ABSTRACT
Fosfomycin has gained attention as a combination therapy for methicillin-resistant Staphylococcus aureus infections. Hence, the detection of novel fosfomycin-resistance mechanisms in S. aureus is important. Here, the minimal inhibitory concentrations (MICs) of fosfomycin in CC1 methicillin-resistant S. aureus were determined. The pangenome analysis and comparative genomics were used to analyse CC1 MRSA. The gene function was confirmed by cloning the gene into pTXΔ. A phylogenetic tree was constructed to determine the clustering of the CC1 strains of S. aureus. We identified a novel gene, designated fosY, that confers fosfomycin resistance in S. aureus. The FosY protein is a putative bacillithiol transferase enzyme sharing 65.9–77.5% amino acid identity with FosB and FosD, respectively. The function of fosY in decreasing fosfomycin susceptibility was confirmed by cloning it into pTXΔ. The pTX-fosY transformant exhibited a 16-fold increase in fosfomycin MIC. The bioinformatic analysis showed that fosY is in a novel genomic island designated RIfosY (for “resistance island carrying fosY”) that originated from other species. The global phylogenetic tree of ST1 MRSA displayed this fosY-positive ST1 clone, originating from different regions, in the same clade. The novel resistance gene in the fos family, fosY, and a genomic island, RIfosY, can promote cross-species gene transfer and confer resistance to CC1 MRSA causing the failure of clinical treatment. This emphasises the importance of genetic surveillance of resistance genes among MRSA isolates.
Introduction
Fosfomycin exhibits broad-spectrum activity against both gram-positive and gram-negative bacteria by interfering with the first committed step of peptidoglycan synthesis [Citation1]. The lack of new antibiotics and the development of resistance in bacteria have revived the interest of clinicians in older drugs such as fosfomycin. In recent years, fosfomycin in combination with other antibiotics has been used to treat infections by methicillin-resistant Staphylococcus aureus (MRSA), an important pathogen [Citation2].
Mutations in the genes encoding fosfomycin transporters such as glpT and uphT or the target enzyme MurA and the acquisition of the fosfomycin-modifying enzymes are two major mechanisms of fosfomycin resistance [Citation3]. The main enzymes that inactivate fosfomycin include three types of metalloenzymes with different substrates and two kinases [Citation4]. In low-GC gram-positive bacteria, fosfomycin-modifying enzymes catalyse the reaction between bacillithiol and fosfomycin [Citation5]. To date, two genes fosB and fosD, encoding bacillithiol-S-transferases, have been identified in staphylococci [Citation6,Citation7].
FosB, first discovered in the plasmids of S. epidermidis in 1990 [Citation6], was later found in S. aureus. FosD, located in the plasmid of S. aureus [Citation7], shows 78.9% nucleotide and 74.1% amino acid sequence identity with the fosfomycin-resistance determinant fosB carried by MRSA strains.
Multilocus sequence type 1 (ST1) MRSA isolates, also known as pulsed-field type USA400, were among the most prominent community-associated (CA) strains in the USA [Citation8]. Recently, ST1 has been sporadically reported in other regions [Citation9], including China [Citation10]. As fosfomycin remains effective against community-onset MRSA [Citation11], it has become an important alternative treatment for CA-MRSA infections. Therefore, the discovery of new resistance mechanisms in S. aureus is important.
In this study, we identified a novel gene, fosY, associated with fosfomycin resistance in genetic mobile elements in CC1 MRSA and characterised the gene function and surrounding structure of fosY. We also constructed a phylogenetic tree to clarify the characteristics of fosY-positive MRSA clone in China.
Materials and methods
Bacterial strains
Staphylococcus aureus isolates and clinical data were collected from 22 tertiary hospitals in 18 provinces and municipalities in China in a multicentre prospective study, and 471 MRSA isolates were previously subjected to next-generation sequencing, de novo assembly and MLST typing [Citation12].
Antimicrobial susceptibility testing
The minimal inhibitory concentrations (MICs) of fosfomycin were determined using the agar dilution method according to CLSI guidelines using Mueller–Hinton agar plates containing 25 mg/L glucose-6-phosphate. Susceptibility to other antibiotics was tested using the broth dilution method. Fosfomycin activity was interpreted based on the European Committee on Antimicrobial Susceptibility Testing [Citation13] whereas the activities of other antibiotics were interpreted in accordance with the Clinical and Laboratory Standards Institute [Citation14]. ATCC 29213 (S. aureus) was used as a quality control strain.
