ABSTRACT
An emerging infectious disease caused by “Anaplasma capra” was reported in a 2015 survey of 477 hospital patients with a tick-bite history in China. However, the morphological characteristics and parasitic location of this pathogen are still unclear, and the pathogen has not been officially classified as a member of the genus Anaplasma. Anaplasma capra-positive blood samples were collected, blood cells separated, and DNA of whole blood cells, erythrocytes, and leukocytes extracted. Multiplex PCR detection assay was used to detect whole blood cell, erythrocytes and leukocytes, DNA samples, and PCR identification, nucleic acid sequencing, and phylogenetic analyses based on A. capra groEL, 16S rRNA, gltA, and msp4 genes. Scanning electron microscopy (SEM), transmission electron microscopy (TEM), Wright–Giemsa staining, chromogenic in situ hybridization (CISH), immunocytochemistry, and indirect immunofluorescence assay (IFA) were used to identify the location and morphological characteristics of A. capra. Multiple gene loci results demonstrated that erythrocyte DNA samples were A. capra-positive, while leukocyte DNA samples were A. capra-negative. Phylogenetic analysis showed that A. capra is in the same clade with the A. capra sequence reported previously. SEM and TEM showed one or more pathogens internally or on the outer surface of erythrocytes. Giemsa staining, CISH, immunocytochemistry, and IFA indicated that erythrocytes were A. capra-positive. This study is the first to identify the novel zoonotic tick-borne Anaplasma sp., “Anaplasma capra,” in host erythrocytes. Based on our results, we suggest revision of Genus Anaplasma and formally name “A. capra” as Anaplasma capra sp. nov.
Key points
First identified the novel zoonotic tick-borne Anaplasma sp. in host erythrocytes.
Suggest formally name the novel erythrocytes pathogen as Anaplasma capra sp. nov.
Introduction
In 2015, an emerging infectious disease caused by “Anaplasma capra,” was reported in a survey of 477 hospital patients with tick-bite history in Mudanjiang Forestry Central Hospital, Heilongjiang Province, China. A total of 28 (6%) of the 477 patients were diagnosed with “A. capra,” and the pathogen was isolated from three cases [Citation1]. Patients positive for this pathogen presented with an undifferentiated influenza-like illness, gastrointestinal symptoms, rash, eschar, regional lymphadenopathy with potential progression to central nervous system involvement, and cerebrospinal fluid pleocytosis [Citation1]. Anaplasma capra was provisionally nominated as a novel tick-borne zoonotic Anaplasma sp. by Li et al. (2015) [Citation1] after being first identified in a goat (Capra aegagrus hircus) [Citation2]. Anaplasma capra has been shown to infect humans, ruminants (sheep, goat, cattle, etc.), pet dogs, and wild animals (e.g. takin, musk deer, goral) [Citation1,Citation3–8], and has been detected in a variety of ticks, e.g. Ixodes persulcatus, Haemaphysalis longicornis, H. concinna, H. qinghaiensis, and Rhipicephalus microplus [Citation9–12]. Anaplasma capra is distributed all over the world, including China, France, Japan, South Korea, and Italy [Citation5,Citation7,Citation13–15], posing a potential health threat to both humans and animals.
The Genus Anaplasma comprises a group of Gram-negative obligate intracellular bacteria consisting of many members, e.g. A. bovis, A. ovis, A. marginale, A. centrale, and A. phagocytophilum [Citation16]. While A. ovis, A. marginale, and A. centrale are known as intraerythrocytic pathogens, A. bovis and A. phagocytophilum infect monocytes and neutrophil granulocytes, respectively [Citation17,Citation18]. Anaplasma phagocytophilum presents as a mild to severe febrile illness, including multiple organ failure and death [Citation1,Citation19–21]. Anaplasma capra is a novel zoonotic Anaplasma sp., included with other Anaplasma species in the list of prokaryotic names with standing in nomenclature (LPSN), but not validly published. Its morphological characteristics and type cells infected are unclear. Although molecular detection and identification based on groEL, 16S rRNA, gltA, msp2, and msp4 genes of A. capra have been performed in the last five years [Citation6], neither morulae nor other forms of the pathogen had been detected so far in peripheral blood smears [Citation1,Citation10]. Hence, studies to determine the host parasite site, morphological characteristics, and pathogenesis of A. capra is needed to distinguish it from other tick-borne Anaplasma infections.
