ABSTRACT
Defence against oxidative stress is crucial for Mycobacterium tuberculosis to survive and replicate within macrophages. Mycobacteria have evolved multilayer antioxidant systems, including scavenging enzymes, iron homeostasis, repair pathways, and metabolic adaptation, for coping with oxidative stress. How these systems are coordinated to enable the physiological adaptation to different intensities of oxidative stress, however, remains unclear. To address this, we investigated the expression kinetics of the well-characterized antioxidant genes at bacteriostatic H2O2 concentrations ranging from 1 mM to 10 mM employing Mycolicibacterium smegmatis as a model. Our results showed that most of the selected genes were expressed in a H2O2 concentration-dependent manner, whereas a subset exhibited sustained induction or repression without dose–effect, reflecting H2O2 concentration-dependent physiological adaptations. Through analyzing the dynamics of the coordinated gene expression, we demonstrated that the expressions of the H2O2 scavenging enzymes, DNA damage response, and Fe–S cluster repair function were strikingly correlated to the intensity of oxidative stress. The sustained induction of mbtB, irtA, and dnaE2 indicated that mycobacteria might deploy increased iron acquisition and error-prone lesion bypass function as fundamental strategies to counteract oxidative damages, which are distinct from the defence tactics of Escherichia coli characterized by shrinking the iron pool and delaying the DNA repair. Moreover, the distinct gene expression kinetics among the tricarboxylic acid cycle, glyoxylate shunt, and methylcitrate cycle suggested that mycobacteria could dynamically redirect its metabolic fluxes according to the intensity of oxidative stress. This work defines the H2O2 concentration-dependent gene expression kinetics and provides unique insights into mycobacterial antioxidant defence strategies.
Introduction
Despite the availability of vaccine and chemotherapy, Mycobacterium tuberculosis (Mtb) remains the most successful bacterial pathogen. Among the factors that contribute to the success of Mtb, the ability to counteract reactive oxygen species (ROS) generated by macrophages is crucial for its survival and replication within the host [Citation1]. In addition, increasing evidence shows that ROS-mediated oxidative stress and DNA damage contribute substantially to cell death caused by most bactericidal antibiotics in mycobacteria, indicating that antioxidant response also affects the antibiotic efficacy [Citation2–7]. Therefore, systematic understanding of the mechanisms deployed by mycobacteria to defence against oxidative stress will promote the development of novel anti-tuberculosis strategies.
The toxicity of ROS mainly stems from the reaction of superoxide (O2–) or H2O2 with iron from mononuclear iron enzymes and the [4Fe–4S] clusters of dehydratases, resulting in inactivation of enzymes involved in various metabolic pathways, including the syntheses of branched-chain and aromatic amino acids, respiration, ribonucleotide reduction, and the tricarboxylic acid (TCA) cycle [Citation8–10]. Auxotrophy for branch-chain amino acids and inability to grow on TCA cycle substrates were observed in Escherichia coli mutant strains deficient in superoxide dismutase (SOD) or catalase/peroxidase [Citation11]. Hydrogen peroxide could also react with the pool of loose iron via the Fenton reaction and form extremely reactive and damaging hydroxyl radicals (OH·), which contributes substantially to ROS-induced DNA damage [Citation12]. Hydroxyl radicals can directly react with the sugar–phosphate backbone and the base moiety within the double helix, leading to double-strand breaks (DSBs) during the process of DNA repair [Citation13]. In addition, due to chelation of Fe2+ by triphosphates, H2O2 could also react with the pool of nucleotides, resulting in DNA damage upon being incorporated into DNA by polymerase [Citation3,Citation11]. Moreover, accumulation of damaged proteins resulted from the oxidation of cysteine and methionine residues was also observed in E. coli exposed to H2O2 [Citation14]. Given the common chemical nature of ROS toxicity on cellular components, it is not surprising that bacteria have evolved a highly conserved multilayer antioxidant systems, including scavenging enzymes, maintenance of iron homeostasis, DNA and protein repair and metabolic adaptation, for coping with oxidative stress [Citation11,Citation15–17].
Despite the well-established role in mycobacterial antioxidant response and pathogenesis, how these antioxidant systems are coordinated to enable the physiological adaptation to different intensities of oxidative stress remains unclear [Citation17,Citation18]. For instance, due to the pivotal role of iron metabolism in the action of ROS, the activity of iron acquisition could not only determine the intracellular iron concentration and the repair of iron-cofactored enzymes, but also positively correlate with the severity of DNA damage [Citation15]. In E.coli, it was proposed that the antioxidant response would shrink the loose-iron pool and thereby alleviate the Fenton reaction and DNA damage [Citation15,Citation16]. It remains unclear that how mycobacteria maintain the balance between iron metabolism and DNA damage under oxidative stress. In addition, although the toxicity of ROS is a dose-dependent event, the physiological states and the adaptation strategies under different intensities of oxidative stress remain elusive. This could be exemplified by a recent study indicating that the regulation of Clp protease activity under oxidative stress was associated with the intensity of oxidative stress [Citation19].
In this study, using Mycolicibacterium smegmatis (Msm) as a model strain, we defined the expression kinetics of the well-characterized antioxidant genes at bacteriostatic H2O2 concentrations ranging from 1 mM to 10 mM. Our real-time RT-PCR (qRT-PCR) results revealed both H2O2 concentration-dependent expression and steadily sustained induction or repression of antioxidant genes regardless of H2O2 concentration, demonstrating H2O2 concentration-dependent physiological transitions and adaptations. Through analyzing the dynamics of coordinated gene expression, we provided several unique insights into mycobacterial antioxidant defence strategies under different intensities of oxidative stress.
Results and discussion
H2O2 can induce bacteriostatic or bactericidal effect, depending on the exposed H2O2 concentrations. We measured the survival of Msm after being exposed to H2O2 concentrations ranging from 1 to 20 mM for 50 min. The H2O2 concentrations used in this study were validated by the peroxide assay. Our results showed that H2O2 at 1–10 mM caused only a slight survival defect (killing of 13–25% Msm cells), while exposure to 15 and 20 mM H2O2 resulted in killing of 50% and 99.6% Msm cells (P < 0.0001), respectively (A). The observed bacteriostatic H2O2 concentration on Msm (≤10 mM) is consistent with the observations on Mtb [Citation20], suggesting that Msm and Mtb may deploy similar mechanisms to adapt to oxidative stress. To minimize the effect of cell death on the gene expression dynamics, H2O2 concentrations ranging from 1 to 10 mM were selected in the qRT-PCR experiments. To interrogate mycobacterial physiological states and adaptation strategies under different intensities of oxidative stress, we measured the expression dynamics of the well-characterized antioxidant genes that carry out the committed antioxidant reactions and were functionally validated in mycobacteria and/or E.coli (Table S1).
