?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.ABSTRACT
Candida auris has globally emerged as a multidrug-resistant fungus linked to healthcare-associated outbreaks. There is still limited evidence on its virulence, pathogenicity determinants, and complex host–pathogen interactions. This study analyzes the in vivo fungal behaviour, immune response, and host–pathogen interactions upon C. auris infection compared to C. albicans and C. parapsilosis in G. mellonella. This was performed by immunolabelling fungal structures and larval plasmatocytes and using a quantitative approach incorporating bioinformatic morphometric techniques into the study of microbial pathogenesis. C. auris presents a remarkably higher immunogenic activity than expected at its moderate degree of tissue invasion. It induces a greater inflammatory response than C. albicans and C. parapsilosis at the expense of plasmatocyte nodule formation, especially in non-aggregative strains. It specifically invades the larval respiratory system, in a pattern not previously observed in other Candida species, and presents inter-phenotypic tissue tropism differences. C. auris filaments in vivo less frequently than C. albicans or C. parapsilosis mostly through pseudohyphal growth. Filamentation might not be a major pathogenic determinant in C. auris, as less virulent aggregative phenotypes form pseudohyphae to a greater extent. C. auris has important both interspecific and intraspecific virulence and phenotype heterogeneity, with aggregative phenotypes of C. auris sharing characteristics with low pathogenic species such as C. parapsilosis. Our work suggests that C. auris owns an important morphogenetic plasticity that distinguishes it from other yeasts of the genus. Routine phenotypic identification of aggregative or non-aggregative phenotypes should be performed in the clinical setting as it may impact patient management.
Introduction
Candida auris is an emergent fungal pathogen which, because of its multi-drug resistance [Citation1–3], difficult identification by conventional microbiology techniques [Citation4], high transmissibility, and environmental adaptability [Citation5,Citation6], has been associated with large outbreaks of healthcare-related invasive infections with a high mortality rate worldwide [Citation2,Citation5,Citation7–9]. Consequently, in 2019, the Centers for Disease Control (CDC) considered it to pose an urgent threat to public health in the context of multi-drug resistant organisms [Citation10,Citation11].
To date, there is still limited evidence on C. auris virulence, pathogenicity determinants, and the complex host–pathogen interactions. Pathogenicity has been recently studied in several animal models such as Galleria mellonella [Citation12–18], Caenorhabditis elegans [Citation18], and mice [Citation15]. However, it appears to be highly diverse, with some authors describing a comparable or even higher degree of pathogenicity to that of C. auris [Citation12,Citation13] contrarily to other reports [Citation14,Citation15]. High interspecific but also intra-species virulence heterogeneity seems to be an inherent characteristic of C. auris infection, as it also differs between aggregative and non-aggregative phenotypes, strains, and clades [Citation16,Citation19,Citation20].
Despite the first insights into its cellular, molecular, and genetic pathogenicity determinants have been provided [Citation13,Citation20–22], histological data on in vivo host–pathogen interactions are also scarce. Some works in both murine and invertebrate animal models have superficially described the tissue fungal burden after infection [Citation15,Citation16,Citation18,Citation19,Citation22–24]. Nevertheless, histological studies afford little evidence on fungal invasiveness, distribution, burden, and morphology, as well as host immune and cell biological response upon C. auris infection. Despite plasmatocytes are the responsible cells for fungal phagocytosis [Citation25], different hemocyte subpopulations have not been analyzed nor quantified. Furthermore, no quantitative data have been reported to date beyond subjective, purely qualitative depictions using a very limited number of strains, individuals, and samples. Morphometry has been widely used in several fields of medical research such as cancer or ischemic heart disease to provide valuable quantitative histological information [Citation26,Citation27]. Indeed, immunofluorescence and immunohistochemistry have improved these techniques, enabling the differentiation of specific cell types.
To tackle this problem, we incorporated for the first time in the study of fungal pathogenesis the body of knowledge of morphometrics to quantitatively analyze the in vivo C. auris specific traits and mechanisms of invasion as well host cell and immune response by immunolabelling fungal structures and larval plasmatocytes in a G. mellonella infection model, compared to Candida albicans and Candida parapsilosis as high and low pathogenicity controls, respectively.
Material and methods
Selection and processing of fungal strains
Two strains of C. auris (CJ175 and CJ101), 1 strain of C. albicans (reference strain ATCC SC5314), and 1 strain of C. parapsilosis (reference strain ATCC 22019) were randomly selected and used for the essays. C. auris strains were isolated from clinical samples obtained from blood cultures of patients admitted to surgical or medical intensive care units (ICU) of the University and Polytechnic La Fe Hospital (Valencia, Spain) who developed candidemia, and kept at −80°C until use.
