ABSTRACT
Serum hepatitis B virus (HBV) pregenomic RNA (pgRNA) is a surrogate marker for reflecting the transcriptional activity of covalently closed circular DNA. However, there is still no standardized assay for the quantitative detection of serum HBV RNA in chronic hepatitis B patients. In this study, quantitative polymerase chain reactions for detecting the preC/C-RNA (preC/C region HBV pgRNA), SF-RNA (splicing variants-free pgRNA) and XR-RNA (X region remained pgRNA) regions were set up. The dynamic changes of serum pgRNA splicing variants and 3′ terminal truncations were analysed in three retrospective cohorts: 35 treatment-naive chronic HBV-infected patients (cohort A), 52 chronic hepatitis B (CHB) patients who received nucleos(t)ide analogs (NAs) therapy for 48 weeks (cohort B) and eight CHB patients who are under long-term NAs treatment (cohort C). The accuracy and sensitivity of HBV RNA detection were assessed by the National Standard of HBV RNA. We confirmed that high proportions of pgRNA splicing variants and 3′ terminal truncations were present and significantly affect the quantitative detection of serum HBV RNA in both treatment-naive and NAs-treated CHB patients. To achieve the higher accuracy and sensitivity on the detection of HBV RNA level, the primers and probes should be designed at the 5′ terminal region of HBV genome and outside the mainly spliced sequence of pgRNA, especially for CHB patients under long-term NAs treatment. This study would help to better understand the significance of the pgRNA splicing variants and 3′ terminal truncations, and further guide the clinical detection of serum HBV RNA.
Introduction
Hepatitis B virus (HBV) infection is a major health problem worldwide with about 30% of the world’s population showing serological evidence of current or past HBV infection [Citation1]. Although the currently used antiviral agents, including nucleos(t)ide analogs (NAs) and pegylated interferon-alpha (Peg-IFN-α)-2a, can efficiently inhibit HBV replication and disease progression for chronic hepatitis B (CHB) patients [Citation2], the covalently closed circular DNA (cccDNA) is still difficult to be eradicated [Citation3]. We and other groups have previously confirmed that serum HBV RNA is mainly the non- or partially reverse-transcribed pregenomic RNA (pgRNA) and present in HBV virion-like particles [Citation4-9]. Since pgRNA can only be produced from cccDNA, serum HBV pgRNA can be a surrogate marker for HBV DNA in reflecting the activity of cccDNA when HBV DNA is lost during the antiviral therapy [Citation10-14]. Meanwhile, monitoring serum HBV RNA can predict the antiviral efficacy [Citation5,Citation15-20], and reflect the risk of HBV viral rebound after drug withdrawal [Citation4,Citation21-24]. Therefore, it is worth detecting and monitoring serum HBV RNA in clinical practice.
It has been reported that serum HBV RNA consists of full length, splicing variant and 3′ terminal truncated pgRNA [Citation8,Citation25-27], or, even the over-genome long HBx RNAs [Citation9,Citation27]. In our previous study, we found that the levels of serum HBV RNA detected by targeting PreC/C, S or X regions of HBV genome were different, and were gradually decreased from PreC/C region, S region to X region [Citation28]. Moreover, as suggested by the experts from 2019 EASL-AASLD HBV Treatment Endpoints Conference Faculty, the assays for serum HBV RNA should be standardized, and the type of HBV RNA, such as pgRNA, total RNA, spliced RNA or truncated RNA, should be delineated during the detection of serum HBV RNA [Citation29]. Therefore, to optimize the detection of serum HBV RNA, the dynamic changes of serum HBV RNA species and their influences on the quantitative detection of HBV RNA are needed to be elucidated.
In this study, three reverse transcription-quantitative polymerase chain reactions (RT-qPCRs) targeting three regions of HBV genome were designed to explore the dynamic changes of HBV pgRNA splicing variants and 3′ terminal truncations, as well as their influences on the quantification of serum HBV RNA in three retrospective cohorts. The accuracy and sensitivity of above three RT-qPCRs were assessed by the National Standard of HBV RNA.
Materials and methods
Patients
Three retrospective cohorts were used in this study. Cohort A consisted of 35 treatment-naive chronic HBV-infected patients, including 15 HBeAg-negative and 20 HBeAg-positive patients who were admitted to Beijing 302 Hospital. Cohort B consisted of 52 baseline HBeAg-positive CHB patients who received the NA-therapy for 48 weeks at NanFang Hospital affiliated to Southern Medical University and Tianjin Second People's Hospital. A total of 199 serum specimens were collected at 0, 12, 24 and 48 weeks post NAs treatment. According to the decreasing range of HBV RNA at 48 weeks, the patients were divided into rapid decline group (≥ 2 log10) and slow decline group (< 2 log10). Cohort C consisted of eight CHB patients who received long-term NA-therapy for 5 years at the Beijing YouAn Hospital affiliated to Capital Medical University. A total of 56 serum specimens were collected at 0, 24, 48 weeks, 2 years, 3 years, 4 years and 5 years post NAs treatment. The clinical characteristics of patients in cohort A, B, and C are shown in . The experimental protocol conformed to the ethical guidelines of the 1975 Declaration of Helsinki. This study was approved by the Ethics Committee of Tianjin Second People’s Hospital. Written informed consent was obtained from each patient.
