as E. stercoris by using 16S rDNA sequencing [99.86% identity to the 16S rRNA gene of E. stercoris strain 994B6 12ER2A (accession no. KP119860)]. Strain ES183 was resistant to amikacin (MIC = 32 mg/L), colistin (MIC = 4 mg/L), and all tetracyclines (MIC = 2-128 mg/L) ().
Table 1. MIC values of antibiotics tested in this study.
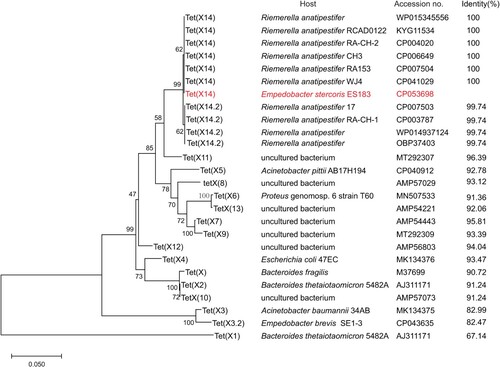
WGS of ES183 was performed to understand the mechanism of resistance to tetracyclines in ES183. Hybrid assembly of short-read (221.1× coverage, average read length 149 bp) and long-read (1064× coverage, average read length 20,327 bp) sequencing data generated a 2.82-Mb chromosome with GC content of 31.89% and 3 plasmids: pES183-1 (10,810 bp; GC content of 24.75%), pES183-2 (2,766 bp; GC content of 33.73%) and pES183-3 (4819 bp; GC content of 25.88%). Five resistance genes were detected in ES183, including a blaEBR-1-like gene (82.17% identity; 99.01% coverage), an aadS gene, two copies of tet(X2) and a novel tet(X) variant with a size of 1167 bp. The novel tet(X) gene encoded a 388-aa protein that displayed 67.14-96.39% identity to reported variants [Tet(X) to Tet(X13)] (). Phylogenetic analysis showed that the novel Tet(X) variant formed a clade separated from the reported Tet(X) variants (). Taken together, the results suggest that a novel member of tet(X) family was identified, designated tet(X14). We additionally noted that the amino-acid sequence of Tet(X10) is identical to Tet(X2), that of Tet(X13) is different from Tet(X6) with one amino acid (L368S), and that of Tet(X9) differs from Tet(X7.2) with two amino acids (I156L and G177 V).
Figure 1. Phylogenetic analysis of the amino acid sequences of Tet(X14) and its homologs. The maximum-likelihood tree was inferred using MEGA X Version 10.0.5 with 1000 bootstraps. Eleven amino acid sequences of Tet(X14) identified in this study and GenBank with the other published Tet(X) variants are included in the analysis. Numbers above each node show the percentage of tree configurations that occurred during 1000 bootstrap trials. The scale bar is in fixed nucleotide substitutions per sequence position. Host strains, accession numbers and identity of each Tet(X) variants relative to Tet(X14) detected in strain ES183 (in red) are listed.
To determine the activity of tet(X14) against tetracyclines, the gene was cloned into pUC19 and the resulted recombinant vector was transferred to E. coli DH5α to construct the transformant DH5α-pUC19-tet(X14). A 16- to 64-fold increase in the MIC of all tested tetracyclines was observed for the tet(X14) transformant (), suggesting that tet(X14) was active against tetracyclines. To compare the activity of tet(X14) with that of other tet(X) variants, we further constructed transformants DH5α-pUC19-tet(X3), DH5α-pUC19-tet(X4), and DH5α-pUC19-tet(X6). The MICs of oxytetracycline, chlortetracycline, demeclocycline, doxycycline and minocycline were comparable among the 4 transformants (2-fold difference), while the MICs of tigecycline and eravacycline were 4- to 8-fold lower for DH5α-pUC19-tet(X14) than the other transformants (). This indicates that Tet(X14) mediated slightly lower level of resistance to tigecycline and eravacycline than Tet(X3), Tet(X4) and Tet(X6).