Figure 1. Transcriptional profiles of genes encoding for scavenging enzymes. (A) Survival of Msm treated with H2O2. Exponential-phase Msm cultures were treated with different H2O2 concentrations for 50 min, survival was measured by tenfold serial dilution plating. (B) Scavenging enzymes that are responsible for degrading ROS. (C–E) Expression of katG, aphC, and sodA in Msm exposed to H2O2 for 50 min. Transcript levels were measured by the qRT-PCR, normalized relative to sigA, and expressed as Log2 fold change from untreated cultures. Data shown are mean ± SE with at least three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
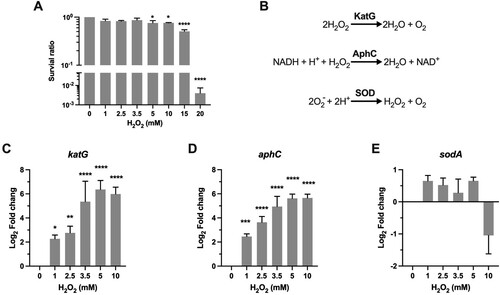
Scavenging enzymes
A hallmark of the aerobic bacteria response to oxidative stress is the induction of scavenging enzymes, which can timely reduce the concentration of ROS and thus prevent the oxidative damages to cellular components (B) [Citation11]. Mycobacterial ROS scavenging enzymes include catalase KatG [Citation21], the alkyl hydroperoxidase AhpC [Citation22], and superoxide dismutases SodA and SodC [Citation23]. All of these genes contribute to mycobacterial resistance against ROS and are well-established virulence factors [Citation21,Citation23–28]. KatG and AhpC can directly target and detoxify H2O2 (B). Our qRT-PCR results demonstrated that the induction of katG and ahpC was steadily elevated from ∼4-fold (P < 0.05) at 1 mM H2O2 to ∼64-fold (P < 0.0001) at 5 mM H2O2, and no further induction was observed at 10 mM H2O2 (C-D). This H2O2 concentration-dependent kinetics of katG and ahpC expression indicates that the committed transcriptional regulation could response sophistically via tittering the concentration of intracellular H2O2.
SodA belongs to the Fe-SOD family and accounts for a major portion of the superoxide dismutase activity of Mtb (B) [Citation29]. Although SOD do not directly react with H2O2, Mtb mutants with reduced sodA expression became extremely sensitive to H2O2 and were severely attenuated in mice [Citation24]. We therefore analyzed the expression of Msm sodA and the results showed that its expression was slightly increased at the H2O2 concentrations ranging from 1 to 5 mM. Moreover, in contrast to the sustained induction of katG and ahpC at 10 mM H2O2, expression of sodA was downregulated ∼2-fold at this H2O2 concentration (E). Overall, the expression pattern of Msm sodA is consistent with the previous studies showing that the expression of sodA and sodC was not remarkably changed in Mtb exposed to H2O2 [Citation20,Citation30].
Iron utilization
The toxicity of ROS mainly stems from the reaction of superoxide (O2–) and H2O2 with iron [Citation8–10]. Therefore, maintaining iron homeostasis plays a pivotal role in bacterial defences against oxidative stress (A)[Citation16]. We examined transcriptional profiles of well-characterized genes representative of iron scavenge (mbtB), import (irtA), regulation (ideR), and Fe–S cluster repair (suf) [Citation31–35].
Figure 2. Transcriptional profiles of genes encoding for iron utilization. (A) Carton illustration of mycobacterial Fe acquisition system and its role in oxidative stress. Ms 1460–1466 are the M. smegmatis homologues of Mtb Rv1460-1466 cluster. (B–E) Expression of mbtB, irtA, ideR, and Ms1640 in Msm exposed to H2O2 for 50 min. Transcript levels were measured by the qRT-PCR, normalized relative to sigA, and expressed as Log2 fold change from untreated cultures. Data shown are mean ± SE with at least three independent experiments. **P < 0.01, ***P < 0.001, ****P < 0.0001.
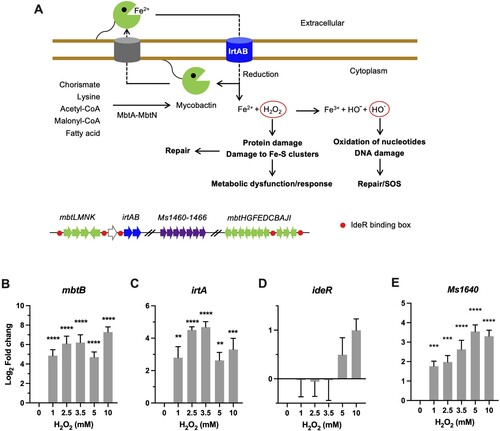
Mycobacteria rely on siderophore molecules called mycobactins for the acquisition of extracellular iron [Citation34]. Biosynthesis of mycobactin is carried out via a system encoded by 14 genes located in two mbt gene clusters (mbtA-J and mbtK-N) (A). The qRT-PCR results demonstrated that the expression of Msm mtbB was markedly induced by 52-fold (P < 0.0001) by 1 mM H2O2 and remained elevated at the tested H2O2 concentrations (B). In line with this, the expression of irtA, which encodes a virulence factor involved in import of Fe-carboxymycobactin [Citation31,Citation36], was also significantly induced 8- to 27-fold (P < 0.01) by the tested H2O2 concentrations (C). Thereafter, these expression profiles appear to argue that mycobacteria tend to upregulate the iron acquisition system upon oxidative stress [Citation20,Citation30]. This is strikingly different from the defence strategy of E. coli against oxidative stress, which was characterized by shrinking the iron pool, shown by the increased sequestration of unincorporated iron and repression of iron uptake [Citation16]. Intriguingly, the expression of ideR, a functional counterpart of the E. coli Fur repressor of iron-dependent genes including irtA and mbt clusters (A) [Citation33,Citation35], was induced by 10 mM H2O2 (∼2-fold) (D). However, it seems the induction of ideR did not result in repression of irtA and mbt (B-D).