Material and methods
Blood samples collection and preparation
Two goats with a history of tick bites were purchased from a goat farmer in Luoyang City, Henan province, central region of China. One goat was A. capra-positive by PCR, and the other was A. capra-negative. EDTA blood samples were collected from the two goats, and the erythrocytes and leukocytes separated from the blood samples using an Erythrocytes Separation Kit (TBD, Tianjin, China). The density gradient of the separation solution separates erythrocytes and leukocytes. Subsequently, DNA of the whole blood cells, erythrocytes and leukocytes, were extracted from the samples using a Blood DNA Kit (OMEGA, Norcross, GA, USA), according to the manufacturer’s recommendations. All DNA samples were stored at −20°C until used.
PCR, gene sequencing and phylogenetic analysis
A multiplex PCR detection assay was used to detect erythrocyte and leukocyte DNA samples targeting the A. capra groEL gene, as previously described [Citation22] (). Anaplasma capra-positive samples were genetically profiled by amplification of the16S rRNA (rrs) gene as well as gltA and msp4 genes as previously described [Citation1] (). Selected, A. marginale, A. platys, and A. centrale genes were used to exclude co-infections in A. capra-positive samples (). As positive controls, a co-infected DNA sample (concurrently infected with “A. capra,” A. bovis, A. ovis, and A. phagocytophilum) and A. marginale-, A. platys-, and A. centrale-positive samples were used, and negative control double distilled H2O was used.
Table 1. Primers used in the PCR assays of Anaplasma spp.
The PCR products were sequenced to confirm the presence of Anaplasma capra DNA. The sequences obtained (GenBank Accession Nos.: groEL MT804297, 16S rRNA MT799937, gltA MT804296, msp4 MT804298) were compared with published sequences in GenBank using BLASTn search (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Phylogenetic analyses were performed and phylogenetic trees constructed based on the sequence distance method using the neighbor-joining (NJ) algorithm with Mega 7.0 software.
Electron microscopy
Anaplasma capra-positive erythrocytes were fixed with a commercial electron microscope fixing solution (Solarbio, Beijing, China). After fixation, the cells were removed by centrifugation. The fixed cells were 0.01 M phosphate buffer solution (PBS) buffer-washed, post-fixed with 2% OsO4, water-washed, and dehydrated in graded ethanol (EtOH) series (30%, 50%, 70%, 80%, 90%, 100%, and 100% at 15-min intervals). For SEM, after the cells were dehydrated in graded EtOH series, the EtOH was replaced with isoamyl acetate, mounted on a metal stub, sputter-coated with gold for 5 min, and examined using a SU8100 electron microscope (HITACHI, Japan) at 3 kV. For TEM observation, the cells were infiltrated with acetone and Pon 812 epoxy resin (SPI, West Chester, USA) (1/1, 2 h; then, 1/2, 12 h) and cured at 60°C for 48 h. The cured resin blocks were trimmed, thin-sectioned, thin sections collected on formvar copper 200 mesh grids, and then post-stained with 2% aqueous uranyl acetate and Reynold’s lead citrate. The sections were examined using a HT7700 electron microscope (HITACHI, Japan) [Citation23]. The morulae of A. capra, following lysis of the erythrocytes and centrifuged at 12,000 × g, were observed by TEM.
Wright–Giemsa staining, CISH, immunocytochemistry, and IFA
Blood cell smears were fixed in methanol for 10 min and stained with Wright–Giemsa stain, and the intracellular morulae observed under light microscopy. To identify the specificity of the intracellular morulae detected in the blood cells, CISH, immunocytochemistry, and IFA were conducted with A. capra-positive samples.
CISH was performed with a commercial kit according to the manufacturer’s protocol (Invitrogen, Carlsbad, USA). Briefly, the blood films were reverse-stained with eosin, visualized using Digital Slice Scanner (Pannoramic MIDI, Hungary), and fixed with a fixative (Solarbio, Beijing, China). The CISH probes were designed specifically for Anaplasma capra (Invitrogen, Carlsbad, USA). To achieve sufficient signal to background ratio, multiple probes were targeted along each individual lncRNA/mRNA sequence of the A. capra sp. nov groEL gene (KM206275). A set of 16 probes covering the entire length of the RNA molecule allowed for optimal signal strength. The probes were labeled with digoxigenin (DIG), and brown-stained cells were considered positive.