Whole genome sequencing, comparative genomics analysis and phylogenetic analysis
The fosfomycin resistance gene detection was performed using web-based Center for Genomic Epidemiology (http://www.genomicepidemiology.org/). The mutations of chromosomal genes were confirmed via allele genes table in cgMLST generated in SeqSphere+ software version 4.1.9 (Ridom GmbH). The comparative genomics analysis of ST1 isolates in this study was performed using panaroo [Citation15] to analyse the pangenome and detect new resistance determinant.
A multisequence alignment of the amino acid sequences of new protein and other fosfomycin-modifying enzymes which have been reported previously (Table S1) was conducted using ClustalX and the phylogenetic tree was constructed via the MEGA X [Citation16] using the maximum likelihood method. The alignment of amino acid sequences was generated using ESPript 3.0 [Citation17].
One of the ST1 strains N12HSA28 was selected randomly for further analysis. The Nanopore sequencing using a MinION sequencer (Oxford Nanopore Technologies, Oxford, UK) was performed. Hybrid assembly was performed using Unicycler with Illumina and Nanopore reads. The assembled contigs were annotated using the RAST server. Further sequence analysis was performed using Mauve and sequencing alignment software BLASTn (https://blast.ncbi.nlm.nih.gov/BlastAlign.cgi), a comparison map was generated using Easyfig2.1.
The maximum likelihood tree of ST1 S. aureus genomes included isolates in this study and those from public Genbank databases was constructed from a core genome alignment using Panaroo [Citation15] with the tree generated by IQ-TREE 2 using the bootstrap method for test with 1000 replicates [Citation18]. The phylogenetic tree was modified via iTOL (https://itol.embl.de/login.cgi). The information of public genomes was in Table S2.
Cloning experiments
The fosY gene from N12HSA28 was amplified using PCR, digested with BamHI and MluI, and ligated into BamHI- and MluI-digested pTXΔ, a plasmid from S. aureus [Citation19]. The recombinant plasmid pTX-fosY was transformed into S. aureus RN4220 via electroporation and successful transformants were selected with tetracycline (12.5 mg/L). The plasmid pTX16 was also introduced into the RN4220 strain as a control.
Nucleotide sequence accession number
The fosY DNA sequence has been deposited in the Nucleotide database in NCBI under accession number MN961674.1. The genome of N12HSA28 was submitted to NCBI under accession number CP091523. The sequence of the resistance island carrying fosY (RIfosY) has been deposited in the genbank database under accession number OM925572.
Results
Identification of fosY in CC1 MRSA
A multicentre prospective study in China identified 11 MRSA strains belonging to STl. The fosfomycin MIC of these strains ranged from 0.5 to 16 mg/L, fosB and fosD were not present, and chromosomal mutations were not detected (). We compared the accessory genome of these strains and found an ORF of 423 bp, annotated as putative metallothiol transferase, in three strains whose fosfomycin MIC was ≥8 mg/L. The 423 bp ORF, encoding a protein of 140 amino acids, was designated as fosY. This novel gene exhibited 75.8% and 80.4% sequence identity with fosB in S. epidermidis [Citation6] and fosD gene in S. aureus [Citation7], respectively.
Table 1. Fosfomycin susceptibility testing of CC1-MRSA in this study.
Further screening of all sequenced isolates revealed another ST5537 strain, which was a single-locus variant of ST1 and classified as complex clone 1 (CC1), that also carried this gene. presents basic information about the 12 strains. The isolates were scattered across six provinces. Most strains were collected from the secretion of SSTI patients or sputum of patients with respiratory infections.