In E.coli, the Suf (mobilization of sulphur) system is induced by OxyR under oxidative stress to assemble and repair Fe–S clusters [Citation11]. In mycobacteria, this system is encoded by the suf operon (Ms1640-1466, represent the counterparts of Mtb Rv1460-1466 cluster) and the Msm mutants with inactivated suf displayed growth deficiency under low-iron conditions (A) [Citation32]. Our qRT-PCR results demonstrated that the expression of Ms1640 steadily increased from 4-fold at 1 mM H2O2 to 13-fold at 5 mM H2O2 (P < 0.0001), and no further induction was observed at 10 mM H2O2 (E).
DNA repair
DNA damage is the major event underlying ROS-induced cell death [Citation12]. H2O2-induced DNA damage is mainly mediated by hydroxyl radicals (OH·) generated via the Fenton reaction, which are extremely reactive with both base and ribose moieties of the DNA (A). One of the main difference in the H2O2 susceptibility between mycobacteria and E.coli appears to be the highly refractory of mycobacteria to DNA damage-dependent, mode-one killing caused by low concentrations of H2O2 (1–2 mM) [Citation20,Citation37]. The mechanism underlying this remains elusive. It was suggested that the thick cell wall contributes to the intrinsic resistance of Mtb to ROS [Citation17]. However, our qRT-PCR showed that the expression of recA, a key DNA damage-induced factor involved in recombination, DNA repair, and induction of SOS response, was significantly induced 5.5-fold (P < 0.001) by 1 mM H2O2 (B), similar to that as observed in E.coli [Citation38]. Moreover, the induction of recA was apparently H2O2 dose-dependent, approaching 27-fold (P < 0.0001) increase at 5 and 10 mM H2O2 (B), suggesting that DNA damage is correlated with the intensity of oxidative stress. These results suggest that, despite the lack of mode-one killing, DNA damage could be induced by low H2O2 concentrations in Msm.
Figure 3. Transcriptional profiles of genes encoding for DNA repair proteins. (A) Illustration of H2O2-mediated damage to DNA and the repair systems. (B–D) Expression of recA, dnaE2, and mazG in Msm exposed to H2O2 for 50 min. Transcript levels were measured by the qRT-PCR, normalized relative to sigA, and expressed as Log2 fold change from untreated cultures. Data shown are mean ± SE with at least three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
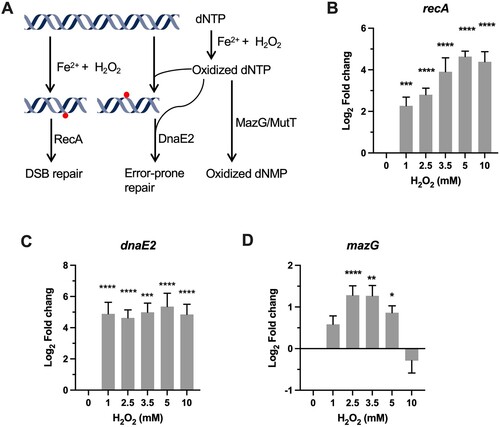
Consistent with the induction of recA, the expression of the SOS gene dnaE2 (the functional homologue of E.coli dinB) encoding the error-prone DNA polymerase was also significantly increased at the tested H2O2 concentrations [Citation39]. However, unlike the expression pattern of recA, the expression level of dnaE2 is independent of H2O2 concentrations, shown by the sustained magnitude of induction (40- to 75-fold, P < 0.0001) at the tested H2O2 concentrations. This is intriguing when considering that the expression of dinB was not markedly increased in E.coli upon exposure to 1 mM H2O2, while the SOS genes (recA, recN, lexA, and dinD) exhibited moderate induction (fold change < 10) [Citation38]. The mechanism underlying the observed difference of transcriptional regulation of error-prone polymerase upon exposure to H2O2 remains unclear. Nevertheless, given that error-prone lesion bypass is critical for preventing lethal DNA damage, the remarkable induction of dnaE2 may contribute to mycobacterial intrinsic resistance to mode-one killing [Citation11,Citation39].
Owing to chelation of Fe2+ by triphosphates, free nucleotides are also frequently oxidized by H2O2, which can lead to mutagenesis and DNA damage upon mis-incorporated into DNA [Citation3,Citation13,Citation40] (A). Our previous study demonstrated that sanitization of oxidized dCTP by MazG contributes to mycobacterial defence against oxidative stress [Citation3,Citation40,Citation41]. The qRT-PCR results showed that mazG exhibited a “bell-curve” pattern of induction at the H2O2 concentrations varying from 1 to 5 mM (1.7- to 3.3-fold, P < 0.05). At 10 mM H2O2, expression of mazG was downregulated (D), indicative of an intricate crosstalk among different transcriptional regulators [Citation42]. Induction of mazG under oxidative stress may alleviate the mutagenesis and DNA damage caused by misincorporation of oxidized dNTPs (A)[Citation3].
Metabolic adaptation
ROS-mediated metabolic deficiency mainly stems from the inactivation of [4Fe–4S] clusters of metabolic enzymes [Citation8–10] (A). Given that the generation of ROS is an inevitable process occurring in normal aerobic metabolism, it is not surprising that bacteria have evolved specialized metabolic enzymes and/or adaptation tactics to deal with oxidative stress. For example, in E.coli exposed to redox-cycling compounds, fumarase C, a non-Fe–S enzyme, and ROS-resistant aconitase A, were induced to replace the highly sensitive housekeeping counterparts to restore the TCA flux [Citation43,Citation44]. While in mycobacteria, accumulating evidence demonstrates that metabolic enzymes contribute to antioxidant defence and pathogenesis [Citation2,Citation5,Citation18,Citation22,Citation45,Citation46].
To probe the metabolic adaptation under different intensities of oxidative stress, we first examined transcriptional profiles of icl1 and prpD, both are well-characterized antioxidant enzymes belonging to glyoxylate shunt and methylcitrate cycle (A) [Citation5,Citation18]. The qRT-PCR data demonstrated that icl1 and prpD exhibited a very similar expression pattern, shown by unchanged expression at 1 and 2 mM H2O2, followed by steadily increased induction by high H2O2 concentrations (B-C). The expression of icl1 was significantly induced 33-fold (P < 0.01) by 5 mM H2O2 and 43-fold (P < 0.0001) by 10 mM H2O2. The induction of prpD began at 3.5 mM H2O2 (6-fold) and approached maximal at 5 mM H2O2 (43-fold, P < 0.0001). These transcriptional profiles indicate that mycobacterial defence against high level of ROS implicates redirection of metabolism by ICL and PrpD.