For immunocytochemistry, the A. capra-positive serum samples collected from the goat were used as the primary antibody and incubated at 4°C overnight. Subsequently, HRP-conjugated Rabbit Anti-Goat IgG (H+L) (Servicebio, Wuhan, China) was applied to the smears and incubated at 37°C for 1 h. The antibody was visualized using 3,3′-diaminobenzidine (Servicebio, Wuhan, China), and the images recorded using a light microscope (Nikon, Tokyo, Japan). As negative controls, the antiserum from the A. capra-negative goat and A. capra-negative blood smears were processed in the same manner and examined.
For IFA, A. capra-positive whole blood cells were processed for the preparation of antigen slides. The A. capra-positive serum samples collected from goats were used as the primary antibody, and Donkey anti-Goat IgG (H+L) Cross-Adsorbed Secondary Antibody (Thermo Fisher Scientific, Carlsbad, USA) used as a secondary antibody [Citation20]. The A. capra-negative whole blood cells smear were used as negative controls. The slides were examined using a Digital Slice Scanner (Pannoramic MIDI, Hungary) and cells with blue fluorescence considered positive.
Results
The multiplex PCR detection assay showed that the whole blood DNA samples were positive for “A. capra,” A. bovis, and A. phagocytophilum. Three erythrocyte DNA samples were A. capra-positive only, while three leukocytes DNA samples were both A. bovis- and A. phagocytophilum-positive (). Therefore, multi-site identification was conducted based on 16S rRNA, gltA, and msp4 genes of A. capra using erythrocyte and leukocyte DNA samples (). The PCR identification results based on multiple gene loci showed that all erythrocyte DNA samples were A. capra-positive and A. marginale-, A. platys-, and A. centrale-negative; leukocyte DNA samples were A. capra-negative; and samples infected with A. bovis, A. ovis, A. marginale, A. platys, and A. centrale were A. capra-negative.
Figure 1. Results of multiplex PCR detection assay for the identification of A. capra sp. nov in whole blood, erythrocyte, and leukocyte DNA samples from the same goat whole blood sample. Lane W: Whole blood DNA sample positive for A. capra sp. nov (874 bp), A. bovis (529 bp), and A. phagocytophilum (172 bp). Lanes E1–E3: Erythrocytes DNA samples positive for A. capra sp. nov (874 bp). Lanes L1–L3: Leukocytes DNA samples positive for A. bovis (529 bp) and A. phagocytophilum (172 bp). Lane P: Positive control of multiplex PCR for detecting A. capra sp. nov, A. bovis, A. ovis (347 bp), and A. phagocytophilum. Lane N: Negative control.
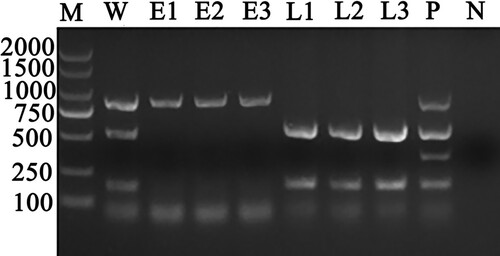
Figure 2. Identification results of different blood cell DNA samples by PCR based on gltA, 16S rRNA and msp4 genes of A. capra. A-C was the result based on gltA, 16S rRNA and msp4 gene, respectively. Erythrocyte DNA samples were A. capra-positive only based on multiplex loci, while leukocytes DNA samples were A. capra-negative.