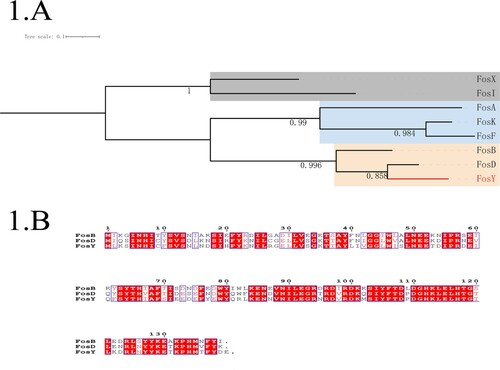
To further clarify the genetic relationship of the novel protein FosY, a phylogenetic tree was constructed using the multiple amino acid sequences of other fosfomycin-modifying enzymes ((A)). The phylogenetic tree showed that these fosfomycin-modifying enzymes could be differentiated into three groups. FosY and two other fosfomycin-modifying enzymes, that were reported in gram-positive bacteria, were in the same clade. The amino acid sequence of FosY shares 65.9% and 77.5% identity with that of FosB [Citation6] and FosD [Citation7], respectively. A detailed comparison of the amino acid sequences of FosB, FosD, and FosY is shown in (B).
Figure 1. Phylogeny of known fosfomycin-modifying enzymes and amino acid sequence alignment of fosfomycin-modifying enzymes in Staphylococcus aureus. (A) Phylogenetic tree obtained for all the identified fos enzymes. Protein sequences were aligned using ClustalW and the tree was generated using MEGA X. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The clade containing fosY is highlighted. (B) Amino acid sequence alignment of fosfomycin-modifying enzymes in Staphylococcus aureus. Sequence alignment was generated by ClustalW and ESPript 3.0. The same amino acid in the three enzymes is highlighted in red.
Function of fosY
Fosfomycin susceptibility testing showed that the MIC of the ST1 strain, N12HSA28, was 16 mg/L. Although this strain remained susceptible to fosfomycin, the MIC was 4–32-fold higher than that of other fosY-negative ST1 isolates (). To further confirm the functionality of fosY and its contribution to fosfomycin resistance, fosY was cloned into pTXΔ and expressed in S. aureus RN4220.
The MIC of fosfomycin for the RN4220 pTX-fosY transformant was 16 mg/L, whereas the MIC for the pTX16 transformant was 1 mg/L. The MICs of other antibiotics were essentially invariant (). Therefore, the RN4220 pTX-fosY transformant exhibited a 16-fold increase in the MIC of fosfomycin, compared with the RN4220 carrying the empty vector, indicating that fosY can decrease fosfomycin susceptibility in S. aureus.
Table 2. MICs for N28HSA12, cloning strains and recipients.
Genetic structure surrounding fosY
The whole genome of N12HSA28 was sequenced to characterise the genetic environment surrounding fosY. The ST1 MRSA strain N12HSA28 contained a 2,833,771-bp circular chromosome in which fosY was located. Further analysis showed that fosY is located on a 27.6-Kb genomic island inserted into the chromosomal radC gene with a 24-bp invert repeat (TCGTTGCACATATAGGTATGAAAA) and a 5-bp direct repeat (AATCA) at both termini (). Consequently, we designated this element a “resistance island carrying fosY” (RIfosY). Further genome mining revealed that fosY, arsM, and a functional operon containing three genes, asrR, asrB, and asrC, colocalised in RIfosY. The genomic structures of the other three fosY-positive strains were compared with the reference RIfos using Mauve, showing that the genetic environments surrounding fosY in all these strains in our study were identical to RIfosY.
Figure 2. Schematic presentation of fosY-carrying genomic island (RIfosY) in comparison with other sequences. Regions of 100% nucleotide sequence identity are marked in dark grey, while dark grey represents region of 85% nucleotide sequence identity. Arrows indicate the positions and orientations of the genes. The genetic environments of fosY in other strains were identical to RIfosY (99.98% nucleotide identity).
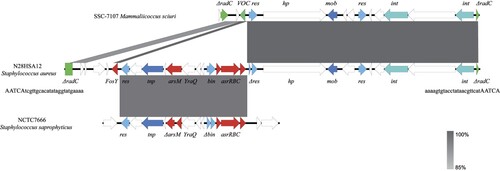
A comparative genomic analysis was warranted to further our understanding of the origin of RIfosY. The 15.7-Kb flanking sequences carrying recombinase, integrase, and functionally unknown genes had the highest similarity to the genomic island of Mammaliicoccus sciuri strain SSC-7107 collected from a human sample in Pakistan (accession number CP071138), which was inserted into the homologous radC gene. The other 9-Kb arsenic-resistance fragment, flanked by recombinases, was similar to the chromosome of Staphylococcus saprophyticus strain NCTC7666 isolated from milk ice cream (accession number LR134089). RIfosY is presumably generated via multiple interspecies recombination events. In addition, fosY was homologous to the vicinal oxygen chelate (VOC) family gene in Mammaliicoccus sciuri strain SSC-7107 (coverage 77%, identity 100%).