Figure 4 . Transcriptional profiles of genes encoding for metabolic enzymes involved in antioxidant defense. (A) Illustration of TCA, glyoxylate shunt, and methylcitrate cycle. Enzymes or enzyme complex involved in antioxidant response were shown as bold. PDH, pyruvate dehydrogenase; CIT, citrate; isoCIT, isocitrate; α-KG, α-ketoglutarate; α-KDH, α-ketoglutarate dehydrogenase; SUC-CoA, succinyl-CoA; SUC, succinic acid; SDH, succinate dehydrogenase; FUM, fumarate; MAL, malic acid; OAA, oxaloacetate; GLO, glyoxylate; PRO-CoA, propionyl-CoA; 2MC, 2 methylcitrate; 2MIC, 2 methylisocitrate. (B–F) Expression of icl, prpD, sdhA, fum, and aceE in Msm exposed to H2O2 for 50 min. Transcript levels were measured by the qRT-PCR, normalized relative to sigA, and expressed as Log2 fold change from untreated cultures. Data shown are mean ± SE with at least three independent experiments. *P < 0.05, **P < 0.01, ****P < 0.0001.
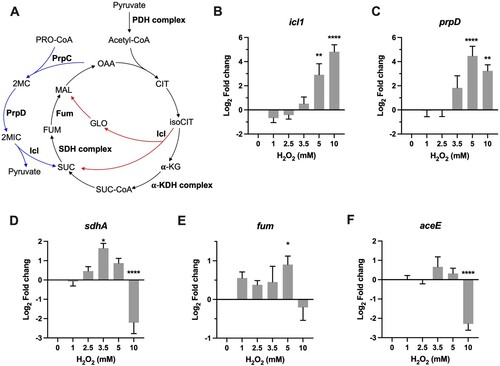
ICL conducts the first committed step in the glyoxylate shunt, which implicates anaplerosis of TCA cycle intermediates and gluconeogenesis. In mycobacteria, ICL also functions as methylisocitrate lyase belonging to the methylcitrate cycle [Citation47], a pathway involved in the metabolism of propionyl-CoA generated through β-oxidation of odd-chain fatty acids or degradation of branched-chain amino acids and cholesterol [Citation18,Citation48]. Because both glyoxylate shunt and methylcitrate cycle can generate succinate, we therefore measured the expression of sdhA and fum, which encode for succinate dehydrogenase A and a non-Fe–S fumarase, respectively (A). As shown in D, sdhA exhibited a “bell-curve” pattern of induction at the H2O2 concentrations varying from 1 to 5 mM, with the maximal induction at 3.5 mM H2O2 (3.2-fold, P < 0.05). Expression of fum was slightly increased (1.3- to 2.0-fold, P < 0.05) and sustained at the H2O2 concentrations varying from 1 to 5 mM (E). In Mtb, Fum deficiency increases susceptibility to H2O2 [Citation45]. Taken together, the induction of sdhA and fum by intermediate H2O2 concentrations may be indicative of maintenance of the TCA flux, a mechanism deployed by E.coli to defence against ROS [Citation43,Citation44].
The most intriguing aspect of these expression profiles was the reciprocal expression pattern between icl1/prpD and sdhA/fum at 10 mM H2O2 (B–E). The maximal induction of icl1/prpD and sudden repression of sdhA/fum at 10 mM H2O2 perhaps reflect metabolic adaptation that may result in increased catabolism of propionyl-CoA or accumulation of succinate. In addition, the repression of sdhA, a FAD-dependent Fe–S-containing respiratory component, could also reduce the generation of endogenous ROS [Citation11]. Previous study demonstrated that Mtb could slow and remodel its TCA cycle to increase production of succinate, which is used to flexibly sustain membrane potential, ATP synthesis, and anaplerosis, in response to varying degrees of O2 limitation [Citation46]. However, the role of succinate in the antioxidant response remains elusive. To further probe whether Msm also slows its TCA cycle under intensified oxidative stress, we measured the expression of aceE, which encodes for a key component of pyruvate dehydrogenase complex (A), and the results showed that the expression of aceE was significantly downregulated by 4.2-fold (P < 0.0001) at 10 mM H2O2 (F). Given that glycerol is the major carbon source for Msm cultured in vitro, the repression of aceE may likely reflect a decreased fuelling of TCA cycle. Together, these results indicate that mycobacterial metabolic adaptation to ROS is strikingly dependent on the intensity of oxidative stress, and mycobacteria tend to shut down the TCA cycle and increase the activities of glyoxylate shunt and methylcitrate cycle to defence against intensified oxidative stress.
Protein repair and degradation
We examined the transcription dynamics of genes linked to the well-characterized protein repair or degradation/recycling pathways to probe how they responded to oxidative stress. We have focused on the genes encoding for peptide methionine sulfoxide reductase (msr) and protease complex including proteasome and Clp system.
Oxidized methionine can be repaired by methionine sulfoxide reductase Msr. Previous studies demonstrated that deletion of msrA in either Msm or E.coli resulted in hypersensitivity to H2O2 [Citation49,Citation50]. However, our qRT-PCR results showed that the expression of msrA was significantly downregulated 1.6- to 4.3-fold (P < 0.05) upon exposure to the tested H2O2 concentrations (A). Intriguingly, transcriptional repression was also observed on the genes encoding for the proteasome system (B-D). Specifically, while expression of unfoldase mpa and prcA, which encodes for the α subunit of the proteolytic component and located in the pup-prcB-prcA operon, was slightly reduced (B), the expression of the Pup (prokaryotic ubiquitin-like protein) ligase PafA were significantly and markedly downregulated (C-D). Given that Mpa and PafA work together to recognize, unfold, and translocate the substrate proteins into the proteolytic chamber [Citation51], these transcriptional profiles are indicative of active reduction of mycobacterial proteasome activity upon exposure to H2O2. According to the previous study showing that the mpa-, pafA-, and prcBA-deficient strains became more resistant than the wild-type Mtb to H2O2 [Citation52], it appears that the observed repression of proteasome components may protect mycobacteria from H2O2. The mechanism underlying the protective effect of diminished proteasome activity remains undefined, however, it might associate with the regulatory role of proteasome on maintaining metal homeostasis (Cu2+, Fe2+, and Zn2+)[Citation51].