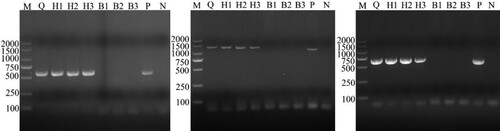
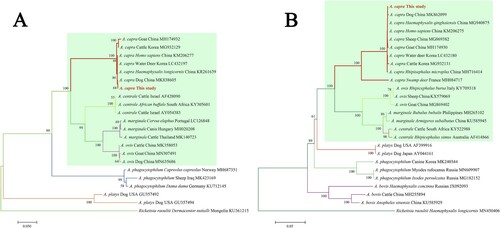
Subsequently, the sequences of groEL (874 bp), 16S rRNA (1261 bp), gltA (594 bp), and msp4 (656 bp) of A. capra were obtained and submitted to GenBank (Accession Nos. MT804297, MT799937, MT804296, and MT804298, respectively). The entire 16S rRNA sequence of the Anaplasma sp. from a goat was 100% homologous to sequences observed in a human (GenBank Accession No. KM206273) and H. longicornis (GenBank Accession No. KY242456). The phylogenetic tree, based on 16S rRNA sequences, showed that the isolated Anaplasma sp. nov is in the same clade with A. capra from R. microplus (MH762071), H. longicornis (KY242456), H. sapiens (KM206273), goat (MG869483), sheep (MF066917), cattle (MF000918), and Korean water deer (LC432092), but clearly separated from other Anaplasma spp. ((A)). Further analyses of gltA, msp4, and groEL sequences showed that Anaplasma sp. nov demonstrated a high homology to A. capra reported previously (KM20627, KM206274, KM206275, KM206277) ( and (A,B)). Phylogenetic tree analysis based on the sequences of gltA, msp4, and groEL genes indicated that the Anaplasma sp. sequence detected in the present study is in the same clade with A. capra sequences previously reported ( and (B)). However, the sequence from the French red deer and swamp deer appeared to have a different genotype, which was in the same cluster with “A. capra,” but on a different branch ( and (B)). Moreover, phylogenetic trees based on the four gene sequences showed that A. capra is in the same cluster with A. marginale, A. centrale, and A. ovis, suggesting that A. capra and the three Anaplasma spp. may have some similar characteristics.
Figure 3. Phylogenetic analysis of A. capra sp. nov identified in this study based on 16S rRNA (1261 bp, A) and gltA (594 bp, B) genes. The trees were constructed using NJ method with MEGA 7.0 software and the numbers on the tree indicate bootstrap values for the branch points. Bootstrap analysis was performed with 1000 replicates. Numbers on the branches indicate percent support for each clade. Red font denotes the sequences obtained in this study. Rickettsia raoultii was used as outgroup. The green background represents intraerythrocytic Anaplasma spp.
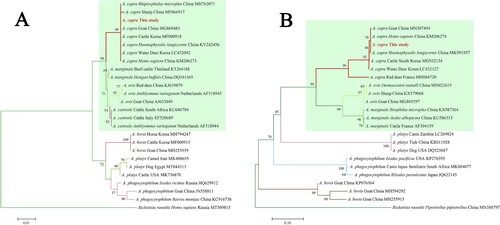
Figure 4. Phylogenetic analysis of A. capra sp. nov identified in this study based on msp4 (656 bp, A) and groEL (874 bp, B) genes. The trees were constructed using NJ method with MEGA 7.0 software and the numbers on the tree indicate bootstrap values for the branch points. Bootstrap analysis was performed with 1000 replicates. Numbers on the branches indicate percent support for each clade. Red font denotes the sequences obtained in this study. Rickettsia raoultii was used as outgroup. The green background represents intraerythrocytic Anaplasma spp.
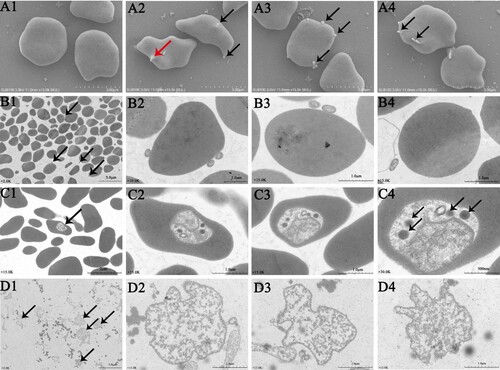
The erythrocytes infected with A. capra were examined using electron microscopy. SEM images demonstrated the presence of one or more A. capra cells (0.2–0.4 µm in diameter) on the outer surface of the erythrocytes ((A2–A4)). Additionally, invasion of the erythrocytes was also observed ((A2)). TEM images showed one or more typical small round to oval morulae (0.2 × 0.4 µm) on the membrane and in multiple erythrocytes ((B1–B4, C1–C4)). The morulae in the cytoplasm of the erythrocytes were about 10 times (0.8 × 1 µm) the size of those on the outside cells ((C1–C4)). The morulae density was not uniform, and electron-dense particles (Lysosomes) were observed in less-dense areas of the pathogen ((C2–C4)). When separated from erythrocytes, the morulae consisted of a membrane-bound vacuole containing dense granular bacterial subunits with a lower pathogen density than intracellular morulae, and the lysosomes were absent ((D1–D4)).