Blood cultures were processed with a BacT/ALERTTM VIRTUOTM automated microbial detection system. Definitive identification of C. auris was performed by internal transcribed spacer (ITS) sequencing using ITS4-ITS5 primers following the protocol used by Bellemain et al. [Citation28] The sequencing technique was performed using GenomeLabTM GeXP kits (Beckman Coulter, Fullerton, CA, USA) and the sequences obtained were compared with those of Microbial Genomes BLAST (http://www.ncbi.nlm.nih.gov/guide/sequence-analysis/) and Mycobank Database. (C. auris CJ175 GenBank accession number: KJ126759.1, 96% similarity with sequence no AB375772. C. auris CJ101 accession number KC692045.1, 95% similarity with sequence no AB375772). For DNA extraction, inoculum was prepared from a 48-h culture grown on Sabouraud agar with chloramphenicol (Oxid Madrid, Spain) by resuspending 2–3 colonies in 800 µL of lysis buffer (Tris HCl 0.06M, EDTA 0.1M, NACl 1M, BrijTM −58 0.5%, sodium deoxycholate 0.2%, Lauryl Sarcosyl 0.5%). Once homogenized, 80 µL of Proteinase K (Qiagen, France) was added and incubated for 45 min at 56°C. Three freeze/thaw cycles were performed at −80°C/95°C for 1 min. Finally, extraction was performed using the DNeasy Blood and Tissue kit (Qiagen) on the QIAsymphony® automated extraction system (Qiagen, France). All isolates were sent for confirmation and complete genome sequencing to the Mycology Reference Laboratory of the National Microbiology Centre of the Carlos III Health Institute (ISCIII). Aggregative or non-aggregative phenotypes were determined by documenting immediate yeast aggregation at 200× magnification after vortexing for 3-minutes 1 mL of sterile saline with a concentration of 108 CFU/mL.
Infection model in G. mellonella
Healthy sixth instar larvae of G. mellonella weighing 250–350 mg (TruLarv™, BioSystems Technology Ltd., UK) were selected. Larvae were decontaminated upon arrival using 70% ethanol and were kept in groups of 10 in Petri dishes at 15°C and dark conditions until inoculation.
Fungal isolates were grown on Sabouraud’s agar for 24 h at 37°C. Individual colonies were collected with sterile plastic loops, washed twice in sterile phosphate-buffered saline (PBS), counted with a TC20TM Automated Cell Counter (BioRad Laboratories, France), and adjusted to 105 CFU/μL in sterile PBS. A standard inoculum of 10 μL of the solution (106 CFU) was chosen [Citation12,Citation16].
Thirty larvae were inoculated with 106 CFU of C. albicans in the left rear proleg using a 10 μL Hamilton syringe with a 26-gauge blunt needle. Another group of 30 larvae was infected with 106 CFU of C. parapsilosis. Eighteen larvae were inoculated with 106 CFU of the C. auris strain CJ101, and 15 larvae with 106 CFU of the C. auris strain CJ175 by intrahemocelic injection of 10 μL of the solution (106 CFU) Groups of 10 infected larvae were placed in Petri dishes and were incubated at 37°C. Further analyses were blinded by tagging every Petri dish with an individual code. Larvae were euthanized after 24 h of incubation following the AVMA Guidelines for the euthanasia of animals. They were firstly anesthetized through immersion in 5% ethanol and later euthanized by immersion in a solution of neutral-buffered 10% formalin. To preserve tissue architecture while allowing formalin cuticular penetration and tissue diffusion, larvae were fixed for 28 days as previously described [Citation16].
Tissue processing and immunohistochemical staining
Each larva was midsagittally sectioned and embedded in paraffin. Histological 3–4 μm thick sections were obtained with the microtome. Immunohistochemistry (IHC) with specific antibodies against Candida spp. and G. mellonella plasmatocytes was performed.
Candida spp. immunostaining
Midsagittal tissue sections were deparaffinized and rehydrated through graded alcohols. All samples were tagged for identification with the number of the larva and Petri dish blinded coding system. The primary murine polyclonal IgG anti-C. albicans antibody (PA1-27158, Thermo Fisher Scientific, Waltham, MA, USA) was chosen to specifically label fungal structures, as it was shown to have cross-reactivity with C. auris and C. parapsilosis strains. Tissue sections were incubated with the primary antibody at a dilution of 1:20,000 at room temperature for 45 min. Amplification of the primary antibody signal was carried out by 45 with polyclonal goat anti-rabbit immunoglobulins/HPR as secondary antibody for 45 min (Thermo Fisher Scientific, Waltham, MA). Tissues were then counterstained with hematoxylin–eosin.
G. mellonella novel plasmatocyte immunostaining
We specifically immunolabeled G. mellonella plasmatocytes. Following deparaffination and rehydration, a murine monoclonal IgG2b MS13 antibody against Manduca sexta plasmatocyte beta-integrin (Developmental Studies Hybridoma Bank, The University of Iowa, Iowa City, IA, USA) was used for G. mellonella plasmatocyte immunostaining. It was shown to cross-react and adequately immunolabel plasmatocytes of G. mellonella larvae. The primary MS13 antibody was incubated at a dilution of 1:20 for 45 min at room temperature. Subsequently, sections were incubated for 45 min with the secondary polyclonal goat anti-rabbit immunoglobulins/HPR and counterstained with hematoxylin–eosin.
Histopathological evaluation and morphometric analysis
All samples were assessed using the optic Leica DMD108 digital microimaging network instrument (Leica Microsystems, Wetzlar, Germany). Entire midsagittal larval sections were analyzed. The whole larval area was mapped by taking photomicrographs at 40× magnification for quantification of fungal density, 100× magnification for plasmatocyte analysis, and 200× up to 630× to evaluate in vivo fungal morphology, distribution, and degree of phagocytosis. The computerized morphometric study was carried out using the software Image Pro-Plus 7.0 (Media Cybernetics, Silver Spring, MD, USA).