Figure 5 . Transcriptional profiles of genes encoding for protein repair and degradation. Expression of msrA, prcA, mpa, pafA, clpP1, clpX, and clpC1 in Msm exposed to H2O2 for 50 min. Transcript levels were measured by the qRT-PCR, normalized relative to sigA, and expressed as Log2 fold change from untreated cultures. Data shown are mean ± SE with at least three independent experiments. *P < 0.05, **P < 0.01, ****P < 0.0001.
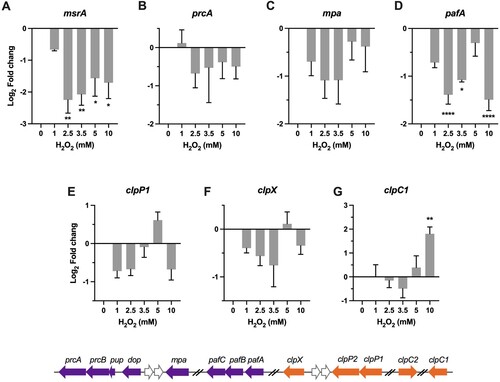
The mycobacterial Clp system is the essential proteolytic system composed of the proteolytic subunits, ClpP1 and ClpP2, and the ATPase adapters, ClpX or ClpC1 [Citation53,Citation54]. A recent study established that the induction of clpSA (ClpA is orthologous to ClpC1) protected E.coli from endogenous H2O2 [Citation19]. As shown in E–G, while the expression of clpP1 and clpX was slightly reduced or unchanged at most of tested H2O2 concentrations, the expression of clpC1 was significantly increased at 10 mM H2O2 (4.2-fold, P < 0.01) (G). Because the ATPase adapter determines substrate specificity of the Clp proteolytic process, the exclusive induction of clpC1 may indicate a selective protein degradation process at high H2O2 concentrations. Previous study demonstrated that silencing of clpC1 in Mtb resulted in accumulation of proteins mainly involved in intermediary metabolism, respiration, and lipid metabolism [Citation55], suggests that the induction of clpC1 under oxidative stress may associate with metabolic adaptation.
Profiles of coordinated gene regulation
To interrogate the physiological states and survival mechanisms under different intensities of oxidative stress, we performed cluster analysis of the H2O2 concentration-dependent dynamics of gene expression. As shown in A, four clusters of coordinated gene expression were identified.
Figure 6. Profiles of coordinated gene regulation. (A) Heatmap showing the unsupervised hierarchical clustering based on the Log2 fold change values of tested genes. Colour shown represents Z-scored expression values. (B-F) H2O2 concentration-dependent expressional patterns of clustered genes. Data shown are mean ± SE with at least three independent experiments.
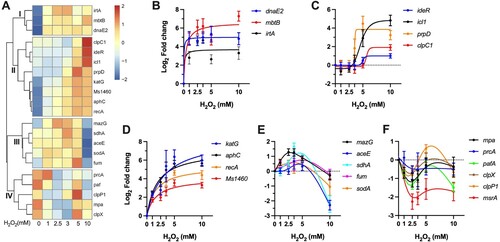
The cluster I includes three genes showing sustained level of induction throughout the tested H2O2 concentrations (B). This subset includes genes involved in error-prone DNA synthesis (dnaE2) and iron acquisition (mbtB, irtA). The sustained expression of these genes at different intensities of oxidative stress is well consistent with the notion that DNA and iron are the primary targets of ROS [Citation11,Citation16]. Moreover, this expression profile also indicates that mycobacteria may deploy increased iron acquisition and error-prone lesion bypass as fundamental strategies to combat oxidative stress, which is distinct from the defence tactics of E. coli, characterized by shrinking the iron pool and delaying the DNA repair [Citation11,Citation16].
The cluster II shows an expression pattern of H2O2 concentration-dependent increasing (A). Based on the dose-dependence, this subset genes can be further grouped into two classes. The first class includes ideR, icl1, prpD, and clpC1(C), which exhibited unchanged expression at low H2O2 concentrations (1–3.5 mM) and sudden induction at high concentrations (5 or/and 10 mM), indicative of responses specific to stimuli induced by intensified oxidative stress. The most intriguing feature of this class is the enrichment of genes involved in metabolic pathways, including methylcitrate cycle (icl1, prpD) and glyoxylate shunt (icl1). In addition, recent study also established that clpC1 mainly targets proteins involved in metabolism [Citation55]. Thereafter, it appears that metabolic remodeling may play an important role in mycobacterial defence against high level of ROS. The second class is characterized by steadily increased expression of katG, aphC, recA, and the Suf system (D), demonstrating that the expressions of the H2O2 scavenging enzymes, DNA damage response, and Fe–S cluster repair function were well correlated to the intensity of oxidative stress.
The cluster III contains a geneset (mazG, aceE, sdhA, fum, and sodA) that exhibited a “bell-curve” pattern of expression (E), suggesting an intricate crosstalk among transcriptional regulators under different intensities of oxidative stress. This profile was characterized by steadily increased induction by H2O2 below 5 mM and sudden repression by 10 mM, suggesting that the bacilli may encounter novel pressures that requires additional bacterial adaptation. This class also showed enrichment of genes involved in metabolism, including TCA cycle (sdhA, fum), pyruvate metabolism (aceE), and nucleotide metabolism (mazG). These data indicate that mycobacteria can sophistically remodel its metabolism to adopt to different intensities of oxidative stress (C,E).
The cluster IV is exclusively composed of genes involved in protein degradation and repair (F), which exhibited repressed (msrA and pafA) or unchanged expression. The repression of pafA might provide an antioxidant effect, given that the mpa-, pafA-, and prcBA-deficient strains were shown to be more resistant than wild-type Mtb to H2O2 [Citation52]. The role of mycobacterial Clp system in antioxidant response remains unclear. However, the remarkable upregulation of clpC1 and baseline expression level of clpP1 and clpX at 10 mM H2O2 imply that mycobacterial Clp system might participate in antioxidant defence at intensified oxidative stress by selective protein degradation [Citation19].
