ABSTRACT
Klebsiella pneumoniae carbapenemase (KPC)-producing Enterobacterales are prevalent worldwide and pose an alarming threat to public health. The incidence and transmission of blaKPC-2 gene via horizontal gene transfer (e.g. transposition) have been well documented. However, the dynamics of transposon structure bearing blaKPC-2 and their exact effects on the evolution and dissemination of blaKPC-2 gene are not well characterized. Here, we collected all 161 carbapenem-resistant Enterobacterales (CRE) isolates during the early stage of CRE pandemic. We observed that the prevalence of KPC-2-producing Enterobacterales was mediated by multiple species and sequence types (STs), and that blaKPC-2 gene was located on three diverse variants of Tn1721 in multi-drug resistance (MDR) region of plasmid. Notably, the outbreak of KPC-2-producing plasmid is correlated with the dynamics of transposon structure. Furthermore, we experimentally demonstrated that replicative transposition of Tn1721 and IS26 promotes horizontal transfer of blaKPC-2 and the evolution of KPC-2-producing plasmid. The Tn1721 variants appearing concurrently with the peak of an epidemic (A2- and B-type) showed higher transposition frequencies and a certain superior ability to propagation. Overall, our work suggests replicative transposition contributes to the evolution and transmission of KPC-2-producing plasmid and highlights its important role in the inter- and intra-species dissemination of blaKPC-2 gene in Enterobacterales.
Introduction
Klebsiella pneumoniae carbapenemase (KPC) is a class A serine β-lactamase that efficiently hydrolyzes most β-lactam antimicrobial agents, including carbapenems, limiting treatment options in infected patients seriously [Citation1]. KPC-producing Enterobacterales, particularly K. pneumoniae, have spread worldwide over the last decade, becoming an urgent public health threat [Citation2]. KPC-2, the most commonly identified variant, is a dominant factor leading to carbapenem resistance in Enterobacterales. The blaKPC-2 gene is typically identified in mobile transposon, which is most often situated on conjugative plasmids [Citation2–4]. Tn4401 is the major vehicle of blaKPC-2 in most countries and regions, such as Europe [Citation5], the United States [Citation6], and Brazil [Citation7]. In Asia, blaKPC-2 is mostly located on diverse variants of Tn1721 and IS26 [Citation8–10].
Horizontal gene transfer (HGT) plays an important role in the evolution of bacteria and the dissemination of antibiotic resistance genes [Citation11]. Tn4401 and Tn1721, typical replicative transposons belonging to Tn3 family, have been proved to mobilize blaKPC-2 at a high transposition frequency, and the latter is capable of transferring blaKPC-2 both internal and external to this element [Citation12,Citation13]. However, few published studies provided comprehensively analysis of epidemic of KPC-2-producing Enterobacterales, the dynamics of genetic structure surrounding blaKPC-2 and transposition mechanism of these elements. Notably, blaKPC-2 is located on diverse variants of Tn1721 that exhibited various transposition frequencies and movement patterns [Citation13], and the mechanism of movement of A2-type (Tn1721-blaKPC-2-IS26) remains undetermined.
Here, we show that outbreak of KPC-2-producing plasmid is correlated with the dynamics of transposon structure in Enterobacterales. The A2- and B-type Tn1721 appearing concurrently with the peak of the epidemic of blaKPC-2-carrying isolates demonstrated higher transposition frequency. Their specific replicative transposition (the IS26 pattern for A2-type and the Tn1721-blaKPC-2-IRL2 pattern for B-type) had a certain superior ability to conjugate into another strain. Thus, replicative transposition contributes to the evolution and dissemination of KPC-2-producing plasmid in Enterobacterales, facilitating the inter- and intra-species dissemination of blaKPC-2.
Materials and methods
Media and growth of strains
Bacteria were routinely cultured at 37°C in Luria–Bertani (LB) medium or on LB agar. Plasmids were constructed in Escherichia coli DH5α (supE44 Δ lacU169 [ϕ 80 lacZ ΔM15] hsdR17 recA1 endA1 gyrA96 thi-1 relA1). HB101 (recA13 F- STRr) and E. coli J53 (AZr) were used as recipient strains in transposition experiments and conjugation experiments, respectively. The following antibiotics were added at the indicated concentrations: imipenem (IPM), 1 mg/L; streptomycin (STR), 25 mg/L; trimethoprim (TMP), 25 mg/L and sodium azide (AZ), 100 mg/L.