Table 1. The clinical characteristics of the chronic HBV-infected patients in cohort A.
Table 2. The clinical characteristics of the CHB patients in cohort B.
Table 3. The clinical characteristics of the CHB patients in cohort C.
Cell culture
Huh-7 cells were obtained from the American Type Culture Collection and cultured by the Dulbecco's modified Eagle medium (DMEM) containing 10% fetal bovine serum (FBS), 100 IU/mL penicillin and 100 μg/mL streptomycin (Gibco, Carlsbad, CA, USA). HepAD38 cell line was maintained in the DMEM containing 10% FBS, 100 IU/mL penicillin and 100 μg/mL streptomycin (Gibco, Carlsbad, CA, USA), 400 μg/mL G418 and 6 μg/mL tetracycline. Tetracycline was removed from the cell culture for at least 9 days when HepAD38 cells were used for HBV collection. All cells were placed at 37°C in 5% CO2 humidified incubators.
National Standard for HBV RNA detection kit
The National Standard (Batch No. 340008-201901) for quantitative detection of HBV RNA was obtained from the China National Institutes for Food and Drug Control, which consist of HBV RNA-positive plasma and was suitable for the quality evaluation and control of HBV RNA detection.
Design and optimization of primers and TaqMan probes
Primers and probes targeting the preC/C-RNA, SF-RNA and XR-RNA regions of the HBV genome for RT-qPCR were designed using the Primer Premier 5.0 software. The sequences of primers and probes are given in .
Table 4. Sequences of primers and probes.
Extraction and quantification of serum HBV RNA
HBV RNA was extracted by the EasyPure Viral RNA Kit (TransGen Biotech, China) according to the manufacture’s instruction, and was treated with DNase I (Thermo Fisher Scientific, USA) at 37°C for 30 min. The levels of HBV RNA were detected by RT-qPCR in Applied Biosystems StepOnePlus Real-Time PCR Systems (Applied Biosystems, Mannheim, USA) with a TaqMan probe method. The RT-qPCR reaction mixture (50 μL) contained 25 μL RT-qPCR mix (Hotgen, Beijing, China), 1.5 μL forward primer (10 μM), 1.5 μL reverse primer (10 μM), 1 μL TaqMan probe (10 μM), 15 μL DNase I-treated HBV RNA and 6 μL ddH2O. The experiments were performed using the following protocol: one cycle at 45 °C for 15 min, one cycle at 95 °C for 5 min, 40 cycles at 95 °C for 10 s and 60°C for 45 s. The standard curves for preC/C-RNA, SF-RNA and XR-RNA primers were drawn and calibrated by the same plasmid standard containing 1.2× wild-type HBV genome which contained one copy preC/C-RNA region, one copy SF-RNA region and two copies XR-RNA region, and was 10-fold serially diluted to prepare the standards with a series of concentrations.
In vitro transcription for full-length pgRNA
Linearized full-length HBV genome was generated by primers with T7 promoter sequence, and then transcribed using the MEGAscript™ T7 Transcription Kit (Invitrogen) according to the manufacturer's protocol. Transcribed RNAs were then treated with DNase I and purified by the QIAmp MinElute Viral Spin Kit (Qiagen), and finally quantified using the NanoDrop ND-2000 (Thermo scientific).
Analysis of splicing variants by PCR and sequencing
The unspliced pgRNA and splicing variants were amplified by nested-PCR, and the corresponding primers are shown in . Next, the PCR products were electrophoresed in 1.0% agarose gel, and were subsequently recovered by the TIANgel Midi Purification Kit (TIANGEN Biotech, Beijing, China). Finally, the purified PCR products were cloned into pEAST-Blunt vector for cloning sequencing.
Isoform-sequencing (Iso-Seq)
Huh-7 cells were co-transfected with prcccDNA and pCMV-Cre plasmids (1:1 in ratio) provided by Prof. Qiang Deng at Fudan University, Shanghai, China. Three days later, the transfected Huh-7 cells were harvested, and total RNA was isolated by TRIzol reagent (Invitrogen, USA). The genomic DNA was removed by DNase I, and then Iso-Seq was performed.
Statistical analyses
Data were shown as mean (standard deviation) or median (range). The paired or unpaired non-parameters Mann–Whitney test were used for statistical analysis with SPSS 24.0 software (IBM, Chicago, IL, USA). Graphs were plotted using GraphPad Prism 7 (GraphPad Software, USA) and MedCalc® Statistical Software version 19.5.2 (MedCalc Software, Mariakerke, Belgium). All tests were two-tailed, and P < 0.05 was considered statistically significant.