Figure 5. Photomicrographs of A. capra sp. nov and infected goat erythrocytes. A1–A4, SEM photomicrographs of erythrocytes and A. capra sp. nov. Normoerythrocytes (A1) and infected erythrocytes. Arrows indicate A. capra sp. nov (A2–A4). A2 shows invading pathogen (red arrowhead). B1–B4 and C1–C4, TEM photomicrographs of erythrocytes and A. capra sp. nov. Uranyl acetate and Reynold's lead citrate stain. Morulae are observed beside and within multiple erythrocytes (arrows). B2–B4 and C2–C4 show higher magnification of the morulae. Electron-dense particles (Lysosomes, arrows) are observed in less-dense areas (C2–C4, arrows). D1–D4, TEM photomicrographs of the separated A. capra sp. nov morulae. D1 (arrows) shows lower magnification of the morulae and D2–D4 present higher magnification of the morulae. No Lysosomes are observed in the morulae.
Wright–Giemsa-stained A. capra-positive erythrocytes smear showed numerous small morulae in the cytoplasm ((A1–A2)). In the CISH assay, microscopic observation demonstrated reddish-brown (DIG) specific probe signals on the surface and inside of A. capra-positive erythrocytes ((B1)). The negative controls showed erythrocytes with non-specific probe signals ((B2)).
Figure 6. Wright–Giemsa, CISH, immunocytochemistry, and IFA analyses of uninfected and infected goat erythrocytes. A1–A2, Wright–Giemsa-stained erythrocytes. B1–B2, CISH assay of the erythrocytes smear. A1–D1 are negative controls. The probe was labeled with DIG. C1–C2 and D1–D2, immunocytochemistry and IFA of erythrocytes incubated with positive goat serum. Black and white arrows denote A. capra sp. nov-positive erythrocytes. Red arrows show A. phagocytophilum-positive neutrophilic granulocytes.
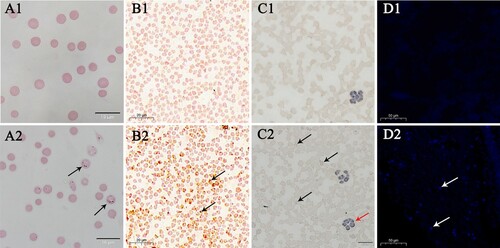
Immunocytochemistry-positive cells were observed in erythrocytes, and appeared as intracytoplasmic aggregates, known as morulae ((C1–C2)). No morulae were observed in the negative controls. In addition, immunocytochemistry-positive cells were occasionally noted in neutrophilic granulocytes, as the whole blood cells were also Anaplasma phagocytophilum-positive.
IFA positive fluorescence was obtained with A. capra antiserum, confirming that the cells contained A. capra parasites ((D2)). In contrast, negative-control erythrocytes showed no fluorescence ((D1)). Thus, owing to its unique morphological characteristics and parasitic site, we propose to revise the Anaplasma genus and formally name “A. capra” as Anaplasma capra sp. nov.
Discussion
Anaplasma capra sp. nov was first identified from seven goats based on the 16S rRNA gene in Southwest China. Sequences from the goats formed two Anaplasma spp. clusters that clearly distinguished them from the clusters of A. marginale, A. centrale, and A. ovis, suggesting that this pathogen could be a potential new Anaplasma sp. [Citation2]. Similarly, Liu et al. [Citation17] (2012) performed molecular analysis of the pathogen based on 16S rRNA gene in goats from Zhejiang, China. A comparable genotype of this Anaplasma sp. was detected in humans with tick-bite history since 2015 based on groEL, 16S rRNA, gltA, msp2, and msp4 genes [Citation1], and was provisionally nominated as “A. capra,” belonging to the Genus Anaplasma. Likewise, we found that A. capra sp. nov exhibited significant differences in groEL, 16S rRNA, gltA, and msp4 genes, when compared with other Anaplasma spp. [Citation5]. Many studies have used this molecular method to identify and characterize A. capra sp. nov in different hosts [Citation6,Citation7,Citation24–26].