Global phylogenetic analysis of fosY-positive CC1 S. aureus
To explore the characteristics of the fosY-positive ST1 clone in China, a phylogenetic tree was constructed that included 12 CC1 strains (11 ST1 and 1 ST5537), identified in this study, and 206 ST1 strains identified worldwide. Global CC1 clone were classified into some clades, which presented different features. The regions and hosts of the CC1 strains were diverse and were included in the outer circle of the tree. In addition, mecA has been identified and labelled in .
Figure 3. A core-genome global phylogenetic tree of ST1 Staphylococcus aureus. The tree is rooted at the midpoint. The labels of fosY-positive strains are marked in red. The branches of the strains isolated in this study are marked in green. Major clades are identified based on the coloured backgrounds of the branches. The inner coloured ring indicates the collection region of all the genomes. The next ring indicates mecA and the hosts of the isolates are shown in the outermost ring.
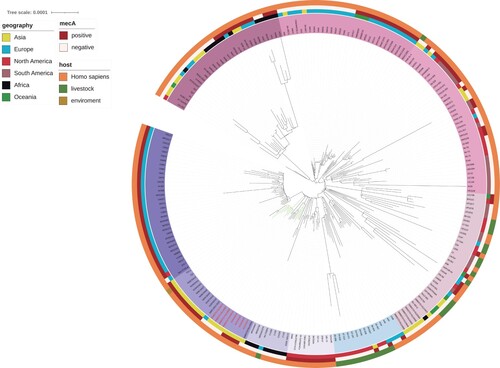
The global phylogenetic tree showed strains isolated in this study clustered into two subgroups (). One of the subgroups included four fosY positive MSSA isolates from Thailand. The intact RIfosY structure carring fosY were found in the genome of these four Thailand strains with 99.98% nucleotide identity. All fosY-positive strains were adjacent to each other in the phylogenetic tree. In addition, all strains collected in this study are included in the Southeast Asia clade, which is close to the European and African clades.
Discussion
Recently, fosfomycin has gained attention for the treatment of MRSA infection. Multicentre clinical and experimental studies have demonstrated the clinical efficacy and safety of fosfomycin, in combination with other antibiotics such as imipenem and daptomycin [Citation2,Citation20,Citation21]. Previous studies have focused on the mechanism of fosfomycin resistance in S. aureus and have suggested that mutations in chromosomes might be the primary underlying mechanism, as genes that encode the fosfomycin-modifying enzymes are rare in fosfomycin-resistant isolates [Citation22,Citation23]. The discovery of the novel fosfomycin-resistance-related gene, fosY, in this study, indicates the importance of fosfomycin-modifying enzyme genes in fosfomycin resistance in S. aureus.
Since the identification of fosfomycin-resistance genes in the plasmid of S. epidermidis almost 30 years ago [Citation6], several fosfomycin-resistance genes in staphylococci have been reported. Wang et al. reported some fosB genes, designated fosB3 to fosB6, in the plasmids of S. aureus and Enterococcus faecium, which differed from the first fosB gene by only 1% to 2% [Citation24,Citation25]. In 2008, the new fosD gene was first reported in pTZ2162 from S. aureus in Japan [Citation7]. These genes encode modifying enzymes that inactivate fosfomycin [Citation3]. In Bacillus species, the homologous FosB enzyme, which belongs to the VOC superfamily [Citation26], catalyses the reaction between bacillithiol and fosfomycin [Citation5], As bacillithiol is also described as a major thiol in staphylococci [Citation27], these acquired fosfomycin-modifying enzymes in S. aureus are further supposed as bacillithiol transferase. In this study, we confirmed the sequence of another fosfomycin-modifying enzyme gene, fosY, in S. aureus. We further showed that fosY diverges from the previously described fosB and fosD genes; meanwhile, the function of fosY gene that decreases fosfomycin susceptibility was confirmed in this study. Using phylogenetic analysis, we also showed that FosY clusters with FosB and FosD in the phylogenetic tree, indicating that they share a similar ancestry and we, consequently, identified FosY as a bacillithiol transferase.