Results
The serum HBV RNA level detected at preC/C-RNA region showed the highest accuracy and sensitivity assessed by the National Standard of HBV RNA
To test whether pgRNA splicing variants or 3′ terminal truncations would affect the quantification of serum HBV RNA that mainly consisted of pgRNA, three sets of RT-qPCR primers and probes were designed. As shown in (A), two pairs of RT-qPCR sets, namely preC/C-RNA and splicing variants-free pgRNA (SF-RNA), respectively, were designed in core region with the same forward primer and probe, whereas the reverse primer of SF-RNA was 92 nt later for that of preC/C-RNA. The preC/C-RNA primers we designed were located in the region that was ahead of 2447 site and only a few kinds of splicing variants, like SP5, SP6, SP7 and SP12 were located ahead of this site. But the abundances of them are extremely low. Thus, the amplified product of total RNA set could represent the authentic level of total pgRNA to some extent. Since the reverse primer of SF-RNA was located in the region that was removed during the formation of main pgRNA splicing variants, but not for the reverse primer of preC/C-RNA, serum HBV RNA detection of SF-RNA did not contain the main pgRNA splicing variants. The third pair of RT-qPCR set was designed at 3′ terminal (X region) of HBV genome, which termed XR-RNA region remained pgRNA set (XR-RNA). Serum HBV RNA detected at this region did not contain the majority of 3′ terminal truncated pgRNA, which was reduced by the RNase H activity of P protein in the reverse transcription process. Besides, the standard curves calculated with serially diluted 1.2× wild-type HBV DNA plasmids and the quality control with full-length pgRNA derived from in vitro transcription were made for the comparison of these RT-qPCR tests and indicated that above three pairs of qPCR primers and probes had almost the same amplification efficiency (Figure S1).
Figure 1. The accuracy and sensitivity of HBV RNA detection at preC/C-RNA, SF-RNA and XR-RNA regions were assessed by the National Standard of HBV RNA. (A) The diagram of HBV pgRNA and the relative locations of preC/C-RNA, SF-RNA and XR-RNA regions targeted by qPCR primers. SP: pgRNA splicing variant; *SP1-3 were the pgRNA splicing variants identified in our previous study.26 (B) The accuracy on the detection of HBV RNA at preC/C-RNA, SF-RNA and XR-RNA regions was assessed by HBV RNA standards. (C) The sensitivity on the detection of HBV RNA at preC/C-RNA, SF-RNA and XR-RNA regions was precisely analysed by HBV RNA standards. The lowest concentration of 95% positive was considered as the LLOD.
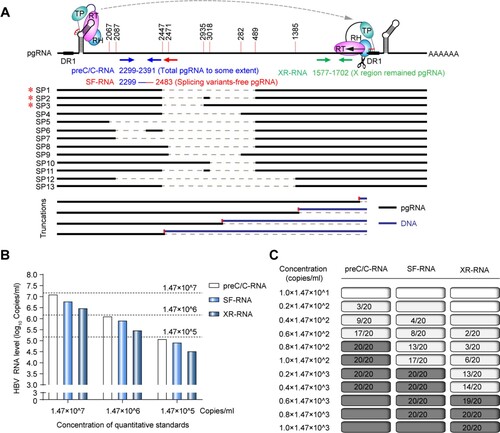
Next, National Standard of HBV RNA was 10-fold serially diluted from 1.47×107 copies/mL to 1.47×105 copies/mL, treated with DNase I at 37°C for 30 min and quantitatively detected at preC/C-RNA, SF-RNA and XR-RNA regions following the RT-qPCR method mentioned above. As a result, the levels of HBV RNA detected at preC/C-RNA region were closest to the concentrations of National Standard of HBV RNA ((B)). Moreover, in a more precisely diluted test, the LLOD of HBV RNA detected at preC/C-RNA, SF-RNA and XR-RNA regions were confirmed to be 1.18×102, 2.94×102 and 8.82×102 copies/mL, respectively, suggesting that the accuracy and sensitivity on the quantitative detection of HBV RNA were highest at preC/C-RNA region, and were lowest at XR-RNA region ((C)).
HBV pgRNA splicing variants and 3′ terminal truncations affect the quantitative detection of serum HBV RNA in treatment-naive chronic HBV-infected individuals
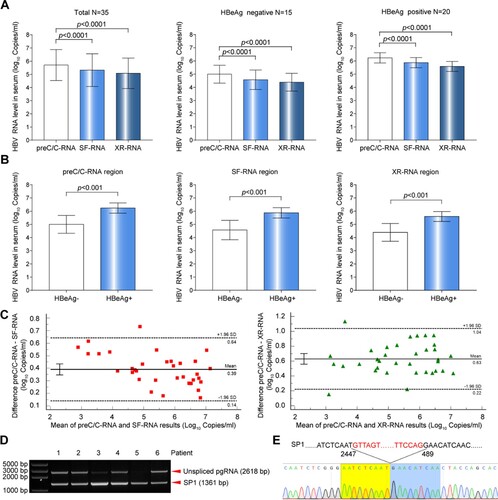
The levels of serum HBV RNA were quantitatively detected in 35 treatment-naive chronic HBV-infected individuals (cohort A). The result revealed that the levels of serum HBV RNA detected at preC/C-RNA region were significantly higher than those of SF-RNA and XR-RNA regions, regardless of HBeAg status ((A)). Furthermore, a slightly higher ratio of spliced pgRNA to total pgRNA [approximately equal to the ratio of (preC/C-RNA–SF-RNA)/preC/C-RNA] was observed in HBeAg-negative group than that in HBeAg-positive group (61.60% vs. 55.55%, P = 0.124), but with no statistical significance. Together with the ratio of 3′ terminal truncated pgRNA to total pgRNA [approximately equal to the ratio of (preC/C-RNA–XR-RNA)/preC/C-RNA] was similar between the two groups (71.49% vs. 72.47%, P = 0.616), suggesting that the ratios of pgRNA splicing variants or 3′ terminal truncations to total HBV RNA were also irrespective of the HBeAg status. However, the levels of serum pgRNA detected by preC/C-RNA, SF-RNA and XR-RNA regions in HBeAg-positive patients were all significantly higher than that in HBeAg-negative patients ((B)). Bland Altman analysis showed that the bias with a 95% confidence interval were 0.39 (0.14–0.64) log10 copies/mL between preC/C-RNA and SF-RNA, and 0.63 (0.22-1.04) log10 copies/mL between preC/C-RNA and XR-RNA in cohort A ((C)). Besides, patients in cohort A were also divided into different phases according to natural history definition. The result revealed that the levels of serum HBV RNA detected at preC/C-RNA region were significantly higher than those of SF-RNA and XR-RNA regions, regardless of the phases of natural history definition (Figure S2). Moreover, PCR and sequencing assays confirmed that unspliced pgRNA and considerable amount of main pgRNA splicing variant (SP1) were present in the sera of six chronic HBV-infected individuals with high HBV RNA level, which further supported that pgRNA splicing variants could affect the quantification of serum HBV RNA ((D,E)).