Clinical isolates
A total of 161 non-duplicated, carbapenem-resistant Enterobacterales (CRE) isolates were collected from August 2006 to December 2010 during routine identification and antimicrobial susceptibility testing by the Microbiology Laboratory, Huashan Hospital, Fudan University (Shanghai, China). This collection comprised all clinical isolates from the first occurrence of KPC-2-producing isolates (K. pneumoniae) to the prevalence of this cabapenemase (Dataset S1). For comparison, a susceptible collection of 112 carbapenem-sensitive K. pneumoniae (CS-KP) were also collected contemporaneously from similar departments (Dataset S2).
Multilocus sequence typing (MLST) and pulsed-field gel electrophoresis (PFGE)
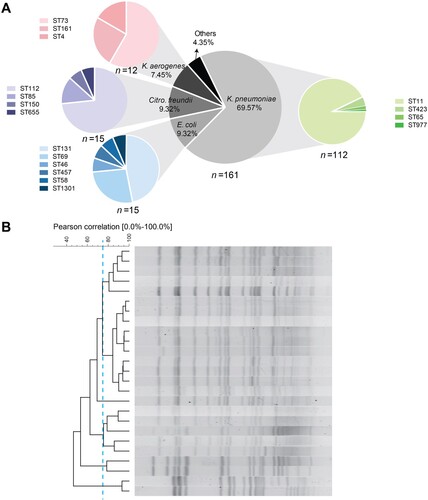
MLST was performed according to the protocol described on the Pasteur Institute MLST website (https://bigsdb.pasteur.fr/klebsiella/klebsiella.html for K. pneumoniae and https://bigsdb.pasteur.fr/ecoli/ecoli.html for E.coli) and PubMLST website (https://pubmlst.org/organisms/citrobacter-spp for Citrobacter freundii and https://pubmlst.org/organisms/klebsiella-aerogenes for Klebsiella aerogenes). Genetic relatedness among the ST11 K. pneumoniae isolates was analyzed by XbaI-PFGE type as described previously [Citation9]. Dendrograms were conducted using the Dice coefficient and the unweighted pair-group method using average linkage clustering [Citation14].
PCR screening of blaKPC-2 and IncFII replicon and analysis of genetic environment of blaKPC-2 gene among clinical isolates
The blaKPC-2 were identified through the amplification and sequence analysis of a 750-bp polymerase chain reaction (PCR) product [Citation15]. IncFII replicon screen was conducted by PCR-based replicon typing using previously reported primers [Citation16]. The blaKPC-2-bearing genetic structures were determined by a series of PCR assays as reported previously [Citation15].
Transformation and conjugation experiments of blaKPC-2-bearing plasmids
Thirty-five blaKPC-2-bearing plasmids were obtained by transformation or conjugation (Dataset S3). Plasmids of clinical isolates were extracted with a Qiagen Plasmid Midi kit (Qiagen, Germany) and examined by agarose gel electrophoresis, and then transformed into E. coli DH5α Electrocompetent cells by electroporation (Micro-Pulser electroporator; Bio-Rad, USA). Conjugation experiment was performed with E. coli J53 (AZr) as the recipient. Transformants and conjugants were selected on MacConkey agar containing IPM (AZ was also used in conjugants selection) and identified by VITEK 2 system (bioMérieux, France) and were further subjected to PCR amplification of IncFII replicons, blaKPC-2 and genetic environment of blaKPC-2 according to our previous operations. For plasmids originated from clinical isolates of E. coli, each transformant was also identified by detecting the deletion of lacY gene as E. coli DH5α lacks the lac operon. Primers are listed in Table S1.
Bioinformatics analysis
All complete genome sequences of Enterobacterales publicly available (5152 in total) and all of the plasmid sequences harbored in these strains (10,507 in total) were downloaded from NCBI database in August 2021 (Chromosomes in Dataset S4 and plasmids in Dataset S5). The blaKPC-2 gene was identified by using nucleotide BLAST. Plasmid incompatibility type was determined by comparing with information in the Plasmid MLST locus/sequence definitions database (https://pubmlst.org/bigsdb?db=pubmlst_plasmid_seqdef& page = sequenceQuery).
Plasmids construction
The primers and plasmids used in this study are listed in Table S1 and Table S2, respectively. pHS10842 (GenBank accession no. KP125892), a vector favorable for use in the exploration of the transposition mechanism of Tn1721-like transposons, has been described previously [Citation13].