Conclusions
Given that bacterial transcriptional profiles could mirror the physiological states and survival mechanisms, this study offers several unique insights into mycobacterial antioxidant defence strategies under different intensities of oxidative stress. The sustained induction of mbtB and irtA by H2O2 indicates that Msm upregulates the iron acquisition upon oxidative stress. Importantly, these transcriptional signatures were in line with the observations in Mtb upon exposure to H2O2 or during infection of mice, shown by upregulation of mbtB and downregulation of bfrA (encoding an iron-storing bacterioferritin) [Citation20,Citation56]. This is strikingly different from the defence strategy of E. coli characterized by shrinking the iron pool and delaying the DNA repair [Citation16]. Given that iron plays a critical role in oxidative damages to DNA, the difference on iron metabolism between mycobacteria and E.coli may substantially affect DNA damage and the SOS response as observed in this study. Although mycobacteria were phenotypically refractory to DNA damage-dependent mode-one killing caused by low concentration of H2O2 (1–2 mM) [Citation20,Citation37], our results indicated that DNA damage occurs under these conditions, as shown by the marked induction of recA by 1–2 mM H2O2. Therefore, it is tempting to speculate that mycobacteria could prevent the killing events downstream of DNA damage by low concentrations of H2O2. Among the various types of DNA damage, DSBs are the major reason accounting for massive cell death. In Mycobacterium, DSBs could be directly repaired through homologous recombination, nonhomologous end-joining, and single-strand annealing (SSA) [Citation57]. Mycobacteria could also deploy translesion DNA polymerase (DnaE2) to prevent DSBs by allowing bypass of lethal replication-blocking lesions [Citation39]. Based on this, it appears that the remarkable and sustained induction of dnaE2 (40- to 75-fold) and recA by H2O2 might contribute to mycobacterial survival of mode-one killing. In this regard, it is worth noting that dinB (functional homologue of dnaE2) was not markedly induced in E.coli exposed to 1 mM H2O [Citation38]. Together, these results suggest that mycobacteria have evolved an adaptative strategy that could simultaneously upregulate the iron acquisition and DNA repair systems to meet the need for repair of Fe-cofactored enzymes and DNA maintenance (refractory to mode-one killing) under oxidative stress.
The dynamic alternations of metabolic gene expression, e.g. the “bell-curve” expression pattern of metabolic genes (aceE, sdhA, fum) and the reciprocal expression of genes belong to different metabolic pathways (TCA vs methylcitrate cycle and glyoxylate shunt), indicate that mycobacterial metabolic response to ROS is strikingly dependent on the intensity of oxidative stress. Given that the Fe–S enzymes (aconitase, SdhA, FumA) of TCA cycle are highly sensitive to ROS, the upregulation of sdhA and fum (encodes a non-Fe–S fumarase C) by intermediate H2O2 concentrations (3.5–5 mM) may reflect a compensatory response to restore the metabolic activities. Induction of fumC and ROS-resistant aconitase A was also observed in E.coli upon exposed to ROS [Citation43,Citation44], suggesting that both species deploy similar strategy to maintain the TCA flux. The reciprocal expression of genes between TCA and methylcitrate cycle/glyoxylate shunt at 10 mM H2O2 suggests that redirection or remodel of metabolic pathways underlies mycobacterial adaptation to intensified oxidative stress. For instance, the remarkable induction of icl1/prpD and the sudden repression of aceE/sdhA/fum by 10 mM H2O2 may reflect the redirection of carbon flux from TCA cycle to methylcitrate cycle and glyoxylate shunt. Induction of icl1 and/or prpD was also observed in Mtb upon exposure to H2O2 or bactericidal antibiotics, as well as during infection of mice [Citation5,Citation20,Citation56], indicating that these conditions could induce similar physiological transition and adaptation in mycobacteria.
Materials and methods
Bacterial strains and growth conditions
Mycolicibacterium smegmatis strain mc2-155 was grown at 37°C, 110 rpm in 7H9 broth (Difco) containing 0.2% (v/v) glycerol and 0.05% (v/v) Tween 80. Three colonies were mixed and inoculated into 2 ml 7H9 and grown at 37°C to reach a cell density of OD600∼2. The seeding culture was diluted 1:100 in 100 ml 7H9 broth (in a 500-ml flask) and grown to exponential phase (OD600 = 0.5–0.6) at 37°C with shaking at 110 rpm. Exponential-phase cultures were split into 10-ml aliquots (in a 15-ml centrifuge tube); one aliquot was left untreated, and the other aliquots were treated with the indicated concentrations of hydrogen peroxide statically at 37°C for 50 min [Citation30,Citation41,Citation58]. The H2O2 concentrations indicated in this study were validated by the Peroxide Assay kit (Sigmaaldrich).
RNA isolation
Culture aliquots were harvested by centrifugation at 3200×g for 5 min at room temperature. The supernatant was discarded, and the cell pellet was immediately resuspended in 1 ml cold TRIzol (Invitrogen), transferred to 2-ml screw cap tubes containing 500 μl of 0.1 mm zirconia-silicate beads. Cells were mechanically disrupted by bead beating (Minilys, Bertin) for five cycles (35 s at maximal speed) with cooling on ice for 1 min between pulses. The cell lysates were centrifuged at 16,000 × g for 5 min at 4 °C, and the supernatant was transferred to a new centrifuge tube containing Phase Lock Gel (TIANGEN). Three hundred microliter of chloroform was added to the supernatant and mixed vigorously for 30 s. The mixture was incubated at room temperature for 10 min, followed by centrifugation at 20,000 × g for 30 min at 4°C. The supernatant was transferred to a new microcentrifuge tube, mixed with an equal volume of isopropanol immediately by gently inverting 10 times, placed on ice for 30 min, followed by centrifugation at 20,000 × g for 30 min at 4°C. The supernatant was discarded, and the pellet was washed by 1 ml of 70% ethanol, followed by centrifugation at 20,000 × g for 10 min at 4°C. The pellet was dried at room temperature. RNA samples were treated with 10 U RNase-free DNaseI (NEB) for 30 min at 37°C, further purified using the GeneJET RNA purification kit (Thermofisher). RNA yields were quantified by Nanodrop (Thermo Scientific, Waltham, MA, USA). RNA quality was assessed by the agarose gel electrophoresis. RNA was subjected to the PCR to confirm lack of residual genomic DNA.
qRT-PCR
cDNA was synthesized using the SuperScript III First Strand kit (Invitrogen) with random hexamer primer, according to the manufacturer’s instructions. qRT-PCR was carried out using TB Green® Premix Ex Taq GC (TaKaRa). Gene expression data were normalized to sigA. Relative gene expression was calculated using the 2−ΔΔCt method. The primers used for the qRT-PCR were described in Table S2.