Figure 2. Comparative analysis of HBV RNA levels detected at preC/C-RNA, SF-RNA and XR-RNA regions in the sera of treatment-naïve chronic HBV-infected individuals. (A) The levels of serum HBV RNA in the sera of 35 treatment-naïve chronic HBV-infected individuals were detected at preC/C-RNA, SF-RNA and XR-RNA regions. (B) The levels of HBV RNA between HBeAg-positive (n = 20) and HBeAg-negative groups (n = 15) were detected at preC/C-RNA, SF-RNA and XR-RNA regions. (C) The bias of serum HBV RNA levels detected at preC/C-RNA and SF-RNA, preC/C-RNA and XR-RNA regions were analysed by Bland Altman plot method. (D) Serum HBV RNA was reverse-transcribed and amplified by nested-PCR. The PCR products were analysed by 1.0% agarose gel electrophoresis. (E) Sequencing analysis of SP1. Statistical significance shown as the p value was evaluated by a One-way ANOVA with Bonferroni’s multiple comparisons and Paired Mann–Whitney U test. eff%: amplification efficiency.
HBV pgRNA splicing variants and 3′ terminal truncations affect the quantitative detection of serum HBV RNA in CHB patients under NAs treatment
To further investigate the influence of pgRNA splicing variants and 3′ terminal truncations on the quantification of serum HBV RNA under NAs treatment, a longitudinal cohort of 52 CHB patients (cohort B) who had received NAs therapy for 48 weeks was enrolled. At 0, 12, 24 and 48 weeks post NAs treatment, the levels of serum HBV RNA detected at preC/C-RNA region were all significantly higher than those at SF-RNA and XR-RNA regions ((A)). Bland Altman analysis showed that the bias with a 95% confidence interval was 0.39 (0.09-0.69) log10 copies/mL between preC/C-RNA and SF-RNA regions, and 0.43 (-0.12-0.98) log10 copies/mL between preC/C-RNA and XR-RNA regions ((B,C)). Furthermore, CHB patients were divided into rapid decline group (≥2 log) and slow decline group (<2 log) according to the decreasing range of serum HBV RNA level at 48 weeks post NAs treatment. The levels of serum HBV RNA detected at preC/C-RNA region were still higher than those of SF-RNA and XR-RNA regions in both groups ((D, E)). Individually, a higher serum HBV RNA levels detected at preC/C-RNA region than those detected at SF-RNA and XR-RNA regions were found in 98.99% (197/199) and 95.98% (191/199) serum specimens, respectively. In 7.69% (4/52) CHB patients, the levels of serum HBV RNA detected at SF-RNA or XR-RNA regions were lower than the lower limit of detection (LLOD), whereas none for those detected at preC/C-RNA region, indicating that detecting HBV RNA at preC/C-RNA region could reduce the false negative rate (Figure S3).
Figure 3. Comparative analysis of HBV RNA levels detected at preC/C-RNA, SF-RNA and XR-RNA regions in the sera of CHB patients under NAs treatment. (A) The levels of HBV RNA were quantitatively detected at preC/C-RNA, SF-RNA and XR-RNA regions in the sera of CHB patients (n = 52) under NAs treatment for 48 weeks. (B) The bias of serum HBV RNA levels detected at preC/C-RNA and SF-RNA regions were analysed by Bland Altman plot method. (C) The bias of serum HBV RNA levels detected at preC/C-RNA and XR-RNA regions were analysed by Bland Altman plot method. The mean difference was given (y axis) for the mean results (x axis). (D) The levels of HBV RNA were quantitatively detected at preC/C-RNA, SF-RNA and XR-RNA regions in rapid decline group and (E) slow decline group. (F) The levels of HBV RNA were detected at preC/C-RNA, SF-RNA and XR-RNA regions in the sera of CHB patients (n = 8) under NAs treatment for 5 years. (G) The levels of serum HBV RNA in each CHB patient under NAs treatment for 5 years were quantitatively detected at preC/C-RNA, SF-RNA and XR-RNA regions. The white block represented the level was lower than LLOD. Statistical significance shown as the p value was evaluated by a Paired Mann–Whitney U test.