Anaplasma capra sp. nov appears to exhibit at least two different genotypes, both are likely zoonotic [Citation4, Citation7]. The sequences of gltA and groEL genes examined in the present study formed two clusters in the phylogenetic tree. In a previous study, sequence analysis based on gltA gene demonstrated a novel A. capra sp. nov genotype in sheep, which was distinct from the isolates identified from patients in northeastern China [Citation27]. Furthermore, our previous study also identified two genotypes based on gltA gene of A. capra sp. nov in sheep and goats from Henan, China [Citation5]. Additionally, it has been reported that sequences based on the 16S rRNA gene from goat strains and the gltA gene from sheep strains formed two distinct A. capra sp. nov clusters [Citation2,Citation28]. Hence, our future research will involve genotyping A. capra sp. nov. As both the genotypes of A. capra sp. nov have been detected in ruminants and hard ticks, they pose a potential health threat to humans. The host range of the zoonotic genotypes needs further study, especially in areas infested with hard ticks.
Anaplasma spp. that have been reported to infect erythrocytes include Anaplasma marginale, A. centrale, and A. ovis [Citation29–31]. A. marginale and A. centrale were first identified in 1910 and 1911, respectively, by Theiler, who observed small morulae of A. marginale that was previously mistakenly as Babesia bigemina at the periphery of stained cattle erythrocytes [Citation18,Citation32]. Anaplasma centrale forms smaller and more central morulae and is closely associated with A. marginale within infected erythrocytes [Citation30], while A. ovis was first detected in the central area of sheep erythrocytes in 1912 [Citation31]. However, there are still no relevant reports on the specific site location of A. capra sp. nov. Similar to other Anaplasma spp., the target cells of this pathogen may be one type of blood cell. Li et al. presumed that A. capra sp. nov most probably infect erythrocytes or endothelial cells in mammals; however, this assumption has not yet been proved [Citation1]. Interestingly, in the present study, A. capra sp. nov was detected inside or at the periphery of erythrocytes that were A. capra sp. nov PCR-positive and A. marginale, A. centrale, and A. ovis PCR-negative, and could infect human erythrocytes. The morphological characteristics of A. capra sp. nov determined in the present study are consistent with those previously reported [Citation1], and different from the other intraerythrocytic Anaplasma spp. [Citation20,Citation23,Citation33–35]. These findings significantly contribute to the research on this novel zoonotic Anaplasma sp.
Erythrocytes infected with intraerythrocytic Anaplasma spp. are destroyed by macrophages, resulting in mild to severe hemolytic anemia [Citation18]. Sheep and goats infected with Anaplasma spp. develop a mild to severe disease, with clinical symptoms such as fever, pale mucous membranes, weight loss, icterus, anorexia, depression, lower milk production, coughing, dyspnea, gastrointestinal signs, abortion, and death [Citation36,Citation37]. Humans infected with A. capra sp. nov exhibit fever, headache, malaise, dizziness, chills, nausea, vomiting, diarrhea, and laboratory abnormalities, e.g. high hepatic aminotransferase concentrations, leucopenia, and thrombocytopenia [Citation1]. The pathogen is carried and may be transmitted to their zoonotic and domestic hosts by many species of hard ticks that are widely distributed [Citation9–12,Citation38–41]. Thus, A. capra sp. nov is a substantial threat to humans and domestic animals, and precautions should be taken to prevent anaplasmosis caused by this novel tick-borne pathogen.
Conclusion
To our knowledge, this was the first study to detect a novel zoonotic tick-borne Anaplasma sp., A. capra, in host erythrocytes. The molecular characteristics, morphological features, and parasitic sites of A. capra were noted to differ from those of other Anaplasma spp. Hence, we propose revising the Genus Anaplasma and formally naming “A. capra” as A. capra sp. nov.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Li H, Zheng YC, Ma L, et al. Human infection with a novel tick-borne Anaplasma species in China: a surveillance study. Lancet Infect Dis. 2015;15(6):663–670.
- Zhou Z, Nie K, Tang C, et al. Phylogenetic analysis of the genus Anaplasma in Southwestern China based on 16S rRNA sequence. Res Vet Sci. 2010;89(2):262–265.
- Shi K, Li J, Yan Y, et al. Dogs as new hosts for the emerging zoonotic pathogen Anaplasma capra in China. Front Cell Infect Microbiol. 2019;9:394.
- Yang J, Liu Z, Niu Q, et al. A novel genotype of “Anaplasma capra” in wildlife and its phylogenetic relationship with the human genotypes. Emerg Microbes Infect. 2018;7(1):210.