For the study of the fungal load, tissue distribution, and in vivo cellular characteristics and host–pathogen interactions, several variables were analyzed:
Fungal density: percentage resulting from the sum of the total area (μm2) occupied by yeast in all analyzed micrographs per larva, divided by the complete area of the larval midsagittal section (μm2).
Fungal distribution: the main tissue distribution of the fungal pathogens -namely the digestive tract, fat tissue, hemolymph, tracheae, respiratory system, and uncharacterizable inflammatory tissue- was recorded.
Fungal morphology: the presence of pseudohyphae or true hyphae was distinctively registered.
Phagocytosis and presence of intracellular yeasts.
Further magnification captures up to 630× magnification were performed for the last two parameters, to better characterize fungal morphology and intracellular structures, whenever present.
The larval immune response was assessed in the whole larval section, based on the plasmatocyte-specific immunolabelling. The following morphometric parameters were studied:
Plasmatocyte density: the number of plasmatocytes was recorded in each 100× magnification photomicrograph. In large immune aggregates, a watershed segmentation algorithm was applied to enable cellular quantification. The sum of the number of plasmatocytes in all micrographs of each larva was divided by the complete larval area. Results were expressed in plasmatocytes per mm2.
Mean plasmatocyte diameter and area per 100× magnification micrograph.
Plasmatocyte roundness: the roundness of each plasmatocyte has been associated with the degree of cellular activation and was determined by the following formula:
Completely circular objects present a roundness = 1; other shapes present a roundness largely > 1 and would correspond to activated cells with an irregularly shaped cell membrane.
Plasmatocyte distribution: the main tissue distribution of plasmatocytes -namely the digestive tract, fat tissue, hemolymph, tracheae, respiratory system, and uncharacterizable inflammatory tissue- was recorded per 100× magnification micrograph.
Plasmatocyte nodulation: nodules were defined as the aggregation of 5 or more plasmatocytes. The presence of nodules was registered in each 100× magnification photomicrograph.
Statistical analysis
The descriptive statistics, graph representations, and hypothesis contrasts were performed with the statistical software R version 4.0.0 (R Development Core Team, 2020). Continuous variables were expressed as mean and standard deviation (SD). Categorical variables were expressed as a percentage. The level considered to indicate statistical significance was p < 0.05. The descriptive statistics for each morphometric parameter were calculated and quantile-quantile (QQ) plots were performed to assess normality for each variable. The Levene test was used to assess the equality of variances in quantitative variables. In the comparison of frequencies of qualitative variables, Pearson’s χ2 test was used, and Bonferroni p-value correction was applied for multiple comparisons. In the case of quantitative variables following normal or non-normal distributions, ANOVA and Kruskal–Wallis tests were chosen, respectively.
Results
The two C. auris isolates were obtained from critically ill patients with candidemia. CJ101 showed an aggregative phenotype while CJ175 was a non-aggregative strain.
Morphometric analysis of the plasmatocyte response to C. auris infection in G. mellonella
C. auris induces a higher tissue density of plasmatocytes than C. albicans and C. parapsilosis at the expense of nodule formation
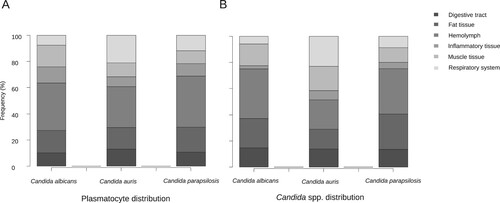
After analyzing more than 1,200 100× magnification micrographs and mapping all the tissue area in larval midsagittal sections, the tissue density of plasmatocytes after infection by C. auris was almost 4-fold higher than after infection by C. albicans and C. parapsilosis (p = 5.22 × 10−9), with a mean of approximately 24 plasmatocytes per mm2 (). No significant differences were observed between larvae infected with C. albicans and C. parapsilosis (p = 1). The presence of inflammatory hemocyte aggregates was significantly higher after infection by C. auris than after infection by C. albicans and C. parapsilosis (p = 8.32 × 10−7). In 62.8% of the micrographs in the C. auris infected group plasmatocytes aggregated forming inflammatory nodules, while in the larvae infected by both C. albicans and C. parapsilosis they barely reached 50% ().
Table 1. Summary of the main morphometric parameters.
Furthermore, the C. auris capability of inducing a larger hemocyte density was independent of the phenotype and in both cases was higher than 20 plasmatocytes per mm2 (26.22 – SD 14.41 and 21.83 – SD 1.11; p = 1). However, their tendency to form nodules was significantly lower in the aggregative strain than in the non-aggregative strain, with a nodulation frequency of 57.3% of the micrographs for aggregative C. auris versus 70.9% for non-aggregative C. auris (p = 0.004) (C and D).
Plasmatocytes of larvae infected by C. auris and C. parapsilosis have a lower size and membrane irregularity than C. albicans
More than 34,500 plasmatocytes were analyzed. On the one hand, significant differences were observed in the mean plasmatocyte area and diameter between C. albicans and C. auris, as well as between C. albicans and C. parapsilosis (p = 6.08 × 10−7 and p = 3.75 × 10−14, respectively). Plasmatocytes were significantly smaller in larvae infected with C. albicans than in those with both C. auris and C. parapsilosis. However, no significant differences were observed when comparing their mean area and diameter between larvae infected with C. auris and C. parapsilosis after Bonferroni p-value adjustment for multiple comparisons (p = 0.061).