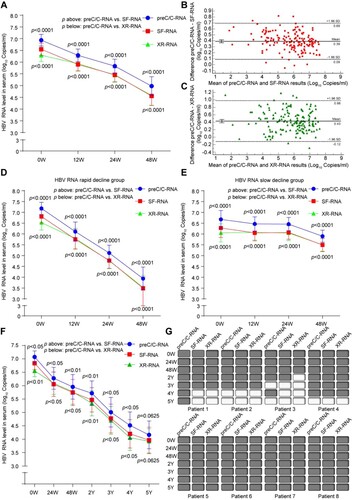
Furthermore, a longitudinal cohort (cohort C) of eight CHB patients under NAs treatment for 5 years were enrolled. Consistently, the levels of HBV RNA detected at preC/C-RNA region were still significantly higher than those of SF-RNA and XR-RNA regions in the sera of CHB patients under long-term NAs treatment ((F)). At 0, 24 weeks, 48 weeks, 2 years, 3 years, 4 years and 5 years post NAs treatment, the average proportion of spliced pgRNA to total pgRNA was 39.75%, 36.31%, 36.93%, 42.95%, 45.22%, 53.03% and 50.99%, respectively. Meanwhile, the average proportion of pgRNA 3′ terminal truncations to total HBV RNA was 68.81%, 33.05%, 33.12%, 56.32%, 54.20%, 69.69%, and 57.80%, respectively. Accordingly, both the ratios of spliced pgRNA and 3′ terminal truncated pgRNA to total HBV RNA decreased after short-term (within 48 weeks) NAs treatment, and subsequently increased after long-term (≥2 years) NAs treatment, suggesting that we should pay more attention to the influences of pgRNA splicing variants and 3′ terminal truncations on HBV RNA detection under long-term NAs treatment. Moreover, the time of serum HBV RNA loss detected at SF-RNA and XR-RNA regions was earlier than that of preC/C-RNA region in some of CHB patients, indicating that the sensitivity of serum HBV RNA detection at preC/C-RNA region was higher than that of SF-RNA and XR-RNA regions ((G)). Besides, we also analysed the differences on the dynamic changes between HBV DNA, preC/C-RNA and HBsAg. Six of the eight CHB patients had serum HBV DNA undetectable, meanwhile, all eight patients remained serum HBV RNA detecting positive after 1 year of ETV treatment. All eight patients had serum HBV DNA undetectable, but only three of them had serum HBV RNA undetectable after 5-year treatment. The decline rate of serum HBV DNA was much faster than serum HBV RNA. None of them lost their serum HBsAg during the 5-year treatment.
The reverse transcription efficiency of pgRNA splicing variants might be lower than that of unspliced pgRNA
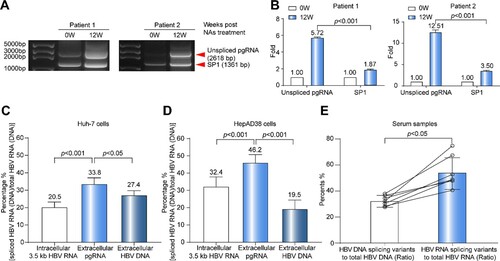
Many viral genomes encompass cryptic, sequence-degenerate and dispersed RNA Packaging Signals (PSs), and it has been reported that some PSs facilitate HBV pgRNA to be packaged in a manner that supports reverse transcription [Citation30]. Given that some PSs are deleted in pgRNA splicing variants, we speculated that its reverse transcription efficiency might be lower than that of unspliced pgRNA. To test this possibility, the levels of serum HBV RNA detected at preC/C-RNA and SF-RNA regions, and the corresponding pgRNA species were analysed in two CHB patients in cohort B. Patient 1 and patient 2 were selected in the rapid decline group and slow decline group, respectively. The unspliced pgRNA and dominant pgRNA splicing variant (SP1) were both detected in the sera of two patients ((A)). The gray value analysis which was calculated with the Image J analysis software showed that, compared to the baseline level, the unspliced pgRNA level in patient 1 and patient 2 increased to 5.72-fold and 12.51-fold, respectively, whereas SP1 level increased to 1.87-fold and 3.50-fold at 12 weeks post NAs treatment, respectively. It suggested that the increasing folds of unspliced pgRNA were higher than that of SP1 under NAs treatment ((B)).
Figure 4. Analysis on the reverse transcription efficiency between pgRNA splicing variants and unspliced pgRNA. (A) HBV RNA in the sera of two CHB patients was reverse-transcribed and amplified by nested-PCR at baseline and 12 weeks post NAs treatment. The PCR products were analysed by 1.0% agarose gel electrophoresis. (B) The gray value analysis on the levels of unspliced pgRNA and SP1 with ImageJ Software. To avoid the difference in band strength due to the difference of amplification efficiency, the comparisons of fold changes between 0 and 12 weeks were conducted in the same sets. (C) The proportions of intracellular 3.5 kb HBV RNA splicing variants, extracellular pgRNA splicing variants and HBV DNA splicing variants were analysed in Huh-7 cells transfected with prcccDNA plasmids and (D) HepAD38 cells. (E) The ratio of splicing variants in serum HBV DNA and RNA in 7 CHB patients of cohort A. SP1: pgRNA splicing variant 1. Statistical significance shown as the p value was evaluated by a Paired Mann–Whitney U test.