- Peng Y, Wang K, Zhao S, et al. Detection and phylogenetic characterization of Anaplasma capra: an emerging pathogen in sheep and goats in China. Front Cell Infect Microbiol. 2018;8:283.
- Shi Y, Yang J, Guan G, et al. Molecular investigation of Anaplasma species in sheep from Heilongjiang province, northeast China identified four Anaplasma species and a novel genotype of Anaplasma capra. Parasitol Int. 2020;76:102072.
- Amer S, Kim S, Yun Y, et al. Novel variants of the newly emerged Anaplasma capra from Korean water deer (Hydropotes inermis argyropus) in South Korea. Parasit Vectors. 2019;12(1):365.
- Jouglin M, Blanc B, de la Cotte N, et al. First detection and molecular identification of the zoonotic Anaplasma capra in deer in France. PLoS One. 2019;14(7):e0219184.
- Seo MG, Ouh IO, Lee H, et al. Differential identification of Anaplasma in cattle and potential of cattle to serve as reservoirs of Anaplasma capra, an emerging tick-borne zoonotic pathogen. Vet Microbiol. 2018;226:15–22.
- Fang LQ, Liu K, Li XL, et al. Emerging tick-borne infections in mainland China: an increasing public health threat. Lancet Infect Dis. 2015;15(12):1467–1479.
- Guo WP, Zhang B, Wang YH, et al. Molecular identification and characterization of Anaplasma capra and Anaplasma platys-like in Rhipicephalus microplus in Ankang, Northwest China. BMC Infect Dis. 2019;19(1):434.
- Yang J, Liu Z, Niu Q, et al. Molecular survey and characterization of a novel Anaplasma species closely related to Anaplasma capra in ticks, northwestern China. Parasit Vectors. 2016;9(1):603.
- Sun XF, Zhao L, Wen HL, et al. Anaplasma species in China. Lancet Infect Dis. 2015;15(11):1263–1264.
- Seo HJ, Jin BC, Kim KH, et al. Molecular detection and phylogenetic analysis of Anaplasma spp. in Korean native goats from Ulsan Metropolitan City, Korea. Vector Borne Zoonotic Dis. 2019;19(10):773–776.
- Rocchigiani G, Ebani V, Nardoni S, et al. Molecular survey on the occurrence of arthropod-borne pathogens in wild brown hares (Lepus europaeus) from central Italy. Infect Genet Evol. 2018;59:142–147.
- Sato M, Nishizawa I, Fujihara M, et al. Phylogenetic analysis of the 16S rRNA gene of Anaplasma species detected from Japanese serows (Capricornis crispus). J Vet Med Sci. 2009;71(12):1677–1679.
- Said M B, Belkahia H, Messadi L. Anaplasma spp. in North Africa: a review on molecular epidemiology, associated risk factors and genetic characteristics. Ticks Tick Borne Dis. 2018;9(3):543–555.
- Liu Z, Ma M, Wang Z, et al. Molecular survey and genetic identification of Anaplasma species in goats from central and southern China. Appl Environ Microbiol. 2012;78(2):464–470.
- Battilani M, De Arcangeli S, Balboni A, et al. Genetic diversity and molecular epidemiology of Anaplasma. Infect Genet Evol. 2017;49:195–211.
- Arraga-Alvarado CM, Qurollo BA, Parra OC, et al. Case report: molecular evidence of Anaplasma platys infection in two women from Venezuela. Am J Trop Med Hyg. 2014;91(6):1161–1165.
- Wei R, Liu HB, Jongejan F, et al. Cultivation of Anaplasma ovis in the HL-60 human promyelocytic leukemia cell line. Emerg Microbes Infect. 2017;6(9):e83.
- Chochlakis D, Ioannou I, Tselentis Y, et al. Human anaplasmosis and Anaplasma ovis variant. Emerg Infect Dis. 2010;16(6):1031–1032.
- Peng Y, Zhao S, Wang K, et al. A multiplex PCR detection assay for the identification of clinically relevant Anaplasma species in field blood samples. Front Microbiol. 2020;11:606.
- Raskin RE, Crosby FL, Jacobson ER. Newly recognized Anaplasma sp. in erythrocytes from Gopher tortoises (Gopherus polyphemus). Vet Clin Pathol. 2020;49:17–22.