Tet(X14) is highly similar with the other Tet(X) variants at the structural level
Alignment of the amino acid sequences of Tet(X14) and the other Tet(X) variants showed that the substrate binding sites and flavin adenine dinucleotide (FAD) binding sites were conserved in all Tet(X) variants with similar secondary structures (Figure S1). The model structure of Tet(X14) was then superposed onto that of other 12 Tet(X) structures [Tet(X2) to Tet(X13)] to perform a homology modelling assay. The overlay of models showed that Tet(X14) shared a high homology with the other Tet(X) variants according to the protein structural architecture ((A)) with RMSD value from 0.003 to 0.055. The data further support that Tet(X14) belongs to Tet(X) family.
Figure 2. Homology modelling and molecular docking of Tet(X14). (A) Cartoon representation of the modelled Tet(X2) (green), Tet(X3) (cyan), Tet(X4) (magenta), Tet(X5) (yellow), Tet(X6) (pink), Tet(X7) (gray), Tet(X8) (tv_blue), Tet(X9) (orange), Tet(X10) (lime green), Tet(X11) (deep teal), Tet(X12) (hot pink), Tet(X13) (yellow orange) and Tet(X14) (violet purple) structure. Predicted binding conformation of tigecycline (B) and tetracycline (C) (green and red) at the substrate-binding site of the modelled Tet(X14) structure with FAD (violet and wheat). The side chains of residues connected with tigecycline or tetracycline with hydrogen bonds are indicated in the enlarged views.
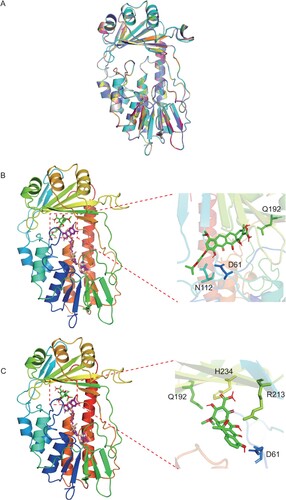
Flexible ligand docking between Tet(X14) and tetracycline-family antibiotics were performed to predict the hydrogen-bond interaction. D61, N112 and Q192 were the predicted residues involved in interactions between Tet(X14) and tigecycline, and E46, R47, R117 and D311 were the binding sites for FAD cofactor ((B)). This is similar with the structure of TetX2-tigecycline complex derived from the crystallization (PDB no. 4A6N) [Citation35]. Potential interaction sites of Tet(X14) with tetracycline were D61, Q192, H234 and R213 ((C)), which were similar with the modelling of Tet(X6) [Citation15]. These results suggest that as the other Tet(X)s, Tet(X14) interacts with tigecycline and tetracycline through a conserved pattern.
Tet(X14) was exclusively detected in Riemerella anatipestifer
To understand the distribution of Tet(X14) in bacteria, the amino acid sequence of Tet(X14) was blasted in GenBank. Ten hits were obtained with identity >99.74% and coverage >97%, including 4 amino-acid sequences and 6 complete genome sequences (). Six hits are identical to the amino acid sequence of tet(X14) identified in this study, and the other 4 hits shared 99.74% similarity with only one amino acid difference (G295D), thus designated Tet(X14.2). This is consistent with the phylogenetic analysis that two subclades were formed by Tet(X14) and Tet(X14.2), respectively (). The Tet(X14) /Tet(X14.2)-positive isolates exclusively belonged to R. anatipestifer. Four Tet(X14) and 2 Tet(X14.2) hits were located on the chromosome of R. anatipestifer strains isolated from ducks in eastern and southern China (). The location of the other 4 hits was undetectable since they were deposited in GenBank as single genes. These results suggest that R. anatipestifer might be the major reservoir of Tet(X14).
Table 2. Strains harbouring tet(X14) in GenBank.