Furthermore, the proportions of intracellular 3.5 kb HBV RNA (consist of pgRNA and preC mRNA) splicing variants, extracellular HBV pgRNA splicing variants and extracellular HBV DNA splicing variants were analysed in HepAD38 cells stably producing HBV particles under the control of tet-off system, and Huh-7 cells transiently transfected with prcccDNA plasmids that can produce HBV cccDNA mimics and virus particles [Citation31]. The result revealed that the ratio of extracellular pgRNA splicing variants to total pgRNA was higher than that of both ratios of intracellular 3.5 kb HBV RNA splicing variants to total 3.5 kb HBV RNA and extracellular HBV DNA splicing variants to total HBV DNA ((C,D)). Meanwhile, isoform-sequencing (Iso-Seq) of HBV RNA in Huh-7 cells revealed that the transcriptome sequencing reads covering the preC/C-RNA region was higher than that covering the SF-RNA region (9920 vs. 8262), and the ratio o f dominant 3.5 kb HBV RNA splicing variants to total 3.5 kb HBV RNA was 16.71%, which was similar with the RT-qPCR result (20.5%) as shown in (C). Consistently, the ratio of splicing variants in serum HBV RNA was higher than that of serum HBV DNA in seven CHB patients of cohort A ((E)). Therefore, we speculated that a reduced reverse transcription efficiency for HBV RNA splicing variants due to the absence of some PSs might lead to the higher ratio of pgRNA splicing variants to total pgRNA than that of HBV DNA.
Discussion
Although it has been proved that serum HBV RNA mainly consist of full-length, splicing variant and 3′ terminal truncated pgRNA [Citation8,Citation26,Citation32], the dynamic changes of different HBV RNA species and their influences on the quantitative detection of serum HBV RNA in CHB patients under NAs treatment have not yet been elucidated. In this study, we investigated the dynamic changes of splicing variants and 3′ terminal truncations of pgRNA in NAs-treated patients at different time-points, and their influences on the quantitative detection of serum HBV RNA in both treatment-naive and NAs-treated patients.
Since serum HBV RNA detected at preC/C-RNA region could avoid the interruption of the main pgRNA splicing variants and 3′ terminal truncations, its level was always significantly higher than that of SF-RNA and XR-RNA regions, suggesting that the detection of serum HBV RNA was affected by the amplified region of HBV genome. Furthermore, high proportions of spliced pgRNA and 3′ terminal truncated pgRNA were observed in treatment-naive patients, irrespective of HBeAg status. In NAs-treated patients, both the ratios of spliced pgRNA to total pgRNA and 3′ terminal-truncated pgRNA to total pgRNA decreased at the early stage of treatment, but increased 2 years post treatment. Therefore, the interference brought by pgRNA splicing variants and 3′ terminal truncations on quantitative detection of serum HBV RNA would be more obvious under long-term NAs treatment. This reminds us that the amplification region should be well-selected. For the decreases in the proportion of spliced pgRNA and 3′ terminal-truncated pgRNA at the early stage of NAs treatment, the possibilities might be that the reverse transcription efficiency of spliced pgRNA was lower than that of unspliced pgRNA, and the degradation of pgRNA mediated by RNase H activity of P protein, especially for the 3′ terminus of pgRNA, could be inhibited by the NAs-mediated inhibition of pgRNA reverse transcription. While for their increases after long-term NAs treatment, the possibility might be that HBV quasispecies with the decreased reverse transcription efficiency were screened out under the pressure of long-term NAs treatment, or the production of pgRNA splicing variants relatively increased after the long-term NAs treatment.
It has been reported that pgRNA splicing variants can also be reverse-transcribed to HBV DNA [Citation33,Citation34]. In this study, we found that the ratio of extracellular pgRNA splicing variants to total pgRNA was higher than that of both intracellular HBV RNA and extracellular HBV DNA, which further supported the above assumption that the reverse transcription efficiency of spliced pgRNA was lower than that of unspliced pgRNA and thus led to less amount of pgRNA splicing variants degraded than that of unspliced pgRNAs during reverse transcription. Consistently, compared to SP1, the higher increase of unspliced pgRNA level at 12 weeks post NAs treatment also indicated that more unspliced pgRNA were not degraded due to the NAs-mediated inhibition of reverse transcription. The mechanism of the lower reverse transcription efficiency of pgRNA splicing variants was not further investigated in our study. We speculated that the trans-packaging mechanism of pgRNA was one of the main reasons for this phenomenon. Further research is needed to understand the specific mechanism. Moreover, the National Standard for HBV RNA detection was first used to assess the accuracy and sensitivity of HBV RNA detection at preC/C-RNA, SF-RNA and XR-RNA regions in this study. Consistent with the results of above cohorts, the level of serum HBV RNA detected at preC/C-RNA region showed a highest accuracy and sensitivity compared to SF-RNA and XR-RNA regions, suggesting that the quantitative detection of serum HBV RNA should avoid the interruption of pgRNA splicing variants and 3′ terminal truncations to improve the accuracy and sensitivity.