On the other hand, plasmatocyte roundness or membrane irregularity was significantly lower after C. albicans infection than after both C. auris and C. parapsilosis infection (p = 2.969 × 10−6 and 3.3 × 10−8, respectively) while it did not differ between the last two species (p = 0.24) (A). The mean roundness values and SD observed for each group are presented in . No differences were observed when comparing aggregative and non-aggregative phenotypes (p = 1).
Tissue tropism of G. mellonella plasmatocytes in response to infection varies depending on the species of Candida
Hemolymphatic plasmatocyte distribution was predominant in all three Candida species (A). More than 30% of photomicrographs mainly showed plasmatocytes within the hemolymph. A significant differential distribution was observed, however, when analyzing other larval tissues. Whilst in larva infected with C. albicans and C. parapsilosis the fat tissue distribution was the second most frequently observed distribution (17.3% and 19.3%, respectively), plasmatocytes of larva einfected with C. auris infected were seen in close relation with the respiratory system in more than 20% of the photomicrographs (A). C. albicans showed remarkable muscle tissue inflammation (16.6%) (A and F). These differences were statistically significant (p = 5.602 × 10−7). Both non-aggregative and aggregative C. auris phenotypes showed similar tissue distribution patterns, with a discrete predominance of peri-intestinal plasmatocytes for aggregative C. auris (15.9% in aggregative C. auris vs. 8% in non-aggregative C. auris) and fewer hemolymphatic plasmatocyte observations (27.2% in aggregative C. auris vs. 37.5% in non-aggregative C. auris), as seen in . The plasmatocyte distribution was partially superimposable to that observed for fungal structures, as seen in , , and .
Figure 2. Detail on fungal invasiveness and plasmatocyte characteristics as a response to Candida spp. infection in G. mellonella. (A) Detail on small nodule of activated irregular plasmatocytes. Anti-MS13 antibody. CJ101 strain. 400× magnification. (B) Large quiescent plasmatocyte containing many phagocyted yeasts. Anti-MS13 antibody. CJ175 strain. 400× magnification. (C) C. auris pseudohyphae in adipose tissue and hemolymph. Anti-C. albicans antibody. CJ101 strain. 630× magnification. (D) C. auris pseudohyphal forms with a peri-respiratory distribution. Anti-C. albicans antibody. CJ101 strain. 630× magnification. (E) C. albicans true hyphae located subcuticularly, Anti-C. albicans antibody. SC5314 strain. 630× magnification. (F) Large yeast aggregate invading muscle tissue. Anti-C. albicans antibody. SC5314 strain. 400× magnification. C, cuticle; D, digestive tissue; F, fat tissue; H, hemolymph; M, muscle tissue; P, plasmatocyte; p, pseudohyphae; R, respiratory tissue; Y, yeasts.
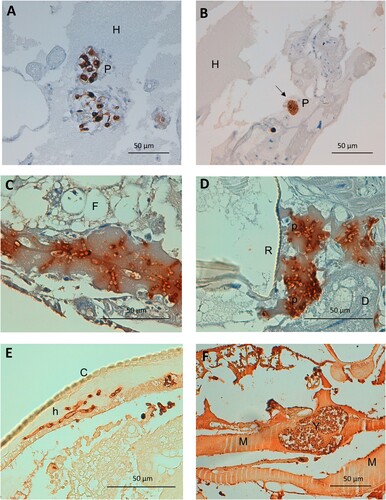
Figure 3. Differences of plasmatocyte and fungal tissue distribution after infection with non-aggregative and aggregative C. auris.
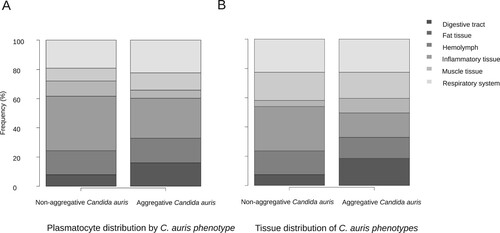
Table 2. Plasmatocyte and Candida spp. in vivo tissue distribution.
Analysis of fungal burden, distribution, and morphology after C. auris infection in G. mellonella
C. auris seems to produce an intermediate fungal burden between C. albicans and C. parapsilosis
A statistically non-significant trend was observed in the mean fungal density (p = 0.058). The mean percentage of the larval midsagittal section area infected was 1.26% (SD 3.00) for C. albicans, 0.51% (SD 0.62) for C. auris, and 0.19 (SD 0.15) for C. parapsilosis. However, this higher pathogenicity was mainly observed for non-aggregative C. auris phenotypes. The fungal density of the aggregative strain CJ101 did not differ from that observed for C. parapsilosis. The mean fungal density for the non-aggregative strain was 0.83% of the larval tissue area (SD 0.81) while for the aggregative strain was 0.25% (SD 0.18).