For pHS10842-ΔtnpATn1721, IRR and tnpR fragments were amplified with primers JP934/JP935 and JP936/JP937, respectively. The IRR fragment share 20-bp sequences with AhdI restriction sites of pHS10842 and the tnpR fragments, respectively. The tnpR fragments also share 20-bp sequences with the IRR fragment and AflII restriction sites of pHS10842, respectively. The two fragments were subcloned into AhdI and AflII restriction sites of pHS10842 by use of the NEBuilder HiFi DNA assembly master mix (New England BioLabs, USA), generating the derivative with the tnpA deletion of Tn1721, pHS10842-ΔtnpATn1721.
The transposase deletion was introduced into IS26 by digesting plasmid pHS10842 with SwaI and XmnI to removal a 560-bp fragment (base 11,366 to base 11,925 in GenBank accession no. KP125892) and generate blunt ends. The DNA was ligated and transformed into competent cells of E. coli DH5α. pHS10842-ΔtnpATn1721Δtnp26 was constructed in a similar strategy with pHS10842-ΔtnpATn1721.
Transposition assays and molecular characterization of transposition events
Transposition assays were performed as described previously [Citation13,Citation17]. Transposition frequency is calculated as the number of IPMr TMPr STRr transconjugants per TMPr STRr transconjugant. For each transposition event, the movement patterns were determined by agarose gel electrophoresis and Southern hybridization [Citation13]. The exact insertion site and target site duplication of Tn1721 and IS26 were determined by using primes specific for regions internal to Tn1721 and IS26 and primers specific for R388 as described previously [Citation13]. Primers are listed in Table S1.
Analysis of the target site consensus sequence
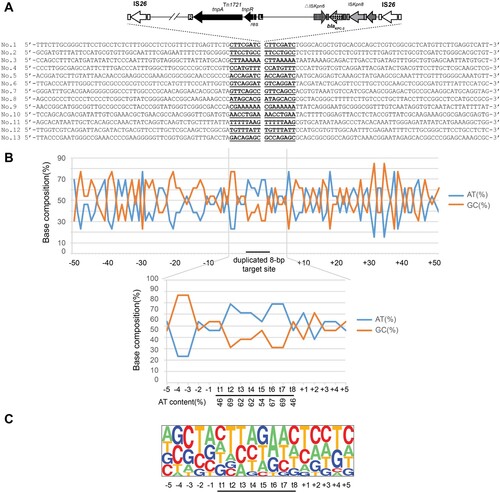
The relative frequencies of the AT and GC contents of the region extending from 50 bp upstream to 50 bp downstream of the duplicated target site for IS26 (8 bp) were calculated and plotted on a line graph. The pictures of the relative frequencies of the bases at each position were generated with the Pictogram program (http://genes.mit.edu/pictogram.html).
Statistics
Statistical significance was assessed by Fisher’s exact test or Chi-square test with Yates’ correction using GraphPad Prism8 software (https://www.graphpad.com/). P < 0.05 was considered statistically significant.
Results
The prevalence of KPC-2-producing Enterobacterales was mediated by multiple species and STs
All of the 161 CRE were identified as blaKPC-2-positive, including K. pneumoniae (112, 69.56%), E. coli (15, 9.32%), Citro. freundii (15, 9.32%), K. aerogenes (12, 7.45%) and other species (7, 4.35%) (A). Among the 112 K. pneumoniae isolates, ST11 was the most prevalent ST, followed by ST423, ST65 and ST977, and PFGE of ST11 K. pneumoniae indicated five diverse subtypes with a criterion of 75% identity (B). For comparison, we collected CS-KP as susceptible controls that were matched by time and ward. As expected, all CS-KP isolates were blaKPC-2-negative and the STs of the susceptible collection were scattered without any dominant STs (Figure S1). E. coli, Citro. freundii and K. aerogenes isolates were comprised of several STs. Together, these results suggested that blaKPC-2 is the chief culprit leading to carbapenem resistance and that the prevalence of blaKPC-2 in Enterobacterales was mediated by multiple species and STs, rather than clonal spread.
blaKPC-2 genes were usually located on three diverse variants of Tn1721 in MDR region of plasmid
The blaKPC-2 genes were usually reported to be located on MDR region of plasmid, and IncFII plasmids contributed significantly to the global prevalence of blaKPC among K. pneumoniae [Citation4,Citation9]. To determine the correlation between IncFII plasmids and blaKPC-2 gene, we conducted a bioinformatic analysis of 5152 complete genome sequences of Enterobacterales publicly available (including 5152 chromosomes and 10,506 plasmids in total, Dataset S4 and Dataset S5). Among these, 311 strains were identified to be blaKPC-2-positive, and the overwhelming majority of blaKPC-2 genes were found on plasmids (300/311, 96.46%). Remarkably, most of the blaKPC-2-bearing plasmids belonged to IncFII group, but this group was less common in blaKPC-2-negative plasmids [174/300 (58%) versus 2195/10206 (21.51%), P < 0.0001; ]. Moreover, IncFII plasmids were significantly over-represented in blaKPC-2-positive K. pneumoniae compared with that in blaKPC-2-negative group [151/204 (74.02%) versus 657/2701 (24.32%), P < 0.0001; ]. These findings suggest that blaKPC-2 genes are mostly located on plasmids in Enterobacterales, and that IncFII is the most common Incompatibility group, especially in K. pneumoniae.