Tet(X14) might be obtained by E. stercoris strain ES183 via recombination
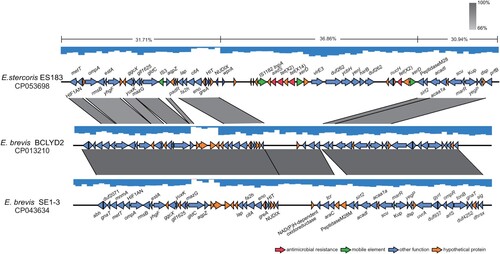
The tet(X14) gene was located at 247668-248834 bp of the chromosome of strain ES183, and an xerD gene was found at upstream of tet(X14) with opposite direction (). It is known that XerD is involved in catalyzing the cutting and rejoining of the bacterial chromosome and plasmid DNA segregation at cell division [Citation37,Citation38]. This adjacency is previously found for plasmid-borne tet(X3) and tet(X5) [Citation17,Citation19]. No predicted genetic mobile elements, like transposons, integrons or integrative and conjugative elements, were found adjacent to tet(X14). To track the source of tet(X14) obtained by ES183, the surrounding region of tet(X14) (210760-289779-bp) was blasted in GenBank, and two best matches with identity > 86% and coverage > 53% were found, including the chromosome of E. brevis BCLYD2 (CP013210) and E. brevis SE1-3 (CP043634). The coverage of the other matches was lower than 17%. The fragment (245019-261642-bp) of ES183 encoding tet(X14) was not found in E. brevis BCLYD2 and E. brevis SE1-3, and the flanking regions were conserved in three isolates (). The GC content of the tet(X14)-encoding region (36.86%) was higher than that of the flanking regions (30.94%-31.71%) and of whole chromosome of ES183 (31.89%). We therefore suppose that the fragment encoding tet(X14) is a genomic island (GEI) inserted at the region between genes encoding NUDIX and peptidase M28, which might be obtained from other species via recombination events.
Figure 3. Identification of a genomic island (GEI) encoding tet(X14) and tet(X2) in ES183 strain. The GEI identified in ES183 inserted between genes encoding NUDIX and peptidase M28. The flanking regions of the GEI are homologous to sequences of two E. brevis genomes (CP013210 and CP043634) (>66% identity) retrieved in GenBank shown by grey shading. GC content of the GEI (36.86%) is higher than that of the flanking regions (30.94% – 31.71%) labeled on the top line. The arrows represent the transcriptional direction of the ORFs. Genes are colour-coded, depending on functional annotations: red, antimicrobial resistance; green, mobile genetic elements; blue, other functions; orange, hypothetical proteins.
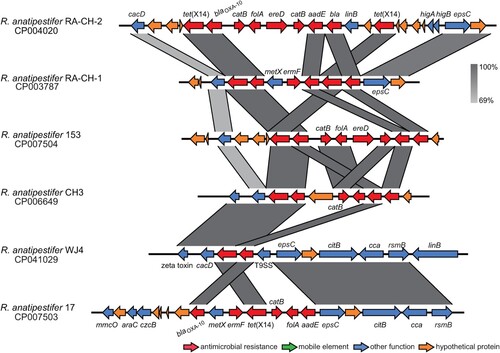
The surrounding environments of tet(X14) identified in R. anatipestifer were fully different from that in ES183 (). The xerD gene was missing in the tet(X14) genetic contexts identified in R. anatipestifer, and the beta-lactamase gene blaOXA-10 adjacent to tet(X14) was common. Additionally, two copies of tet(X14) with multiple resistance genes were found in most R. anatipestifer strains (), implying that tet(X14) might be encoded on antimicrobial resistance islands (ARIs). Comparative genomics study using R. anatipestifer strain ATCC 11845 as the reference identified various tet(X14)-encoded ARIs in three genomes of R. anatipestifer (CP004020, CP007503, and CP007504) (Figure S2). The tet(X14)-encoded ARIs could not be determined in the other genomes due to the lack of suitable reference.
Figure 4. Genomic context of Tet(X14) identified in R. anatipestifer strains. The arrows represent the transcriptional direction of the ORFs. Regions of >69% homology are shown by grey shading. Genes are colour-coded, depending on functional annotations: red, antimicrobial resistance; blue, other functions; orange, hypothetical proteins.