Statistical analyses
Significance tests were performed in the GraphPad Prism 9 (GraphPad Software, San Diego, CA, USA) using a one-way analysis of variance (ANOVA) and a Dunnett posttest. A statistical difference between the control (untreated) and another is marked above the column (*P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001).
Supplemental Material
Download MS Word (15.3 KB)Supplemental Material
Download MS Word (18.1 KB)Acknowledgements
M.W. and W.S. performed experiments and analyzed the data; L.-D.L. and G.-P.Z. designed this study; L.-D.L. analyzed the data, wrote the manuscript, and supervised the project. All authors discussed the results and commented on the manuscript.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Ehrt S, Schnappinger D. Mycobacterial survival strategies in the phagosome: defence against host stresses. Cell Microbiol. 2009;11(8):1170–1178.
- Tiwari S, van Tonder AJ, Vilchèze C, et al. Arginine-deprivation-induced oxidative damage sterilizes Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 2018;115(39):9779–9784.
- Fan XY, Tang B-K, Xu Y-Y, et al. Oxidation of dCTP contributes to antibiotic lethality in stationary-phase mycobacteria. Proc Natl Acad Sci USA. 2018;115(9):2210–2215.
- Vilcheze C, Hartman T, Weinrick B, et al. Enhanced respiration prevents drug tolerance and drug resistance in Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 2017;114(17):4495–4500.
- Nandakumar M, Nathan C, Rhee KY. Isocitrate lyase mediates broad antibiotic tolerance in Mycobacterium tuberculosis. Nat Commun. 2014;5:4306.
- Vilcheze C, Hartman T, Weinrick B, et al. Mycobacterium tuberculosis is extraordinarily sensitive to killing by a vitamin C-induced Fenton reaction. Nat Commun. 2013;4:1881.
- Grant SS, Kaufmann BB., Chand NS, et al. Eradication of bacterial persisters with antibiotic-generated hydroxyl radicals. Proc Natl Acad Sci USA. 2012;109(30):12147–12152.
- Jang S, Imlay JA. Micromolar intracellular hydrogen peroxide disrupts metabolism by damaging iron-sulfur enzymes. J Biol Chem. 2007;282(2):929–937.
- Flint DH, Tuminello JF, Emptage MH. The inactivation of Fe-S cluster containing hydro-lyases by superoxide. J Biol Chem. 1993;268(30):22369–22376.
- Anjem A, Imlay JA. Mononuclear iron enzymes are primary targets of hydrogen peroxide stress. J Biol Chem. 2012;287(19):15544–15556.
- Imlay JA. The molecular mechanisms and physiological consequences of oxidative stress: lessons from a model bacterium. Nat Rev Microbiol. 2013;11(7):443–454.
- Park S, You X, Imlay JA. Substantial DNA damage from submicromolar intracellular hydrogen peroxide detected in Hpx- mutants of Escherichia coli. Proc Natl Acad Sci USA. 2005;102(26):9317–9322.
- Khanna KK, Jackson SP. DNA double-strand breaks: signaling, repair and the cancer connection. Nat Genet. 2001;27(3):247–254.
- Leichert LI, Gehrke F, Gudiseva HV, et al. Quantifying changes in the thiol redox proteome upon oxidative stress in vivo. Proc Natl Acad Sci USA. 2008;105(24):8197–8202.
- Sen A, Imlay JA. How microbes defend themselves from incoming hydrogen peroxide. Front Immunol. 2021;12:667343.
- Khademian M, Imlay JA. How microbes evolved to tolerate oxygen. Trends Microbiol. 2021;29(5):428–440.
- Cumming BM, Lamprecht DA, Wells RM, et al. The physiology and genetics of oxidative stress in mycobacteria. Microbiol Spectr. 2014;2(3):2
- Ehrt S, Schnappinger D, Rhee KY. Metabolic principles of persistence and pathogenicity in Mycobacterium tuberculosis. Nat Rev Microbiol. 2018;16(8):496–507.
- Sen A, Zhou Y, Imlay JA. During oxidative stress the Clp proteins of Escherichia coli ensure that iron pools remain sufficient to reactivate oxidized metalloenzymes. J Bacteriol. 2020;202(18):e00235.
- Voskuil MI, Bartek IL, Visconti K, et al. The response of Mycobacterium tuberculosis to reactive oxygen and nitrogen species. Front Microbiol. 2011;2:105.
- Ng VH, Cox JS, Sousa AO, et al. Role of KatG catalase-peroxidase in mycobacterial pathogenesis: countering the phagocyte oxidative burst. Mol Microbiol. 2004;52(5):1291–1302.
- Bryk R, Lima CD, Erdjument-Bromage H, et al. Metabolic enzymes of mycobacteria linked to antioxidant defense by a thioredoxin-like protein. Science. 2002;295(5557):1073–1077.
- Piddington DL, Fang FC., Laessig T, et al. Cu,Zn superoxide dismutase of Mycobacterium tuberculosis contributes to survival in activated macrophages that are generating an oxidative burst. Infect Immun. 2001;69(8):4980–4987.
- Edwards KM, Cynamon MH, Voladri RKR, et al. Iron-cofactored superoxide dismutase inhibits host responses to Mycobacterium tuberculosis. Am J Respir Crit Care Med. 2001;164(12):2213–2219.
- Li Z, Kelley C, Collins F, et al. Expression of katG in Mycobacterium tuberculosis is associated with its growth and persistence in mice and guinea pigs. J Infect Dis. 1998;177(4):1030–1035.
- Manca C, Paul S, Barry CE, et al. Mycobacterium tuberculosis catalase and peroxidase activities and resistance to oxidative killing in human monocytes in vitro. Infect Immun. 1999;67(1):74–79.
- Master SS, Springer B, Sander P, et al. Oxidative stress response genes in Mycobacterium tuberculosis: role of ahpC in resistance to peroxynitrite and stage-specific survival in macrophages. Microbiology (Reading). 2002;148(Pt 10):3139–3144.
- Springer B., Master S, Sander P, et al. Silencing of oxidative stress response in Mycobacterium tuberculosis: expression patterns of ahpC in virulent and avirulent strains and effect of ahpC inactivation. Infect Immun. 2001;69(10):5967–5973.
- Harth G, Horwitz MA. Export of recombinant Mycobacterium tuberculosis superoxide dismutase is dependent upon both information in the protein and mycobacterial export machinery. A model for studying export of leaderless proteins by pathogenic mycobacteria. J Biol Chem. 1999;274(7):4281–4292.