Table 1. Incompatibility group analysis of plasmids with or without blaKPC-2 obtained from NCBI database.
To evaluate this correlation in clinical isolates, we first performed IncFII replicon screening on all clinical isolates. Significantly, the detection rate of IncFII replicon was 84.82% in K. pneumoniae and the rate was slightly lower for E. coli (73.33%) and K. aerogenes (66.67%) (Table S3). None of Citro. freundii isolates was detected to IncFII-positive. However, the rate was much lower in carbapenem-susceptible collection in comparison with either CR-KP or CRE collection (49.11% vs. 84.82% for CR-KP, P < 0.0001; 49.11% vs. 72.05% for CRE, P = 0.0001, A). Next, the linkage between IncFII plasmids and blaKPC-2 gene was determined in thirty-five transformants and conjugants containing blaKPC-2-positive plasmid by PCR amplification of IncFII replicons and genetic environment of blaKPC-2 (see Dataset S3 for details). Interestingly, 80% of plasmids carrying blaKPC-2 gene (28/35) belong to IncFII group. Finally, five plasmids in various sizes (two IncFII-negative and four IncFII-positive, Dataset S3) were selected for complete sequence analysis to furtherly confirm the correlation between IncFII plasmids and blaKPC-2 gene.
Figure 2. IncFII screen of clinical isolates and genetic structures surrounding blaKPC-2. (A) IncFII-positive rates of blaKPC-2-positive and blaKPC-2-negative groups. (B) Schematic representation of blaKPC-2-bearing genetic elements classified as A1-, A2- and B-type. Genes are depicted as arrows according to the direction of transcription. blaKPC-2 is shown as spotted arrows. Inverted repeats are indicated by rectangles in different colors: Tn1721 (black), Tn3 (dark gray), ISKpn8 (light gray), and IS26 (white). Regions sharing identical sequences across plasmids are indicated by gray shading between the different representations of the plasmids. The GenBank accession numbers for pHS10505, pHS062105, and pHS10842 are KF826292, KF623109, and KP125892, respectively.
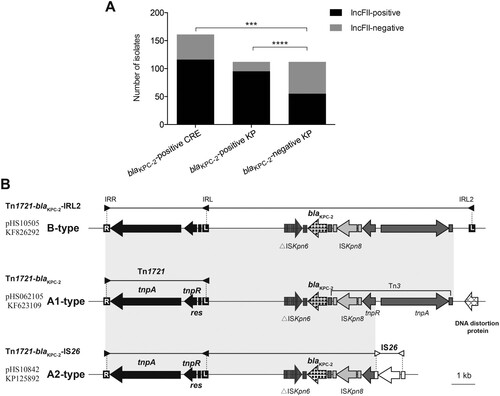
Moreover, our analysis of genetic environment of blaKPC-2 gene among 161 CRE identified three diverse variants of Tn1721 bearing blaKPC-2 (designated A1-, A2- and B-type) (B). All of them possess a core element (Tn1721) that consists of two 38-bp terminal inverted repeats (IRs; the right inverted repeat [IRR] and the left inverted repeat [IRL]), transposase (TnpA), resolvase (TnpR) and resolution site (res). The B-type is composed of Tn1721, blaKPC-2, an additional inverted repeat (IRL2), as well as several unrelated elements (Tn3, ISKpn6, and ISKpn8). The sequence of A1-type (Tn1721-blaKPC-2) shares 100% identity with that of B-type, but it does not contain the additional IRL, IRL2. In the A2-type, Tn3 was disrupted by an IS26, forming the chimera, Tn1721-blaKPC-2-IS26. The A2-type Tn1721 accounted for more than half the samples (59.01%, 95/161), and A1- and B-type accounted for 19.25% (31/161) and 18.63% (30/161), respectively ( and B).
Table 2. Detection rate of Tn1721-like transposons bearing blaKPC-2 on 161 CRE.