- Qin X, Han F, Luo L, et al. Anaplasma species detected in Haemaphysalis longicornis tick from China. Ticks Tick Borne Dis. 2018;9(4):840–843.
- Lu M, Tian JH, Yu B, et al. Extensive diversity of rickettsiales bacteria in ticks from Wuhan, China. Ticks Tick Borne Dis. 2017;8(4):574–580.
- Han R, Yang JF, Mukhtar MU, et al. Molecular detection of Anaplasma infections in ixodid ticks from the Qinghai-Tibet Plateau. Infect Dis Poverty. 2019;8(1):12.
- Yang J, Han R, Niu Q, et al. Occurrence of four Anaplasma species with veterinary and public health significance in sheep, northwestern China. Ticks Tick Borne Dis. 2018;9(1):82–85.
- Henker LC, Lorenzett MP, Fagundes-Moreira R, et al. Bovine abortion, stillbirth and neonatal death associated with Babesia bovis and Anaplasma sp. infections in southern Brazil. Ticks Tick Borne Dis. 2020;11(4):101443.
- Theiler A. Further investigations into anaplasmosis of South African cattle. First Report of the Director of Veterinary Research; 1911:7–46.
- Bevan LLEW. Anaplasmosis of sheep. Vet J. 1912;68:400–401.
- Theiler A. Gall-sickness of South Africa (anaplasmosis of cattle). J Comp Pathol. 1910;23:98–115.
- Vanstreels RET, Yabsley MJ, Parsons NJ, et al. A novel candidate species of Anaplasma that infects avian erythrocytes. Parasit Vectors. 2018;11(1):525.
- Kocan KM, de la Fuente J, Guglielmone AA, et al. Antigens and alternatives for control of Anaplasma marginale infection in cattle. Clin Microbiol Rev. 2003;16(4):698–712.
- Potgieter FT, Rensburg LV. Tick transmission of Anaplasma centrale. Onderstepoort J Vet Res. 1987;54(1):5–7.
- Yasini S, Khaki Z, Rahbari S, et al. Hematologic and clinical aspects of experimental ovine anaplasmosis caused by Anaplasma ovis in Iran. Iran J Parasitol. 2012;7(4):91–98.
- Said M B, Belkahia H, Messadi L. Anaplasma spp. in North Africa: a review on molecular epidemiology, associated risk factors and genetic characteristics. Ticks Tick Borne Dis. 2018;9(3):543–555.
- White SA, Bevins SN, Ruder MG, et al. Surveys for ticks on wildlife hosts and in the environment at Asian longhorned tick (Haemaphysalis longicornis)-positive sites in Virginia and New Jersey. Transbound Emerg Dis. 2020: 1–10. DOI:https://doi.org/10.1111/tbed.13722.
- Egizi A, Bulaga-Seraphin L, Alt E, et al. First glimpse into the origin and spread of the Asian longhorned tick, Haemaphysalis longicornis, in the United States. Zoonoses Public Health. 2020;67:637–650.
- Segura J, Isaza J, Botero L, et al. Assessment of bacterial diversity of Rhipicephalus microplus ticks from two livestock agroecosystems in Antioquia, Colombia. PLoS One. 2020;15(7):e0234005.
- Paulauskas A, Sakalauskas P, Kaminskienė E, et al. First record of Haemaphysalis concinna (Acari: Ixodidae) in Lithuania. Ticks Tick Borne Dis. 2020;11(5):101460.
- Yang J, Liu Z, Niu Q, et al. A novel zoonotic Anaplasma species is prevalent in small ruminants: potential public health implications. Parasit Vectors. 2017;10(1):264.
- de la Fuente J, Lew A, Lutz H, et al. Genetic diversity of Anaplasma species major surface proteins and implications for anaplasmosis serodiagnosis and vaccine development. Anim Health Res Rev. 2005;6(1):75–89.
- Guimarães A, Raimundo JM, Peixoto MP, et al. Molecular detection, characterization of Anaplasma spp. in domestic cats from Rio de Janeiro state. Acta Trop. 2019;191:239–242.
- Khumalo ZTH, Brayton KA, Collins NE. Evidence confirming the phylogenetic position of Anaplasma centrale (ex Theiler 1911) Ristic and Kreier 1984. Int J Syst Evol Microbiol. 2018;68(8):2682–2691.