Two copies of tet(X2) were found at 226213-227379-bp and 249873-251039-bp of the chromosome of strain ES183 with the same transcriptional direction with tet(X14). They with tet(X14) were located at the same GEI identified above (). The downstream copy of tet(X2) was followed by aminoglycosides resistance gene aadS (). This adjacency is similar with that of the firstly reported tet(X2) that an aadS gene was at upstream of tet(X2) in CTnDOT in B. thetaiotaomicron 5482A [Citation13]. No Tn structures were found adjacent to tet(X2) in strain ES183.
Discussion
The widespread of CRE represents a large threat to the public health network globally. Currently, tigecycline and colistin are two last-resort antibiotics frequently used to combat lethal infections caused by CRE. However, the wide use of these antibiotics has resulted in the global emergence of resistance in the clinical setting, which significantly compromises their efficacy. Of more concern, the recently identified plasmid-borne resistance genes for colistin (mcr) and tigecycline [tet(X)] highly challenge the resistance control. At present, the mcr variants have already been extended from mcr-1 to mcr-10, and have disseminated globally [Citation39,Citation40]. It therefore is reasonable to raise the concern that Tet(X) family tigecycline resistance determinants are with potential large-scale dissemination. Indeed, a most recent study dramatically extends the members of Tet(X) family from 7 to 14 [Citation14,Citation15,Citation23]. This study highlights that numerous Tet(X) variants have already circulated in various environmental ecosystem. Monitoring the spread of such resistance genes in the context of One Health (including clinical, animal and environmental sectors) is one of efficient strategies to combat antimicrobial resistance. Identification of new resistance determinants is crucial for fulfilling the strategies and can further aid to improve the current control measures.
In this study, we identified a novel tigecycline resistance gene variant, tet(X14), on the chromosome of an E. stercoris strain recovered from a pig fecal sample in China. Tet(X14) shows the highest amino acid identity with Tet(X11) (96.39%). Overexpression of tet(X14) in E. coli DH5α confers 16-fold and 64-fold increase in MIC of tigecycline and eravacycline, respectively. This demonstrated that tet(X14) is a novel tigecycline resistance gene. Tet(X14) shows a similar affinity for tetracyclines with the other Tet(X) variants, and their tetracycline-binding sites are conserved. We suppose that the evolutionary pattern of Tet(X) family is of restricted amino acid substitutions with defined limits resulting in a functional consistency against tetracyclines. The resistance activity of Tet(X14) against tigecycline is lower than that of plasmid-mediated Tet(X3), Tet(X4) and Tet(X6) (). In the crystallization complex of tigecycline and Tet(X2), Q192 and R213 are identified as the common binding sites in four monomers [Citation35]. These two binding sites are conserved in Tet(X4), while hydrogen bonds have been predicted at R211 but not at Q in Tet(X3). This is consistent with the higher tigecycline resistance conferred by Tet(X4) than Tet(X3) in a mouse model [Citation17]. Only Q192 but no any R was identified in the docking complex of tigecycline and Tet(X14) ((B)). This may explain the lower activity of tigecycline resistance conferred by Tet(X14). However, further study should be performed to validate the prediction results. Of note, Tet(X14) was exclusively identified on the chromosome in this study, Tet(X3) and Tet(X4) were frequently detected on plasmids in various species [Citation17,Citation22,Citation41], and Tet(X6) was almost equally distributed on the chromosome and plasmid based on the data available currently [Citation15,Citation42]. A potential correlation was noted for the tigecycline resistance activity and the location of Tet(X)s that the plasmid-borne Tet(X3) and Tet(X4) showed the highest activity (16 mg/L), and the chromosome-encoding Tet(X14) showed the lowest activity (2 mg/L). The activity of Tet(X6) was between them (8 mg/L). However, more data are needed to determine the correlation in the future.