Outbreak of KPC-2-producing plasmids is correlated with the dynamics of transposon structure
In order to clarify the progression of blaKPC-2 outbreak, we comprehensively analyzed all data obtained in molecular epidemiological investigation. provides an overview of the evolution of blaKPC-2 bearing structure in Enterobacterales. Samples are plotted by month of isolation, wards, species, STs of most common resistant specie (K. pneumoniae) and transposable elements carrying blaKPC-2 gene. As shown in , blaKPC-2 genes that were located in three distinct Tn1721-like transposons on plasmid, originated from K. pneumoniae, then became increasingly prevalent in this specie and spread in Enterobacterales further. The epidemic of blaKPC-2-carrying plasmid reported here had three stages, designated as (i) distributed period (August 2006–April 2009, 19 isolates), (ii) epidemic period (May 2009–February 2010, 50 isolates), and (iii) mixed epidemic period (March–December 2010, 92 isolates). In the distributed period, blaKPC-2 appeared sporadically in K. pneumoniae without any predominate STs, and all Tn1721-like transposons belonged to A1-type. At the second epidemic stage, KPC-2-producing plasmid were prevalent in K. pneumoniae ST11 that consisted of several different subtypes, and spread into other Enterobacterales. A2-type Tn1721 was the dominant structure carrying blaKPC-2. In the mixed epidemic period, KPC-2-producing isolates increased furtherly. This stage involved three distinct Tn1721 variants, and the number of emerging B-type was almost equal to that for A2-type during the same period.
Figure 3. Overview of 161 CRE clinical isolates collected from Huashan Hospital. K. pneumoniae isolates are shown as solid signs: squares (ST423), circles (ST977), triangles (ST65), and asterisks (ST11). Other Enterobacterales isolates are shown as empty signs: circles (E. coli) and triangles (others). Isolates are plotted based on ward (vertical axis) and time of collection (horizontal axis). Color indicates distinct Tn1721 variants carrying blaKPC-2: A1-type (green), A2-type (red), B-type (blue) and undefined (black).
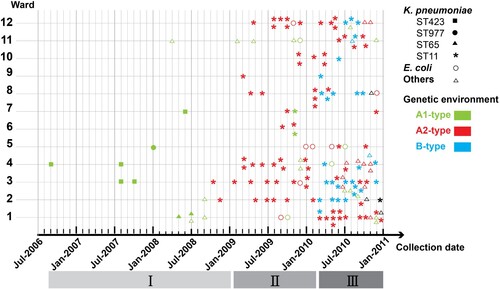
Altogether, outbreak of KPC-2-producing plasmid was correlated with the dynamics of blaKPC-2-bearing transposon structure, and A2- and B-type Tn1721 appeared concurrently with the peak of blaKPC-2 epidemic.
Replicative transposition promotes horizontal transfer of blaKPC-2
A critical step in the dissemination process of blaKPC-2 is the HGT of mobile genetic elements (MGEs) surrounding this determinant. According to our comprehensive analysis of epidemiological data, it was presumed that replicative transposons, Tn1721 and IS26, can mobilize blaKPC-2 through transposition, promoting the dissemination of blaKPC-2 in Enterobacterales. In our previous study, A1- and B-type Tn1721 have been shown to transfer blaKPC-2 both internal and external to this element, and target transposition into 5-bp region that gradually exhibits a degenerated degree of AT-rich regions from both sides to the middle and that is immediately flanked by GC-rich regions [Citation13]. Here, we characterized the movement and target site of A2-type Tn1721 (Tn1721-blaKPC-2-IS26).
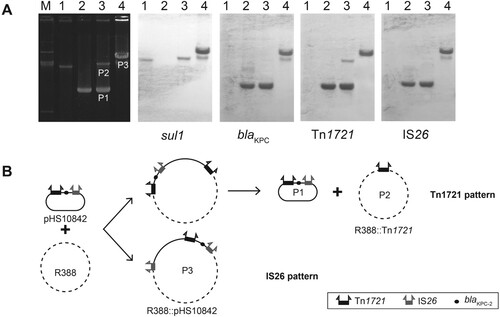
Two distinct patterns of movement mediated by Tn1721 and IS26 existed in this chimera. Tn1721 pattern was the same as the one detected in A1- and B-type Tn1721 previously, including cointegrate forming and resolving steps [Citation13]. In addition to having the Tn1721 pattern, a different IS26 pattern via replicative transposition was also detected in several cases of A2-type Tn1721 (B). Only one plasmid (P3) was obtained from the transconjugant. The donor (pHS10842) and target (R388) plasmids generated this cointegrate (P3) in which both plasmids fused together by directly repeated copies of IS26. This result was supported by a series of data (A), as follows. (i) the size of P3 was larger than that of R388 and P2. (ii) Southern blot analysis showed that P3 contained blaKPC-2, Tn1721, sul1, IS26. (iii) A series of PCRs confirmed that P3 had the same blaKPC-2-bearing genetic structure (Tn1721-blaKPC-2-IS26) as that of pHS10842, and the junctions between donor and target in each case were amplified and sequenced with primers specific for regions internal to IS26 and primers specific for R388.