- Schnappinger D, Ehrt S, Voskuil MI, et al. Transcriptional adaptation of Mycobacterium tuberculosis within macrophages: insights into the phagosomal environment. J Exp Med. 2003;198(5):693–704.
- Rodriguez GM, Smith I. Identification of an ABC transporter required for iron acquisition and virulence in Mycobacterium tuberculosis. J Bacteriol. 2006;188(2):424–430.
- Huet G, Daffe M, Saves I. Identification of the Mycobacterium tuberculosis SUF machinery as the exclusive mycobacterial system of [Fe-S] cluster assembly: evidence for its implication in the pathogen's survival. J Bacteriol. 2005;187(17):6137–6146.
- Rodriguez GM, Voskuil MI, Gold B, et al. Ider, An essential gene in Mycobacterium tuberculosis: role of IdeR in iron-dependent gene expression, iron metabolism, and oxidative stress response. Infect Immun. 2002;70(7):3371–3381.
- De Voss JJ, Rutter K, Schroeder BG, et al. The salicylate-derived mycobactin siderophores of Mycobacterium tuberculosis are essential for growth in macrophages. Proc Natl Acad Sci U S A. 2000;97(3):1252–1257.
- Dussurget O, Rodriguez M, Smith I. An ideR mutant of Mycobacterium smegmatis has derepressed siderophore production and an altered oxidative-stress response. Mol Microbiol. 1996;22(3):535–544.
- Arnold FM, Weber MS, Gonda I, et al. The ABC exporter IrtAB imports and reduces mycobacterial siderophores. Nature. 2020;580(7803):413–417.
- Imlay JA, Linn S. Bimodal pattern of killing of DNA-repair-defective or anoxically grown Escherichia coli by hydrogen peroxide. J Bacteriol. 1986;166(2):519–527.
- Zheng M, Wang X, Templeton LJ, et al. DNA microarray-mediated transcriptional profiling of the Escherichia coli response to hydrogen peroxide. J Bacteriol. 2001;183(15):4562–4570.
- Boshoff HI, Reed MB, Barry CE, et al. Dnae2 polymerase contributes to in vivo survival and the emergence of drug resistance in Mycobacterium tuberculosis. Cell. 2003;113(2):183–193.
- Lyu LD, Tang B-K, Fan X-Y, et al. Mycobacterial MazG safeguards genetic stability via housecleaning of 5-OH-dCTP. PLoS Pathog. 2013;9(12):e1003814.
- Lu LD, Sun Q, Fan X-y, et al. Mycobacterial MazG is a novel NTP pyrophosphohydrolase involved in oxidative stress response. J Biol Chem. 2010;285(36):28076–28085.
- Shi KX, Wu Y-K, Tang B-K, et al. Housecleaning of pyrimidine nucleotide pool coordinates metabolic adaptation of nongrowing Mycobacterium tuberculosis. Emerg Microbes Infect. 2019;8(1):40–44.
- Varghese S, Tang Y, Imlay JA. Contrasting sensitivities of Escherichia coli aconitases A and B to oxidation and iron depletion. J Bacteriol. 2003;185(1):221–230.
- Liochev SI, Fridovich I. Fumarase C, the stable fumarase of Escherichia coli, is controlled by the soxRS regulon. Proc Natl Acad Sci USA. 1992;89(13):5892–5896.
- Ruecker N, Jansen R, Trujillo C, et al. Fumarase deficiency causes protein and metabolite succination and intoxicates Mycobacterium tuberculosis. Cell Chem Biol. 2017;24(3):306–315.
- Eoh H, Rhee KY. Multifunctional essentiality of succinate metabolism in adaptation to hypoxia in Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 2013;110(16):6554–6559.
- Gould TA, van de Langemheen H, Munoz-Elias EJ, et al. Dual role of isocitrate lyase 1 in the glyoxylate and methylcitrate cycles in Mycobacterium tuberculosis. Mol Microbiol. 2006;61(4):940–947.
- Dong W, Nie X, Zhu H, et al. Mycobacterial fatty acid catabolism is repressed by FdmR to sustain lipogenesis and virulence. Proc Natl Acad Sci USA. 2021;118(16):e2019305118.
- Douglas T, Daniel DS, Parida BK, et al. Methionine sulfoxide reductase A (MsrA) deficiency affects the survival of Mycobacterium smegmatis within macrophages. J Bacteriol. 2004;186(11):3590–3598.
- John G. S., Brot N, Ruan J, et al. Peptide methionine sulfoxide reductase from Escherichia coli and Mycobacterium tuberculosis protects bacteria against oxidative damage from reactive nitrogen intermediates. Proc Natl Acad Sci U S A. 2001;98(17):9901–9906.
- Bode NJ, Darwin KH. The pup-proteasome system of mycobacteria. Microbiol Spectr. 2014;2(5):667.
- Darwin KH, Ehrt S, Gutierrez-Ramos J-C, et al. The proteasome of Mycobacterium tuberculosis is required for resistance to nitric oxide. Science. 2003;302(5652):1963–1966.
- Schmitz KR, Sauer RT. Substrate delivery by the AAA+ ClpX and ClpC1 unfoldases activates the mycobacterial ClpP1P2 peptidase. Mol Microbiol. 2014;93(4):617–628.
- Raju RM, Unnikrishnan M, Rubin DHF, et al. Mycobacterium tuberculosis ClpP1 and ClpP2 function together in protein degradation and are required for viability in vitro and during infection. PLoS Pathog. 2012;8(2):e1002511.
- Lunge A, Gupta R, Choudhary E, et al. The unfoldase ClpC1 of Mycobacterium tuberculosis regulates the expression of a distinct subset of proteins having intrinsically disordered termini. J Biol Chem. 2020;295(28):9455–9473.
- Timm J, Post FA, Bekker L-G, et al. Differential expression of iron-, carbon-, and oxygen-responsive mycobacterial genes in the lungs of chronically infected mice and tuberculosis patients. Proc Natl Acad Sci U S A. 2003;100(24):14321–14326.
- Gupta R, Barkan D, Redelman-Sidi G, et al. Mycobacteria exploit three genetically distinct DNA double-strand break repair pathways. Mol Microbiol. 2011;79(2):316–330.
- Milano A, Forti F, Sala C, et al. Transcriptional regulation of furA and katG upon oxidative stress in Mycobacterium smegmatis. J Bacteriol. 2001;183(23):6801–6806.