Mobile genetic elements can disseminate resistance genes and promote intracellular DNA mobility [Citation28]. A genomic island is a distinct region of a bacterial chromosome that has been acquired via horizontal transfer [Citation28]. It has been reported that transposons, such as Tn554 and Tn559, integrate in the chromosomal radC gene in S. aureus [Citation29]. In this study, we describe the insertion of a novel 27.6-kb genomic island, RIfosY, carrying the fosY gene. The structure of the island and the fosY gene were found to be similar to that in other species, such as M. sciuri and S. saprophyticus associated with livestock [Citation30–32], implying that RIfosY may originate and recombine in the microbiome in livestock and then integrate into CC1 S. aureus. Therefore, RIfosY facilitates cross-species gene transfer, and the transfer of RIfosY to other clones may also be possible. Moreover, a resolvase located upstream of fosY may confer the capacity for recombination of this gene. In addition to fosfomycin resistance, the S-adenosylmethionine methyltransferase ArsM and the As(III) efflux system ArsB, which is employed by several bacteria for arsenic tolerance [Citation33,Citation34], are also found in RIfosY. Arsenic has been found in manure, soil, and effluents of livestock farms [Citation35]. Therefore, the acquisition of RIfosY may enhance the competitiveness of S. aureus to survive in livestock.
FosY was identified in the USA400/ST1/SCCmec IV lineage, the most prominent community-associated clone in the USA 20 years ago [Citation8]. To date, ST1 remains one of the most important S. aureus lineages in the clinical setting because of its high virulence. Meanwhile, the ST1 clone has been reported to occur sporadically worldwide and has emerged in the clinical setting and livestock in Asia [Citation9,Citation36]. In the global CC1 phylogenetic tree, different clades exhibit different features. Some MRSA strains are clustered in the same clades with some MRSA and MSSA strains distributed crossly. Isolates in some clades show regional aggregation, whereas those in other clades disseminate between continents. Most MSSA strains are located in the intercontinental clades. This indicates that the important lineage ST1 may have various evolutions.
The ST1 strains in our study belong to the same clade, which were mostly collected in Southeast Asia. The phylogenetic tree indicates that this clade has the most recent common ancestor with the European MRSA clade and African MSSA clade. Eight fosY-positive strains with similar genetic backgrounds were obtained from Sichuan and Hainan Provinces in Southern China and Thailand, implying that fosY-positive strains may be transmitted in these regions. The structures of the fosY resistance island in eight strains were basically identical, further proving the dissemination of this fosY-positive MRSA clone in Southeast Asia. The fosY-positive strains in Thailand were all MSSA, indicating that the acquisition of fosY-carrying genomic islands may occur earlier than the mecA gene. It is worth noting that the novel fosY gene may increase the resistance to fosfomycin in both MSSA and MRSA and disseminate between regions.
Furthermore, some studies have reported ST1 in commercial pig farms, abattoirs, and in companion animals globally [Citation37,Citation38]. In addition, several isolates collected from livestock were included in the evolutionary tree. These livestock-associated strains were almost all MSSA and distributed worldwide and were interspersed with human strains in the same clades, although some studies have reported that the livestock-associated ST1 strains emerged in China [Citation10,Citation39,Citation40]. However, the lack of complete genome information is a limitation to further analyse the relationship between the livestock-associated ST1 in China and the clinical ST1 in our study. All fosY-positive strains in our study contained RIfosY, which is associated with a higher fosfomycin and arsenic resistance. The possibility of these strains infecting livestock must be monitored.
In conclusion, we identified a novel fosfomycin-modifying enzyme-encoding gene, fosY located in RIfosY and originating from other species, in CC1 S. aureus, which provides a transferable source of acquired resistance among isolates. Our findings show that a new member of the Fos family confers a decreased susceptibility to fosfomycin in S. aureus. As fosfomycin is currently considered a treatment of choice for staphylococcal infections, strategies should be implemented to monitor the spread of these resistant factors.