Figure 4. Transposition patterns in A2-type (Tn1721-blaKPC-2-IS26). (A) Southern hybridization of plasmids in typical transconjugants. The electrophoretic profiles of the plasmids and hybridization with the sul1-specific probes (sul1 is a genetic marker of R388), blaKPC-2-specific probes, Tn1721-specific probes, and IS26-specific probes are shown. (B) Two transposition patterns (the Tn1721 pattern and the IS26 pattern) in Tn1721-blaKPC-2-IS26 structure.
Consistent with previous reports [Citation18], 8-bp target site duplication was evidenced for each transposition events (). At the target sites, the AT content for regions from t2 to t7 stabilize at 60-70%, while the AT content at t1 and t8 was slightly lower (46%). Nucleotide composition analysis revealed the t2 and t3 positions were found to be predominantly T residues (54% and 38%, respectively), while t6 and t7 positions were mostly A residues with the same percentage as that for t2 and t3, respectively. It is noteworthy that the insertion sites mostly carried one or even more AA or/and TT nucleotide tandems. These data suggested that IS26 preferentially targets AT-rich regions with AA and TT nucleotide tandems.
Figure 5. Target site analysis of IS26. (A) Sequence alignment of 13 transposition events of IS26 in R388 plasmid. The duplicated 8-bp target site sequence is underlined. The 50 nucleotides upstream and downstream of the target sites are shown. (B) Statistical analysis of the nucleotides at the 13 transposition sites. The percentages of AT and GC residues at each position from 50 nucleotides upstream to 50 nucleotides downstream of the target site are shown. The 8-bp duplication of target site, here named t1, t2, t3, t4, t5, t6, t7, and t8 are designated by a black bar. The percentage of AT and GC residues in the region spanning positions −5 to 5 are also indicated in the lower graphs. (C) Pictogram showing the relative frequencies of each A, T, C, and G residue at the target site. The data were deduced from the 13 experimental transposition events shown in panel A.
The transposition frequency of A2-type Tn1721 was measured to be 3.8 × 10−6 (). This represented a 4-fold increase in efficiency compared to that of A1-type, a blaKPC-2-bearing structure lacking IS26, and 30% of that for B-type Tn1721 containing an additional left inverted repeat (IRL2). The tnpA deletion of either Tn1721 or IS26 decreased half of the frequency, and both tnpA deletions led to a functional inability to mobilize blaKPC-2. Hence, blaKPC-2 embedded between Tn1721 and IS26 was transferred at an apparently higher frequency owing to the existence of both elements.
Table 3. Transposition frequency of Tn1721-like transposons and derivativesTable Footnotea.
Taken together, our findings indicate that all three transposons were capable of transferring blaKPC-2 through replicative transposition, and that A2- and B-type Tn1721 showed higher transposition frequencies and had a certain superior ability to propagation.
Replicative transposition of Tn1721 contributes to the evolution and dissemination of KPC-2-producing plasmid
Given that Tn1721 variants have been shown various capacities of transferring blaKPC-2 via replicative transposition, an attractive hypothesis is that the A2- and B-type Tn1721 occasionally appearing in the late-stage stood out from the rest through selection for their ability to facilitate dissemination of blaKPC-2. As discussed above, the outbreak of KPC-2-producing plasmid was correlated with the dynamics of blaKPC-2-bearing Tn1721 structure, and A2- and B-type Tn1721 arising concurrently with the peak of blaKPC-2 epidemic showed apparently higher transposition frequencies. Such an association and different movement capabilities imply that replicative transposition of Tn1721 variants contributes to the evolution and dissemination of KPC-2-producing plasmid in Enterobacterales. This mechanism could enable blaKPC-2 gene to search for suitable host spontaneously, dealing with the antibiotic pressure from the environment.