Tet(X14) was exclusively detected in R. anatipestifer through blasting in GenBank ( and ). R. anatipestifer is a Gram-negative bacterium belonging to the family Flavobacteriaceae. Intriguingly, E. stercoris also belongs to the same family, suggesting that the members of the family Flavobacteriaceae, especially R. anatipestifer, might be the major reservoir of tet(X14). Moreover, all Tet(X14)-producing strains were exclusively detected in China except for one strain with unknown source (), indicating that Tet(X14) might emerge locally. Of note, R. anatipestifer is an important poultry pathogen which primarily causes infection in domestic ducks [Citation43], and E. stercoris is livestock associated, which is firstly isolated from a mixed manure sample [Citation44]. We suppose that the emergence of Tet(X14) could be caused by the heavy utilization of tetracycline antibiotics in the animal feed, like tetracycline, oxytetracycline, chlortetracycline, and doxycycline [Citation19]. It is reasonable to predict that such resistance gene would jump into the clinical setting through the food chain and/or zoonosis with high possibilities in the future.
We determined the genetic context of tet(X14) to estimate how the gene was captured by the isolates detected here. The surrounding environments of tet(X14) identified here were different from those of the other tet(X) variants that no any known mobile genetic elements were found. Moreover, the genetic contexts of tet(X14) identified in R. anatipestifer were completely different from that identified in E. stercoris, suggesting that tet(X14) was captured by the members of the family Flavobacteriaceae individually, and inter-species transmissions might have not occurred yet. An xerD gene was found at the upstream of tet(X14) identified in E. stercoris. It has been reported that XerD is able to mediate the integration of mobile genetic elements (e.g. phages) into the chromosome via homologous recombination [Citation45]. The gene has frequently been found adjacent to other tet(X) variants [Citation17,Citation19], It thus would be interesting to validate whether XerD is involved in the mobilization of tet(X)s in the future. Additionally, the tet(X14) gene was identified on GEIs in E. stercoris and on ARIs in R. anatipestifer (Figure S2), and the GC content of the tet(X14)-encoding fragment carried by E. stercoris was different from the flanking regions. Together, the data imply the heterologous insertions of tet(X14) via recombination events. Currently, the limited genomic data largely impedes us to track the source and origin of tet(X14).
A limitation of this study is that we could not determine the colistin resistance mechanism for the E. stercoris isolate. No mcr genes were found in the isolate, and it is unknown which genes are involved in the colistin resistance. The species is recently identified and has rarely been studied, therefore very limited data are available for us to analyze its resistance mechanism for colistin. Further study should be performed to identify the underlying mechanism.
In summary, we report the discovery of a novel chromosome-encoding tigecycline resistance gene, tet(X14), in a tigecycline-resistant and colistin-resistant E. stercoris strain. The convergence of resistance to two last-resort antibiotics would largely threaten the global public health system. Tet(X14) has a similar function and structure to other Tet(X) variants, and confers lower tetracycline/glycylcycline MICs than the plasmid-borne Tet(X)s. Recombination may play an important role in the transmission of tet(X14). The expanded members of Tet(X) highlights the potential large-scale dissemination and the necessity of continuous surveillance for tet(X)-mediated tigcycline resistance.
Supplementary_Figure_2._Antibiotic_resistance_islands__ARIs__encoding_tet_X14__identified_in_R._anatipestifer-1_final.jpg
Download ()Supplementary_Figure_1._The_secondary_structure_of_Tet_X14__and_its_homologs-1_final.jpg
Download ()Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- WHO. Antimicrobial resistance: global report on surveillance. Geneva: World Health Organization; 2014.
- Aguilera-Alonso D, Escosa-Garcia L, Saavedra-Lozano J, et al. Carbapenem-resistant gram-negative bacterial infections in children. Antimicrob Agents Chemother. 2020 Feb 21;64:3.
- Tomczyk S, Zanichelli V, Grayson ML, et al. Control of carbapenem-resistant Enterobacteriaceae, Acinetobacter baumannii, and Pseudomonas aeruginosa in healthcare facilities: a systematic review and reanalysis of quasi-experimental studies. Clin Infect Dis. 2019 Feb 15;68(5):873–884.
- Pankey GA. Tigecycline. J Antimicrob Chemother. 2005 Sep;56(3):470–480.