C. auris specifically invades G. mellonella respiratory tissue, with non-aggregative phenotypes mainly located in the hemolymph and aggregative phenotypes showing a more homogeneous distribution of intestinal predominance
Large significant differences were observed regarding the tissue tropism of C. auris compared to C. albicans and C. parapsilosis (p = 2.2 × 10−6). The tissue invasion of C. albicans and C. parapsilosis was homogeneous, with most of the fungal load distributed in the hemolymph, followed by the larval adipose tissue and the intestinal tract. However, C. auris substantially invaded the larval respiratory system (B and D), as seen in . Significant differences were observed between C. albicans and C. parapsilosis after Bonferroni p-value adjustment (p = 0.0048). C. albicans showed a muscle distribution more frequently than C. parapsilosis. Moreover, important differences were observed in the behaviour of different phenotypes (p = 2.2 × 10−13), as depicted in . Although both aggregative and non-aggregative C. auris strains affected tracheal structures in 22.6% and 22.7% of micrographs (D and B), respectively, non-aggregative C. auris was predominantly distributed through the larval hemolymph (30.5% of 594 micrographs) (B), followed by tracheae, muscle cells (19.2%), fat tissue (16%), and intestinal tract (7.6%). For the aggregative strain, digestive structures were predominantly affected after respiratory tissue (18.6% of 758 micrographs) (A), followed by muscle tissue (17.7%), hemolymph (16.8%), and adipose tissue (14.2%) (). The tissue distribution of both phenotypes significantly differed from C. albicans and C. parapsilosis (p < 10−12).
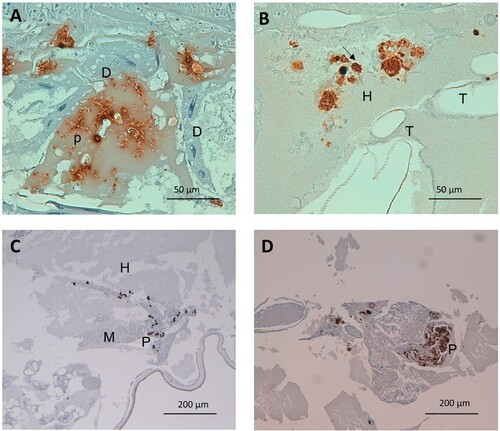
Figure 4. Detail on fungal and plasmatocyte characteristics as a response to G. mellonella infection by aggregative and non-aggregative phenotypes of C. auris. (A) C. auris pseudohyphae invading larval digestive epithelium, after infection with the aggregative strain CJ101. Anti-C. albicans antibody. 400× magnification. (B) Larval hemolymph containing yeasts of C. auris. Note the presence of intracellular yeasts within infected and plasmatocytes (arrow) undergoing cell lysis. Non-aggregative strain CJ175. Anti-C. albicans antibody. 400× magnification. (C) and (D) Small and large plasmatocyte nodules after infection with aggregative (CJ101) and non-aggregative (CJ175) strains of C. auris, respectively. Anti-MS13 antibody. 100× magnification. D, digestive tissue; H, hemolymph; P, plasmatocyte; T, respiratory tracheal system.
C. auris can filament in vivo to a lesser extent than both C. albicans and C. parapsilosis and mainly in the form of pseudohyphal structures
Of 482 micrographs, 259 (35%) showed some degree of filamentation in larvae infected by C. albicans. In larvae with C. auris and C. parapsilosis infection, 16.3% (206/1267) and 27.6% (157/569) of micrographs presented filamented fungal structures (C and D). These differences were statistically significant (p = 3.88 × 10−21).
On the contrary, no significant differences were observed in the degree of filamentation between aggregative and non-aggregative C. auris phenotypes, despite observing filamented forms in 23.2% of micrographs in the aggregative group versus 10.6% in the non-aggregative group.
Yeast form structures were the most frequently found in vivo in all three species, as seen in . Nevertheless, highly significant differences were seen in the cellular morphology (p = 1.97 × 10−64). C. albicans usually presented both pseudohyphal and true hyphal forms (E) (17.5% and 17.3% of the photomicrographs, respectively), followed by C. parapsilosis, which mainly only developed pseudohyphae (26.5%). Finally, C. auris presented a lower frequency of filamentation and a majority of pseudohyphal forms (16.1% of pseudohyphae vs. 0.6% of true hyphae) both in aggregative and non-aggregative phenotypes (C, 2D and A).
Table 3. Fungal morphology characteristics.
C. auris non-aggregative phenotypes and C. albicans show a lower frequency of intracellular forms than aggregative C. auris strains and C. parapsilosis
The frequency of micrographs showing intracellular and intrahemocyte fungal forms was significantly higher in C. parapsilosis (35.1% of 569 micrographs), followed by C. auris (27.5% of 1,267 micrographs) and finally C. albicans (19.3% of 741 micrographs) (p = 8.467 × 10−10). These results run in parallel to the mean plasmatocyte diameter and area and may express larger sizes for those hemocytes with intracellular fungal forms. Furthermore, these differences were observed at the expense of a lower number of intracellular cells in C. auris non-aggregative phenotypes. Furthermore, plasmatocytes after infection with the non-aggregative phenotype strain usually presented a disrupted appearance (B). The frequency of intracellular forms in the aggregative strain was similar to that observed in the C. parapsilosis group (36.1% of 758 micrographs).
Discussion
This study has shown that C. auris behaves differently from other species of Candida in the invertebrate host, but also presents a remarkable inter-phenotypic heterogeneity. In this work we have analyzed the in vivo fungal behaviour, host cell and immune response, and host–pathogen interactions upon C. auris infection in G. mellonella by specifically immunolabelling fungal structures and plasmatocytes, through a novel quantitative approach incorporating morphometric techniques into the study of microbial pathogenesis.