Supplemental Material
Download MS Excel (19.8 KB)Supplemental Material
Download MS Word (27 KB)Acknowledgements
The authors thank Editage (www.editage.cn) for their linguistic assistance during the preparation of this manuscript.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Additional information
Funding
References
- Kahan FM, Kahan JS, Cassidy PJ, et al. The mechanism of action of fosfomycin (phosphonomycin). Ann N Y Acad Sci. 1974;235:364–386.
- Pujol M, Miro JM, Shaw E, et al. Daptomycin plus fosfomycin versus daptomycin alone for methicillin-resistant Staphylococcus aureus bacteremia and endocarditis. A randomized clinical trial. Clin Infect Dis. 2020;72:1517–1525.
- Falagas ME, Vouloumanou EK, Samonis G, et al. Fosfomycin. Clin Microbiol Rev. 2016;29:321–347.
- Silver LL. Fosfomycin: mechanism and resistance. Cold Spring Harbor Perspect Med. 2017;7:a025262.
- Chandrangsu P, Loi VV, Antelmann H, et al. The role of bacillithiol in gram-positive firmicutes. Antioxid Redox Signaling. 2018;28:445–462.
- Etienne J, Gerbaud G, Fleurette J, et al. Characterization of staphylococcal plasmids hybridizing with the fosfomycin resistance gene fosB. FEMS Microbiol Lett. 1991;68:119–122.
- Nakaminami H, Noguchi N, Nishijima S, et al. Characterization of the pTZ2162 encoding multidrug efflux gene qacB from Staphylococcus aureus. Plasmid. 2008;60:108–117.
- DeLeo FR, Otto M, Kreiswirth BN, et al. Community-associated meticillin-resistant Staphylococcus aureus. Lancet (London, England). 2010;375:1557–1568.
- Manara S, Pasolli E, Dolce D, et al. Whole-genome epidemiology, characterisation, and phylogenetic reconstruction of Staphylococcus aureus strains in a paediatric hospital. Genome Med. 2018;10:82.
- Yang X, Zhang J, Yu S, et al. Prevalence of Staphylococcus aureus and methicillin-resistant Staphylococcus aureus in retail ready-to-eat foods in China. Front Microbiol. 2016;7:816.
- Wu D, Chen Y, Sun L, et al. Prevalence of fosfomycin resistance in methicillin-resistant Staphylococcus aureus isolated from patients in a university hospital in China from 2013 to 2015. Jpn J Infect Dis. 2018;71:312–314.
- Chen Y, Sun L, Ba X, et al. Epidemiology, evolution and cryptic susceptibility of methicillin-resistant Staphylococcus aureus in China: a whole-genome-based survey. Clin Microbiol Infect. 2021;28:85–92.
- EUCAST. The European Committee on Antimicrobial Susceptibility Testing. Breakpoint tables for interpretation of MICs and zone diameters. Version 90. 2019. Available from: http://www.eucast.org
- CLSI. Performance standards for antimicrobial susceptibility testing. 29th ed. CLSI supplement M100. Wayne (PA): Clinical and Laboratory Standards Institute. 2019.
- Tonkin-Hill G, MacAlasdair N, Ruis C, et al. Producing polished prokaryotic pangenomes with the Panaroo pipeline. Genome Biol. 2020;21:180.
- Kumar S, Stecher G, Li M, et al. MEGA x: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35:1547–1549.
- Robert X, Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014;42:W320–W324.
- Minh BQ, Schmidt HA, Chernomor O, et al. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 2020;37:1530–1534.
- Wang R, Braughton KR, Kretschmer D, et al. Identification of novel cytolytic peptides as key virulence determinants for community-associated MRSA. Nat Med. 2007;13:1510–1514.
- del Río A, Gasch O, Moreno A, et al. Efficacy and safety of fosfomycin plus imipenem as rescue therapy for complicated bacteremia and endocarditis due to methicillin-resistant Staphylococcus aureus: a multicenter clinical trial. Clin Infect Dis. 2014;59:1105–1112.
- del Río A, García-de-la-Mària C, Entenza JM, et al. Fosfomycin plus β-lactams as synergistic bactericidal combinations for experimental endocarditis due to methicillin-resistant and glycopeptide-intermediate Staphylococcus aureus. Antimicrob Agents Chemother. 2016;60:478–486.
- Fu Z, Ma Y, Chen C, et al. Prevalence of fosfomycin resistance and mutations in murA, glpT, and uhpT in methicillin-resistant Staphylococcus aureus strains isolated from blood and cerebrospinal fluid samples. Front Microbiol. 2015;6:1544.