- Moore IF, Hughes DW, Wright GD. Tigecycline is modified by the flavin-dependent monooxygenase TetX. Biochemistry. 2005 Sep 6;44(35):11829–11835.
- Doan TL, Fung HB, Mehta D, et al. Tigecycline: a glycylcycline antimicrobial agent. Clin Ther. 2006 Aug;28(8):1079–1106.
- Linkevicius M, Sandegren L, Andersson DI. Potential of tetracycline resistance proteins to evolve tigecycline resistance. Antimicrob Agents Chemother. 2016 Feb;60(2):789–796.
- Grossman TH. Tetracycline antibiotics and resistance. Cold Spring Harb Perspect Med. 2016 Apr 1;6(4):a025387.
- EUCAST. The European Committee on Antimicrobial Susceptibility Testing. Breakpoint tables for interpretation of MICs and zone diameters. Version 9.0. 2019.
- EUCAST. The European Committee on Antimicrobial Susceptibility Testing. Breakpoint tables for interpretation of MICs and zone diameters Version 10.0. 2020.
- Speer BS, Bedzyk L, Salyers AA. Evidence that a novel tetracycline resistance gene found on two Bacteroides transposons encodes an NADP-requiring oxidoreductase. J Bacteriol. 1991 Jan;173(1):176–183.
- Peng K, Li R, He T, et al. Characterization of a porcine Proteus cibarius strain co-harbouring tet(X6) and cfr. J Antimicrob Chemother. 2020 Jun 1;75(6):1652–1654.
- Whittle G, Hund BD, Shoemaker NB, et al. Characterization of the 13-kilobase ermF region of the Bacteroides conjugative transposon CTnDOT. Appl Environ Microbiol. 2001 Aug;67(8):3488–3495.
- Liu D, Zhai W, Song H, et al. Identification of the novel tigecycline resistance gene tet(X6) and its variants in Myroides, Acinetobacter and Proteus of food animal origin. J Antimicrob Chemother. 2020 Jun 1;75(6):1428–1431.
- He D, Wang L, Zhao S, et al. A novel tigecycline resistance gene, tet(X6), on an SXT/R391 integrative and conjugative element in a Proteus genomospecies 6 isolate of retail meat origin. J Antimicrob Chemother. 2020 May 1;75(5):1159–1164.
- Zeng Y, Dong N, Zhang R, et al. Emergence of an Empedobacter falsenii strain harbouring a tet(X)-variant-bearing novel plasmid conferring resistance to tigecycline. J Antimicrob Chemother. 2020 Mar 1;75(3):531–536.
- He T, Wang R, Liu D, et al. Emergence of plasmid-mediated high-level tigecycline resistance genes in animals and humans. Nat Microbiol. 2019 Sep;4(9):1450–1456.
- Sun J, Chen C, Cui CY, et al. Plasmid-encoded tet(X) genes that confer high-level tigecycline resistance in Escherichia coli. Nat Microbiol. 2019 Sep;4(9):1457–1464.
- Wang L, Liu D, Lv Y, et al. Novel plasmid-mediated tet(X5) gene conferring resistance to tigecycline, eravacycline and omadacycline in clinical Acinetobacter baumannii. Antimicrob Agents Chemother. 2019 Dec 20;64(1):e01326–19.
- Cui CY, Chen C, Liu BT, et al. Co-occurrence of plasmid-mediated tigecycline and carbapenem resistance in Acinetobacter spp. from waterfowls and their neighboring environment. Antimicrob Agents Chemother. 2020 Apr 21;64:5.
- Sun C, Cui M, Zhang S, et al. Plasmid-mediated tigecycline-resistant gene tet(X4) in Escherichia coli from food-producing animals, China, 2008-2018. Emerg Microbes Infect. 2019;8(1):1524–1527.
- Li R, Liu Z, Peng K, et al. Co-occurrence of two tet(X) variants in an Empedobacter brevis of shrimp origin. Antimicrob Agents Chemother. 2019 Sep 30;63(12):e01636–19.