Discussions
KPC-producing Enterobacterales that have spread extensively throughout the world, are an important cause of nosocomial infections, especially urinary tract infections, respiratory tract infections, and bloodstream-associated infections [Citation1]. During the last decade, several studies have described of diverse transposable elements surrounding blaKPC-2 gene [Citation9,Citation10,Citation12]. However, little is known about the exact impacts of these genetic environment change and its correlation with epidemic of blaKPC-2 gene. Our investigation of clinical CRE provided a detailed description of the whole process from blaKPC-2 gene first occurrence, progress to final prevalence. We observed that the prevalence of KPC-2-producing Enterobacterales was mediated by multiple species and STs, and that blaKPC-2 gene was located on three diverse variants of Tn1721 in multi-drug resistance (MDR) region of plasmid. IncFII was the most common Incompatibility group. Further study indicated that replicative transposition contributed to the evolution and dissemination of KPC-2-producing plasmid.
Transposable elements play an important role in the genetic variation and evolution of bacteria. The blaKPC-2 gene is mostly located on transposable elements, such as Tn4401, Tn1721, and IS26 [Citation8,Citation13]. To date, eight variants of Tn4401 (Tn4401a to Tn4401h) have been identified, with Tn4401a and Tn4401b being the most widespread [Citation1,Citation12,Citation19,Citation20]. Notably, these isoforms demonstrate lacking tnpA or/and tnpR, or have deletions in the noncoding region upstream of blaKPC leading to enhanced or reduced expression of this carbapenemase [Citation21]. It is widely reported that Tn1721 variants are the dominant structures in Asia, particularly in China [Citation8–10,Citation15,Citation22]. Given that the structure of Tn1721 variants in current blaKPC-2 epidemic is too complicated and diverse to track the dissemination process of this gene, we focus on the early stage of blaKPC-2 epidemic. In addition to the dynamics of blaKPC-2-bearing transposon structure raised by this study, host and plasmid factors as well as antibiotic pressure from environment were also worth further study.
Our findings indicated that all three Tn1721 variants were capable of mobilizing blaKPC-2 via replicative transposition, and A2- and B-type Tn1721 exhibited higher transposition frequencies than A1-type. Such various capacities are presumably associated with cointegrate resolution of Tn1721 in transposition assay. For the transconjugants with the Tn1721 pattern, only the cointegrate could be conjugated into the recipient (E. coli HB101, STR resistant [STRr]) and screened on LB agar containing IPM, TMP, and STR in the transposition assay. Once the cointegrate resolved in donor strain, neither plasmid generated here could survive. Because P1 lacked the TMP resistance (TMPr) and the ability to conjugate into recipient (STRr), even though it possessed IPM resistance (IPMr), while P2 was an opposite case. In contrast, both the cointegrate and plasmid resolved from the cointegrate with Tn1721-blaKPC-2-IRL2 pattern possessed IPMr and TMPr, as well as had the ability to conjugate to recipient (STRr). For the transposition via IS26, there is no resolution process. It means that the cointegrate (P3) with IS26 pattern possessed IPMr and TMPr, as well as had the ability of conjugal transfer. Consequently, it survived on LB agar containing IPM, TMP, and STR. In other words, the plasmid with Tn1721-blaKPC-2-IRL2 pattern (in B-type) or IS26 pattern (in A2-type) had a certain superior ability of conjugal transfer and became more widespread. These results strongly supported our molecular epidemiological findings that the two peaks of blaKPC-2 epidemic followed the appearance of A2- and B-type Tn1721, reflecting an important role for replicative transposition of Tn1721 and IS26 in the evolution and transmission of blaKPC-2-carrying plasmid in Enterobacterales.
In conclusion, our work demonstrates replicative transposition facilitates the evolution and transmission of KPC-2-producing plasmid in Enterobacterales, and highlights its important role in the dissemination of antibiotic resistance genes between pathogenic bacterial species.
Supplemental Material
Download MS Excel (411.5 KB)Supplemental Material
Download MS Word (334.1 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Munoz-Price LS, Poirel L, Bonomo RA, et al. Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect Dis. 2013;13(9):785–796.
- Mathers AJ, Peirano G, Pitout JD. The role of epidemic resistance plasmids and international high-risk clones in the spread of multidrug-resistant enterobacteriaceae. Clin Microbiol Rev. 2015;28(3):565–591.
- Shen P, Zhang Y, Tang Y, et al. Molecular dissection of blaKPC-2-bearing plasmids evolving in Klebsiella pneumoniae isolated at one teaching hospital in shanghai, China. FEMS Microbiol Lett. 2016;363(15):fnw142.