- Xu W, Chen T, Wang H, et al. Molecular mechanisms and epidemiology of fosfomycin resistance in Staphylococcus aureus isolated from patients at a teaching hospital in China. Front Microbiol. 2020;11:1290.
- Fu Z, Liu Y, Chen C, et al. Characterization of fosfomycin resistance gene, fosB, in methicillin-resistant Staphylococcus aureus isolates. PLoS One. 2016;11, e0154829.
- Xu X, Chen C, Lin D, et al. The fosfomycin resistance gene fosB3 is located on a transferable, extrachromosomal circular intermediate in clinical Enterococcus faecium isolates. PLoS One. 2013;8, e78106.
- Thompson MK, Keithly ME, Harp J, et al. Structural and chemical aspects of resistance to the antibiotic fosfomycin conferred by FosB from Bacillus cereus. Biochemistry. 2013;52:7350–7362.
- Perera VR, Newton GL, Pogliano K. Bacillithiol: a key protective thiol in Staphylococcus aureus. Expert Rev Anti-Infect Ther. 2015;13:1089–1107.
- Partridge SR, Kwong SM, Firth N, et al. Mobile genetic elements associated with antimicrobial resistance. Clin Microbiol Rev. 2018;31:e00088–17.
- Kadlec K, Schwarz S. Identification of the novel dfrK-carrying transposon Tn559 in a porcine methicillin-susceptible Staphylococcus aureus ST398 strain. Antimicrob Agents Chemother. 2010;54:3475–3477.
- Schauer B, Szostak MP, Ehricht R, et al. Diversity of methicillin-resistant coagulase-negative Staphylococcus spp. and methicillin-resistant Mammaliicoccus spp. isolated from ruminants and new world camelids. Vet Microbiol. 2021;254: Article 109005.
- Karki AB, Neyaz L, Fakhr MK. Comparative genomics of plasmid-bearing Staphylococcus aureus strains isolated from various retail meats. Front Microbiol. 2020;11: Article 574923.
- Liu BH, Lei CW, Zhang AY, et al. Colocation of the multiresistance gene cfr and the fosfomycin resistance gene fosD on a novel plasmid in Staphylococcus arlettae from a chicken farm. Antimicrob Agents Chemother. 2017;61:e01388–17.
- Garbinski LD, Rosen BP, Chen J. Pathways of arsenic uptake and efflux. Environ Int. 2019;126:585–597.
- Yang HC, Rosen BP. New mechanisms of bacterial arsenic resistance. Biomed J. 2016;39:5–13.
- Ji X, Shen Q, Liu F, et al. Antibiotic resistance gene abundances associated with antibiotics and heavy metals in animal manures and agricultural soils adjacent to feedlots in Shanghai, China. J Hazard Mater. 2012;235-236:178–185.
- Witte W. Community-acquired methicillin-resistant Staphylococcus aureus: what do we need to know? Clin Microbiol Infect. 2009;15(Suppl 7):17–25.
- Taniguchi Y, Koide S, Maeyama Y, et al. Predominance of methicillin-resistant Staphylococcus aureus SCCmec type II-CC5 and SCCmec type IV-CC1/CC8 among companion animal clinical isolates in Japan: findings from phylogenetic comparison with human clinical isolates. J Glob Antimicrob Resist. 2020;20:253–259.
- Rodríguez-López P, Filipello V, Di Ciccio PA, et al. Assessment of the antibiotic resistance profile, genetic heterogeneity and biofilm production of methicillin-resistant Staphylococcus aureus (MRSA) isolated from the Italian swine production chain. Foods (Basel, Switzerland). 2020;9:1141.
- Zhang P, Liu X, Zhang J, et al. Prevalence and characterization of Staphylococcus aureus and methicillin-resistant Staphylococcus aureus isolated from retail yak butter in Tibet, China. J Dairy Sci. 2021;104:9596–9606.
- Dan M, Yehui W, Qingling M, et al. Antimicrobial resistance, virulence gene profile and molecular typing of Staphylococcus aureus isolates from dairy cows in Xinjiang Province, northwest China. J Glob Antimicrob Resist. 2019;16:98–104.