- Gasparrini AJ, Markley JL, Kumar H, et al. Tetracycline-inactivating enzymes from environmental, human commensal, and pathogenic bacteria cause broad-spectrum tetracycline resistance. Commun Biol. 2020 May 15;3(1):241.
- He T, Wei R, Li R, et al. Co-existence of tet(X4) and mcr-1 in two porcine Escherichia coli isolates. J Antimicrob Chemother. 2020 Mar 1;75(3):764–766.
- Xu Y, Liu L, Feng Y. A New tet(X6) Tigecycline Resistance Determinant Co-carried with mcr-1 by A Single Plasmid. [bioRxiv preprint]. 2020.
- Cheng Y, Chen Y, Liu Y, et al. Silent dissemination of plasmid-borne tigecycline resistance gene tet(X6) in livestock-associated Acinetobacter towneri. [bioRxiv preprint]. 2020.
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing. 29th ed. CLSI supplement M100. Wayne, PA: Clinical and Laboratory Standards Institute; 2019.
- Wick RR, Judd LM, Gorrie CL, et al. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol. 2017 Jun;13(6):e1005595.
- Zankari E, Hasman H, Cosentino S, et al. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother. 2012 Nov;67(11):2640–2644.
- Sullivan MJ, Petty NK, Beatson SA. Easyfig: a genome comparison visualizer. Bioinformatics. 2011 Apr 1;27(7):1009–1010.
- Zhou K, Ferdous M, de Boer RF, et al. The mosaic genome structure and phylogeny of Shiga toxin-producing Escherichia coli O104:H4 is driven by short-term adaptation. Clin Microbiol Infect. 2015 May;21(5):468 e7–18.
- Kumar S, Stecher G, Li M, et al. MEGA x: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018 Jun 1;35(6):1547–1549.
- Robert X, Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014 Jul;42:W320–W324.
- Waterhouse A, Bertoni M, Bienert S, et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018 Jul 2;46(W1):W296–W303.
- Volkers G, Damas JM, Palm GJ, et al. Putative dioxygen-binding sites and recognition of tigecycline and minocycline in the tetracycline-degrading monooxygenase TetX. Acta Crystallogr D Biol Crystallogr. 2013 Sep;69(9):1758–1767.
- Trott O, Olson AJ. Autodock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010 Jan 30;31(2):455–461.
- Arciszewska LK, Sherratt DJ. Xer site-specific recombination in vitro. EMBO J. 1995 May 1;14(9):2112–2120.
- Colloms SD, McCulloch R, Grant K, et al. Xer-mediated site-specific recombination in vitro. EMBO J. 1996 Mar 1;15(5):1172–1181.
- Wang C, Feng Y, Liu L, et al. Identification of novel mobile colistin resistance gene mcr-10. Emerg Microbes Infect. 2020;9(1):508–516.
- Crofts TS, Gasparrini AJ, Dantas G. Next-generation approaches to understand and combat the antibiotic resistome. Nat Rev Microbiol. 2017 Jul;15(7):422–434.
- Li R, Lu X, Peng K, et al. Deciphering the structural diversity and classification of the mobile tigecycline resistance gene tet(X)-bearing plasmidome among bacteria. mSystems. 2020 Apr 28;5:2.
- Li R, Peng K, Li Y, et al. Exploring tet(X)-bearing tigecycline-resistant bacteria of swine farming environments. Sci Total Environ. 2020 May 11;733:139306.
- Hu Q, Han X, Zhou X, et al. Ompa is a virulence factor of Riemerella anatipestifer. Vet Microbiol. 2011 Jun 2;150(3-4):278–283.
- Schauss T, Busse HJ, Golke J, et al. Empedobacter stercoris sp. nov., isolated from an input sample of a biogas plant. Int J Syst Evol Microbiol. 2015 Oct;65(10):3746–3753.
- Midonet C, Das B, Paly E, et al. XerD-mediated FtsK-independent integration of TLCvarphi into the Vibrio cholerae genome. Proc Natl Acad Sci U S A. 2014 Nov 25;111(47):16848–16853.