- Peirano G, Bradford PA, Kazmierczak KM, et al. Importance of clonal complex 258 and IncFK2-like Plasmids among a global collection of Klebsiella pneumoniae with blaKPC. Antimicrob Agents Chemother. 2017;61(4).
- Naas T, Cuzon G, Villegas MV, et al. Genetic structures at the origin of acquisition of the beta-lactamase bla KPC gene. Antimicrob Agents Chemother. 2008;52(4):1257–1263.
- Chen L, Chavda KD, Melano RG, et al. Complete sequence of a bla(KPC-2)-harboring IncFII(K1) plasmid from a Klebsiella pneumoniae sequence type 258 strain. Antimicrob Agents Chemother. 2013;57(3):1542–1545.
- Pereira PS, de Araujo CF, Seki LM, et al. Update of the molecular epidemiology of KPC-2-producing Klebsiella pneumoniae in Brazil: spread of clonal complex11 (ST11, ST437 and ST340). J Antimicrob Chemother. 2013;68(2):312–316.
- Octavia S, Kalisvar M, Venkatachalam I, et al. Klebsiella pneumoniae and Klebsiella quasipneumoniae define the population structure of blaKPC-2Klebsiella: a 5 year retrospective genomic study in Singapore. J Antimicrob Chemother. 2019;74(11):3205–3210.
- Fu P, Tang Y, Li G, et al. Pandemic spread of blaKPC-2 among Klebsiella pneumoniae ST11 in China is associated with horizontal transfer mediated by IncFII-like plasmids. Int J Antimicrob Agents. 2019;54(2):117–124.
- Zhang W, Zhu Y, Wang C, et al. Characterization of a Multidrug-Resistant porcine Klebsiella pneumoniae sequence type 11 strain coharboring bla KPC-2 and fosA3 on Two Novel hybrid plasmids. mSphere. 2019 Sep 11;4(5).
- Woods LC, Gorrell RJ, Taylor F, et al. Horizontal gene transfer potentiates adaptation by reducing selective constraints on the spread of genetic variation. Proc Natl Acad Sci USA. 2020;117(43):26868–26875.
- Cuzon G, Naas T, Nordmann P. Functional characterization of Tn4401, a Tn3-based transposon involved in blaKPC gene mobilization. Antimicrob Agents Chemother. 2011;55(11):5370–5373.
- Tang Y, Li G, Liang W, et al. Translocation of carbapenemase gene blaKPC-2 both internal and external to transposons occurs via Novel structures of Tn1721 and exhibits distinct movement patterns. Antimicrob Agents Chemother. 2017;61(10).
- Yang J, Ye L, Guo L, et al. A nosocomial outbreak of KPC-2-producing Klebsiella pneumoniae in a Chinese hospital: dissemination of ST11 and emergence of ST37, ST392 and ST395. Clin Microbiol Infect. 2013;19(11):E509–E515.
- Shen P, Zhang Y, Li G, et al. Characterization of the genetic environment of the blaKPC-2 gene among Klebsiella pneumoniae isolates from a Chinese hospital. Braz J Infect Dis. 2016;20(4):384–388.
- Villa L, Garcia-Fernandez A, Fortini D, et al. Replicon sequence typing of IncF plasmids carrying virulence and resistance determinants. J Antimicrob Chemother. 2010;65(12):2518–2529.
- Harmer CJ, Moran RA, Hall RM. Movement of IS26-associated antibiotic resistance genes occurs via a translocatable unit that includes a single IS26 and preferentially inserts adjacent to another IS26. mBio. 2014;5(5):e01801–14.
- He S, Hickman AB, Varani AM, et al. Insertion sequence IS26 reorganizes plasmids in clinically isolated multidrug-resistant bacteria by replicative transposition. mBio. 2015 Jun 9;6(3):e00762.
- Baraniak A, Izdebski R, Fiett J, et al. KPC-Like Carbapenemase-producing Enterobacteriaceae colonizing patients in Europe and Israel. Antimicrob Agents Chemother. 2016;60(3):1912–1917.
- Pecora ND, Li N, Allard M, et al. Genomically informed surveillance for carbapenem-resistant Enterobacteriaceae in a health care system. mBio. 2015;6(4):e01030.
- Cheruvanky A, Stoesser N, Sheppard AE, et al. Enhanced Klebsiella pneumoniae carbapenemase expression from a Novel Tn4401 deletion. Antimicrob Agents Chemother. 2017;61(6).
- Chen YT, Lin JC, Fung CP, et al. KPC-2-encoding plasmids from Escherichia coli and Klebsiella pneumoniae in Taiwan. J Antimicrob Chemother. 2014;69(3):628–631.