The main findings of this work can be summarized as follows: (i) C. auris owns a remarkably higher immunogenic activity than expected at its moderate degree of tissue invasion, inducing greater inflammatory response than C. albicans and C. parapsilosis at the expense of plasmatocyte nodule formation, especially in non-aggregative strains. (ii) C. auris specifically invades the larval respiratory system and presents inter-phenotypic tropism differences, with a more homogeneous distribution of intestinal predominance for aggregative strains resembling other Candida species. (iii) C. auris filaments in vivo less frequently than C. albicans or C. parapsilosis mostly through pseudohyphal growth. Filamentation might not be a major pathogenic determinant in C. auris, as known less virulent aggregative phenotypes tend to form pseudohyphae to a greater extent. (iv) Hemocyte immune response is superimposable to fungal tissue distribution. Plasmatocytes phagocyte less frequently non-aggregative C. auris phenotypes and C. albicans, for which they present a higher degree of activation and membrane irregularity than aggregative C. auris and C. parapsilosis.
C. auris morphogenetic plasticity and distinct expression of virulence factors may account for its remarkable inter and intraspecific heterogeneity [Citation16,Citation22,Citation29,Citation30]. Several genetic and molecular pathogenicity determinants have been defined for C. auris, such as expression of hemolysin, lipases, secreted aspartyl proteinases (SAPs), phospholipases, adhesins, integrins, or biofilm-related genes (IFF4, CSA1, PGA26, among others) [Citation31]. However, morphological plasticity plays a critical role in fungal virulence [Citation30], namely by biofilm formation and hyphal growth for Candida species [Citation32]. In the unique case of C. auris, in vivo phenotypic yeast aggregation and yet uncertain filamentation ability are thought to partly explain this diversity. Despite the existence of discordant data on the species pathogenicity depending on strains and possibly clades, showing both higher [Citation12,Citation13] and lower virulence compared to C. albicans [Citation14–17,Citation19], evidence is congruent on aggregative strains being less virulent. However, it was not until recently that we began to have data on the C. auris ability to produce pseudohyphae. Previously considered unable to filament [Citation12], recent studies have described phenotypic switching to pseudohyphal forms after genotoxic stress [Citation33], temperature changes and passage through mammalian [Citation22], invertebrate organisms [Citation13,Citation16,Citation30], or even in vitro [Citation30].
To date, there is scarce histopathological information on C. auris pathogenesis, and limited evidence is biased owing to works using only subjective approaches with a very limited number of strains, individuals, and samples. Moreover, there are no in vivo studies devoted to analyzing fungal pathogeny and host immune and cell biological responses upon C. auris infection. Computerized morphometric techniques provide a mathematical description of fungal and immune cell forms and densities, offering a more direct understanding of host–pathogen interactions and behaviour upon infection.
Through this approach, we have identified C. auris as highly immunogenic when compared to C. albicans and C. parapsilosis, while showing an intermediate fungal density. This finding contrasts with previous studies of hemocyte count in hemolymph extracts, in which an inverse relationship with infectious fungi and hemocyte density was described [Citation34]. This may be explained due to elevated solid tissue tropism and aggregation into large nodules. As hereby demonstrated and in concordance with histological studies in murine models [Citation19], these nodules run in parallel to the fungal load distribution, as hereby demonstrated. Previous reports on neutrophil and macrophage inflammatory infiltrate in mammal organs upon C. auris infection might imply a common pathway for the innate immune response induction in invertebrates or superior organisms including humans [Citation19,Citation23].
Moreover, the particular traits of the tissue distribution of C. auris described in this work are in line with known highly frequent septic metastatic complications in patients with C. auris infection [Citation6]. We have quantitatively demonstrated its previously suggested specific respiratory tropism in G. mellonella [Citation16], which significantly differed between phenotypes. Although both phenotypes predominantly invaded larval tracheae, aggregative strains similarly reproduced C. albicans and C. parapsilosis distributions. However, we did not observe such an intestinal majority of fungal elements in C. albicans as previously reported in a qualitative manner [Citation34].
On this line, several findings of this work advocate the lower virulence seen in aggregative phenotypes both in invertebrate and vertebrate animal models [Citation12–16,Citation19]. Aggregating C. auris forms induced plasmatocyte nodules to a lesser extent, but these immune cells did not appear to be less active than those seen upon non-aggregating C. auris infection. However, the tissue fungal burden of the aggregative phenotype was lower than in the non-aggregative phenotype and did not differ from that observed in C. parapsilosis: a known lower pathogenicity species of the genus [Citation16,Citation35,Citation36]. Additionally, the aggregative phenotype presents a greater frequency of intracellular forms which, concerning its lower virulence, might imply either increased immune evasion mechanisms [Citation31], lack of phagocytosis, or enhanced yeast “dumping” as yeast release after intracellular phagocyte killing [Citation37] for non-aggregating strains. However, another striking finding of this work is the notably higher frequency of filamented forms in the aggregative group, which has also been previously described [Citation30]. Filamentous forms have traditionally been associated with increased invasiveness and virulence [Citation32]. Nevertheless, these findings make us hypothesize whether pseudohyphal growth in C. auris is not one of the main pathogenic determinants upon infection, especially after hyphal-related proteins implied in pathogenesis such as candidalysin (ECE1) have not been detected in some works [Citation15]. These data raise an important matter for clinical practice. Usually, phenotypic characterization is not routinely determined in hospital-based clinical microbiology laboratories, but we suggest its routine characterization and providing the information to clinicians, as knowing the aggregating phenotype might have implications in patient management and treatment beyond antifungal sensitivity testing.