HBV RNAs produced by integrated HBV genes could also be an interference in serum HBV RNA detection. Theoretically, the integrated HBV DNA could express the viral–human fusion transcripts driven either by SP1/SP2 or XP promoters. However, due to the lack of CP promoter, the integrated HBV DNA was unable to express precore mRNA or pgRNA. Therefore, the preC/C-RNA had the advantage to reflect transcriptional activity of cccDNA that XR-RNA didn’t, because of the ability to avoid the interference of HBV transcript from integrated HBV DNA. Besides, although good correlations could be observed among preC/C-RNA, SF-RNA and XR-RNA assays, we believed that the highest sensitivity of the preC/C-RNA assay would make it more reliable in both predicting treatment response and guiding drug withdrawal decision. However, more studies are still needed to address above questions.
In conclusion, this study first confirmed that the proportions of pgRNA splicing variants and 3′ terminal truncations to total pgRNA were high enough to affect quantitative detection of serum HBV RNA, and were irrespective of HBeAg status and NAs treatment. Since the ratios of pgRNA splicing variants and 3′ terminal truncations could both relatively increase with the extension of NAs treatment, their interruptions on the quantitative detection of serum HBV RNA would be increased under long-term NAs treatment. To achieve the higher accuracy and sensitivity on the detection of HBV RNA level, the primers and probes should be designed at the 5′ terminal region of HBV genome and outside the mainly spliced sequence of pgRNA, especially for CHB patients under long-term NAs treatment. This study would help to better understand the significance on the pgRNA splicing variants and 3′ terminal truncations, and further guide the clinical detection of serum HBV RNA.
Supplementary_data
Download MS Word (1.3 MB)Disclosure statement
One patent which belongs to Peking University of China is relevant to this work. GY, RC, YL, JZ, ZG, JW and FL are employees of Peking University.
Additional information
Funding
References
- Trépo C, Chan HL, Lok A. Hepatitis B virus infection. Lancet (London, England). 2014 Dec 6;384(9959):2053–2063.
- Revill PA, Chisari FV, Block JM, et al. A global scientific strategy to cure hepatitis B. The Lancet Gastroenterol Hepatol. 2019 Jul;4(7):545–558.
- Lucifora J, Xia Y, Reisinger F, et al. Specific and nonhepatotoxic degradation of nuclear hepatitis B virus cccDNA. Science (New York, NY). 2014 Mar 14;343(6176):1221–1228.
- Wang J, Shen T, Huang X, et al. Serum hepatitis B virus RNA is encapsidated pregenome RNA that may be associated with persistence of viral infection and rebound. J Hepatol. 2016 Oct;65(4):700–710.
- Jansen L, Kootstra NA, van Dort KA, et al. Hepatitis b virus pregenomic RNA Is present in Virions in plasma and is associated with a response to pegylated interferon alfa-2a and nucleos(t)ide analogues. J Infect Dis. 2016 Jan 15;213(2):224–232.
- Bai L, Zhang X, Kozlowski M, et al. Extracellular Hepatitis B virus RNAs are heterogeneous in length and circulate as capsid-antibody complexes in addition to Virions in chronic Hepatitis B patients. J Virol. 2018 Dec 15;92(24):e00798.
- Prakash K, Rydell GE, Larsson SB, et al. High serum levels of pregenomic RNA reflect frequently failing reverse transcription in hepatitis B virus particles. Virol J. 2018 May 15;15(1):86.
- Shen S, Xie Z, Cai D, et al. Biogenesis and molecular characteristics of serum hepatitis B virus RNA. PLoS Pathog. 2020 Oct;16(10):e1008945.
- Stadelmayer B, Diederichs A, Chapus F, et al. Full-length 5'RACE identifies all major HBV transcripts in HBV-infected hepatocytes and patient serum. J Hepatol. 2020 Jul;73(1):40–51.
- Huang H, Wang J, Li W, et al. Serum HBV DNA plus RNA shows superiority in reflecting the activity of intrahepatic cccDNA in treatment-naïve HBV-infected individuals. Journal of Clin Virol: The Off Publ Pan Amen Soc Clin Virol. 2018 Feb-Mar;99-100:71–78.
- Giersch K, Allweiss L, Volz T, et al. Serum HBV pgRNA as a clinical marker for cccDNA activity. J Hepatol. 2017 Feb;66(2):460–462.
- Butler EK, Gersch J, McNamara A, et al. Hepatitis b virus serum DNA andRNA levels in nucleos(t)ide analog-treated or untreated patients during chronic and acute infection. Hepatology. 2018 Dec;68(6):2106–2117.
- Huang Q, Zhou B, Cai D, et al. Rapid turnover of Hepatitis B virus covalently closed circular DNA indicated by monitoring emergence and reversion of signature-mutation in treated chronic Hepatitis B patients. Hepatology. 2021 Jan;73(1):41–52.
- Mak LY, Cloherty G, Wong DK, et al. HBV RNA profiles in chronic hepatitis B patients under different disease phases and anti-viral therapy. Hepatology. 2020 Nov 6;73(6):2167–2179.
- van Bömmel F, Bartens A, Mysickova A, et al. Serum hepatitis B virus RNA levels as an early predictor of hepatitis B envelope antigen seroconversion during treatment with polymerase inhibitors. Hepatology. 2015 Jan;61(1):66–76.
- Luo H, Tan N, Kang Q, et al. Hepatitis B virus pregenomic RNA status can reveal the long-term prognoses of chronic hepatitis B patients treated with nucleos(t)ide analogues. J Viral Hepat. 2020 Mar;27(3):323–328.
- Farag MS, van Campenhout MJH, Pfefferkorn M, et al. Hepatitis b virus RNA as early predictor for response to PEGylated interferon Alfa in HBeAg Negative chronic Hepatitis B. Clin Infect Dis: An Off Publ Infect Dis Soc America. 2020 Jan 8;72(2):202–211.
- Zhang M, Li G, Shang J, et al. Rapidly decreased HBV RNA predicts responses of pegylated interferons in HBeAg-positive patients: a longitudinal cohort study. Hepatol Int. 2020 Mar;14(2):212–224.
- van Campenhout MJH, van Bömmel F, Pfefferkorn M, et al. Serum hepatitis B virus RNA predicts response to peginterferon treatment in HBeAg-positive chronic hepatitis B. J Viral Hepat. 2020 Jun;27(6):610–619.
- Lin N, Ye A, Lin J, et al. Diagnostic value of detection of pregenomic RNA in sera of Hepatitis B virus-infected patients with different clinical outcomes. J Clin Microbiol. 2020 Jan 28;58(2):e01275-19.
- Wang J, Chen X, Wu Y, et al. Serum HBV RNA is a potential predictor of Hepatitis B surface Antigen reversion. Hepatol Commun. 2018 Oct;2(10):1168–1171.
- Fan R, Zhou B, Xu M, et al. Association between negative results from tests for HBV DNA and RNA and durability of response after discontinuation of nucles(t)ide analogue therapy. Clin Gastroenterol Hepatol: The Off Clin Pract J Amen Gastroenterol Assoc. 2020 Mar;18(3):719–727. e7.
- Fan R, Peng J, Xie Q, et al. Combining Hepatitis B virus RNA and Hepatitis B core-related antigen: Guidance for safely stopping nucleos(t)ide analogues in Hepatitis B e antigen-positive patients With chronic Hepatitis B. J Infect Dis. 2020 Jul 23;222(4):611–618.
- Carey I, Gersch J, Wang B, et al. Pregenomic HBV RNA and Hepatitis B core-related antigen predict outcomes in Hepatitis B e antigen-negative chronic Hepatitis B patients suppressed on nucleos(T)ide analogue therapy. Hepatology. 2020 Jul;72(1):42–57.
- Lam AM, Ren S, Espiritu C, et al. Hepatitis b virus capsid assembly modulators, but not nucleoside analogs, inhibit the production of extracellular pregenomic RNA and spliced RNA variants. Antimicrob Agents Chemother. 2017 Aug;61(8):e00680-17.
- Wang J, Sheng Q, Ding Y, et al. HBV RNA virion-like particles produced under nucleos(t)ide analogues treatment are mainly replication-deficient. J Hepatol. 2018 Apr;68(4):847–849;76(1):234–236.
- Guan G, Zou J, Zhang T, et al. A global survey of alternative splicing of HBV transcriptome using long-read sequencing. J Hepatol. 2021 Jul 29;76(1):234–236.
- Liu Y, Liu H, Hu Z, et al. Hepatitis b virus Virions produced under nucleos(t)ide analogue treatment are mainly not infectious because of irreversible DNA chain termination. Hepatology. 2020 Feb;71(2):463–476.
- Cornberg M, Lok AS-F, Terrault NA, et al. Guidance for design and endpoints of clinical trials in chronic hepatitis B - report from the 2019 EASL-AASLD HBV Treatment Endpoints conference‡. J Hepatol. 2020 2020/03/01/;72(3):539–557.
- Patel N, White SJ, Thompson RF, et al. HBV RNA pre-genome encodes specific motifs that mediate interactions with the viral core protein that promote nucleocapsid assembly. Nature Microbiol. 2017 Jun 19;2:17098.
- Qi Z, Li G, Hu H, et al. Recombinant covalently closed circular hepatitis B virus DNA induces prolonged viral persistence in immunocompetent mice. J Virol. 2014 Jul;88(14):8045–8056.
- Zhang P, Liu F, Guo F, et al. Characterization of novel hepadnaviral RNA species accumulated in hepatoma cells treated with viral DNA polymerase inhibitors. Antiviral Res. 2016 Jul;131:40–48.
- Soussan P, Pol J, Garreau F, et al. Expression of defective hepatitis B virus particles derived from singly spliced RNA is related to liver disease. J Infect Dis. 2008 Jul 15;198(2):218–225.
- Günther S, Sommer G, Iwanska A, et al. Heterogeneity and common features of defective hepatitis B virus genomes derived from spliced pregenomic RNA. Virology. 1997 Nov 24;238(2):363–371.