Some limitations of the study deserve mention. Despite analyzing more than 4,500 different magnification micrographs and covering the whole tissue area, around 30 larvae per group were included and this sample size could ideally be increased. Likewise, the most important limitation is the use of only 2 strains of C. auris, especially in the highly diverse context of C. auris, as previously discussed. G. mellonella has been identified as an interesting model due to its complex innate immune system with functional and anatomical similarities to those found in more complex organisms [Citation38], but results must be carefully interpreted, and extrapolation might be limited. Further analyses with a greater number of strains, even randomly selecting tissue areas for experimental feasibility, should be performed. Furthermore, these findings should be correlated with genomic analyses, RT–PCR analysis of virulence gene mRNA expression, further phenotypic changes, phospholipase activity detection, and biofilm formation ability, among others, to fully explain its wide morphogenetic plasticity.
In conclusion, we have documented differential traits in fungal invasiveness, distribution, burden, and morphological plasticity as well as host immune and cell biological response for C. auris aggregative and non-aggregative phenotypes compared to other species by incorporating the methodology of morphometrics. C. auris is a potent immune inductor at the expense of nodule formation, with specific respiratory tropism and filamentation capability through pseudohyphal growth. However, filamentation does not seem to be as decisive for its pathogenesis as previously assumed. C. auris owns important both interspecific and intraspecific virulence and phenotype heterogeneity with aggregative phenotypes of C. auris sharing characteristics with low pathogenic species such as C. parapsilosis. Hence, our work suggests that C. auris owns an important morphogenetic plasticity that distinguishes it from other yeasts of the genus. Routine phenotypic identification of aggregative or non-aggregative phenotypes should be performed in the clinical setting as it may impact patient management. Further studies are needed to completely untangle the molecular pathways underlying this plasticity that endow this emergent pathogen with an unusual complexity.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Du H, Bing J, Hu T, et al. Candida auris: epidemiology, biology, antifungal resistance, and virulence. PLoS Pathog. 2020;16(10):e1008921.
- Chowdhary A, Sharma C, Meis JF. Candida auris: a rapidly emerging cause of hospital-acquired multidrug-resistant fungal infections globally. PLoS Pathog. 2017;13:e1006290.
- Ku TSN, Walraven CJ, Lee SA. Candida auris: disinfectants and implications for infection control. Front Microbiol. 2018;9:726.
- Chakrabarti A, Sood P. On the emergence, spread and resistance of Candida auris: host, pathogen and environmental tipping points. J Med Microbiol. 2021;70(3):001318.
- Eyre D, Sheppard A, Madder H, et al. A Candida auris outbreak and its control in an intensive care setting. N Engl J Med. 2018;379:1322–1331.
- Ruiz-Gaitán A, Moret AM, Tasias-Pitarch M, et al. An outbreak due to Candida auris with prolonged colonisation and candidaemia in a tertiary care European hospital. Mycoses. 2018;61(7):498–505.
- Ruiz-Gaitán A, Martínez H, Moret AM, et al. Detection and treatment of Candida auris in an outbreak situation: risk factors for developing colonization and candidemia by this new species in critically ill patients. Expert Rev Anti Infect Ther. 2019;17(4):295–305.
- Lockhart SR, Etienne KA, Vallabhaneni S, et al. Simultaneous emergence of multidrug-resistant Candida auris on 3 continents confirmed by whole-genome sequencing and epidemiological analyses. Clin Infect Dis. 2017;64(2):134–140. Erratum in: Clin Infect Dis. 2018;67(6):987.
- Schelenz S, Hagen F, Rhodes J, et al. First hospital outbreak of the globally emerging Candida auris in a European hospital. Antimicrob Resist Infect Control. 2016;5:5.
- Centers for Disease Control (CDC). Antibiotic resistance threats in the United States. Centers for Disease Control; December 2019. Available from: https://www.cdc.gov/drugresistance/pdf/threats-report/2019-ar-threats-report-508.pdf
- Kadri SS. Key takeaways from the U.S. CDC’s 2019 antibiotic resistance threats report for frontline providers. Crit Care Med. 2020;48(7):939–945.
- Borman AM, Szekely A, Johnson EM. Comparative pathogenicity of United Kingdom isolates of the emerging pathogen candida auris and other key pathogenic Candida species. mSphere. 2016;1(4):e00189–16.
- Sherry L, Ramage G, Kean R, et al. Biofilm-Forming capability of highly virulent, multidrug-resistant Candida auris. Emerg Infect Dis. 2017;23(2):328–331.
- Romera D, Aguilera-Correa JJ, García-Coca M, et al. The Galleria mellonella infection model as a system to investigate the virulence of Candida auris strains. Pathog Dis. 2020;78(9):ftaa067.
- Muñoz JE, Ramirez LM, Dias LDS, et al. Pathogenicity levels of Colombian strains of Candida auris and Brazilian strains of Candida haemulonii species complex in both murine and Galleria mellonella experimental models. J Fungi (Basel). 2020;6(3):104.
- Garcia-Bustos V, Ruiz-Saurí A, Ruiz-Gaitán A, et al. Characterization of the differential pathogenicity of Candida auris in a Galleria mellonella infection model. Microbiol Spectr. 2021: e0001321.
- Carvajal SK, Alvarado M, Rodríguez YM, et al. Pathogenicity assessment of Colombian strains of Candida auris in the Galleria mellonella invertebrate model. J Fungi (Basel). 2021;7(6):401.
- Hernando-Ortiz A, Mateo E, Perez-Rodriguez A, et al. Virulence of Candida auris from different clinical origins in Caenorhabditis elegans and Galleria mellonella host models. Virulence. 2021;12(1):1063–1075.
- Forgács L, Borman AM, Prépost E, et al. Comparison of in vivo pathogenicity of four Candida auris clades in a neutropenic bloodstream infection murine model. Emerg Microbes Infect. 2020;9(1):1160–1169.
- Larkin E, Hager C, Chandra J, et al. The emerging pathogen Candida auris: growth phenotype, virulence factors, activity of antifungals, and effect of SCY-078, a novel glucan synthesis inhibitor, on growth morphology and biofilm formation. Antimicrob Agents Chemother. 2017;61(5):e02396–16.
- Singh R, Kaur M, Chakrabarti A, et al. Biofilm formation by Candida auris isolated from colonising sites and candidemia cases. Mycoses. 2019;62(8):706–709.
- Yue H, Bing J, Zheng Q, et al. Filamentation in Candida auris, an emerging fungal pathogen of humans: passage through the mammalian body induces a heritable phenotypic switch. Emerg Microbes Infect. 2018;7(1):188.
- Torres SR, Pichowicz A, Torres-Velez F, et al. Impact of Candida auris infection in a neutropenic murine model. Antimicrob Agents Chemother. 2020;64(3):e01625–19.
- Fakhim H, Vaezi A, Dannaoui E, et al. Comparative virulence of candida auris with candida haemulonii, candida glabrata and candida albicans in a murine model. Mycoses. 2018;61(6):377–382.
- Charroux B, Royet J. Elimination of plasmatocytes by targeted apoptosis reveals their role in multiple aspects of the Drosophila immune response. Proc Natl Acad Sci U S A. 2009;106(24):9797–9802.
- Ruiz-Saurí A, García-Bustos V, Granero E, et al. Distribution of vascular patterns in different subtypes of renal cell carcinoma. A morphometric study in two distinct types of blood vessels. Pathol Oncol Res. 2018;24(3):515–524.
- Garcia-Bustos V, Sebastian R, Izquierdo M, Rios-Navarro C, Bodí V, Chorro FJ, Ruiz-Sauri A. Changes in the spatial distribution of the Purkinje network after acute myocardial infarction in the pig. PLoS One. 2014;14(2):e0212096.
- Bellemain E, Carlsen T. Antinociceptive effect of geranylgeraniol and 6α, 7β-dihydroxyvouacapan-17β-oate methyl ester isolated from Pterodon pubescens benth. BMC Pharmacol. 2010;10:1–9.
- Bentz ML, Sexton DJ, Welsh RM, et al. Phenotypic switching in newly emerged multidrug-resistant pathogen Candida auris. Med Mycol. 2018;57(5):636–638.
- Fan S, Yue H, Zheng Q, et al. Filamentous growth is a general feature of Candida auris clinical isolates. Med Mycol. 2021;59(7):734–740.
- ElBaradei A. A decade after the emergence of Candida auris: what do we know? Eur J Clin Microbiol Infect Dis. 2020;39(9):1617–1627.
- Mba IE, Nweze EI. Mechanism of Candida pathogenesis: revisiting the vital drivers. Eur J Clin Microbiol Infect Dis. 2020;39:1797–1819.
- Ruiz G B, Ross ZK, Gow NAR, et al. Pseudohyphal growth of the emerging pathogen Candida auris is triggered by genotoxic stress through the S phase checkpoint. mSphere. 2020;5(2):e00151–20.
- Rossoni RD, de Barros PP, Mendonça IDC, et al. The postbiotic activity of Lactobacillus paracasei 28.4 against Candida auris. Front Cell Infect Microbiol. 2020;10:397.
- Gago S, García-Rodas R, Cuesta I, et al. Candida parapsilosis, Candida orthopsilosis, and Candida metapsilosis virulence in the non-conventional host Galleria mellonella. Virulence. 2014;5(2):278–285.
- García-Carnero LC, Clavijo-Giraldo DM, Gómez-Gaviria M, et al. Early virulence predictors during the Candida species-Galleria mellonella interaction. J Fungi (Basel). 2020;6(3):152.
- Essig F, Hünniger K, Dietrich S, et al. Human neutrophils dump Candida glabrata after intracellular killing. Fungal Genet Biol. 2015;84:37–40.
- Asai M, Sheehan G, Li Y, et al. Innate immune responses of Galleria mellonella to mycobacterium bovis BCG challenge identified using proteomic and molecular approaches. Front Cell Infect Microbiol. 2021;11:619981. http://doi.org/10.3389/fcimb.2021.619981