ABSTRACT
A previously unknown Nocardia species was isolated from the lung tissue and bronchoalveolar lavage fluid (BALF) of a 58-year-old woman with bronchiectasis and recurrent pneumonia. This Nocardia (GZ2020T), which grew readily in Columbia blood agar and could induce pneumonia in a mouse model, represents a novel Nocardia species, and its closest known relatives are Nocardia anaemiae NBRC 100462T, Nocardia pseudovaccinii NBRC 100343T and Nocardia vinacea NBRC 16497T. However, unlike all previously known species, GZ2020T is the first genus of Nocardia spp. that is not susceptible to multiple drugs but does show susceptibility to linezolid and moxifloxacin, and thus, GZ2020T potentially represents a substantial health threat to patients with bronchiectasis and immunocompromised individuals. Although the original pathogen source and method of spreading remain uncertain, a mode of transmission from the environment to humans could exist. Vigilance with respect to its spread in the population and the transmission of antibiotic resistance genes in the environment should be maintained.
Introduction
Nocardia spp. are filamentous, aerobic, gram-positive, weakly acid-fast, bacilliform, branched bacteria that were first isolated by Edmond Nocard in 1888 from samples of a bovine farcy case [Citation1]. Over the past several hundred years, Nocardia infections have been continuously reported worldwide [Citation2]. Compared with other pathogens, these bacteria are common pathogens in immunosuppressed patients and can sometimes cause high mortality [Citation3,Citation4]. However, in recent years, an increasing number of reports have indicated that patients with chronic lung diseases (CLDs), such as chronic obstructive pulmonary disease (COPD) and bronchiectasis, can be affected [Citation5,Citation6]. Due to the increasing number of people at risk and the development of molecular diagnostic methods, a gradual increase in the incidence of Nocardia infection has been observed [Citation5,Citation7]. More than 100 species have been identified to date; many of them (over 30 strains) are thought to be associated with human diseases, and the majority of strains exhibit susceptibility to first-line antibiotic treatment [Citation8]. Here, we report a confirmed case of a novel, highly drug-resistant community-acquired Nocardia infection as well as its diagnosis and treatment processes.
Case report
In March 2019, a 58-year-old woman presented with fever, cough, and expectoration. She was diagnosed with pneumonia with Nocardia infection (Nocardia spp.; without antimicrobial susceptibility testing (AST)) at a local hospital ( and A). Subsequently, trimethoprim-sulfamethoxazole (SMZ/TMP 400/80 mg; 0.96 g, po, q8 h) was administered empirically for six months and discontinued upon symptom improvement and partial absorption of lung lesions. Unfortunately, the above mentioned symptoms reoccurred and deteriorated in April 2020. A computed tomography (CT) scan of the chest revealed old expanded lesions, new-onset solid foci in the left lung, and bronchiectasis in both lungs (B). A Nocardia spp. was identified as the primary pathogen in BALF culture, and SMZ/TMP (0.96 g, po, q8 h) was empirically administered again. The patient’s symptoms showed slight improvements after two months of treatment: the fever resolved, but the cough and expectoration were not completely eliminated. She was then discharged with continued outpatient antibiotic therapy. However, 4 days after discharge (June 20, 2020), the previous symptoms recurred again. A chest CT scan indicated that the lesions were similar to those imaged previously, and Nocardia spp. growth was observed in a sputum culture. Subsequently, the patient received combination therapy with meropenem (0.5 g, iv, q8 h) plus amikacin (0.4 g, iv, qd) from June 23 to July 1 and minocycline (100 mg, po, q12 h) plus SMZ/TMP (0.96 g, po, q8 h) from July 2–7, but the symptoms were not completely resolved. On July 7, 2020, she was transferred to our hospital for further treatment ().
Figure 1. Timeline of the patient’s clinical course. Clinical course of the patient’s symptoms and treatment according to the day of illness and day of hospitalization from March 15, 2019, to August 24, 2021. BALF, bronchoalveolar lavage fluid; mNGS, metagenomic next-generation sequencing; AST, antimicrobial susceptibility testing. a: Detection of viruses, fungi, tuberculosis bacteria and other bacteria. Additional details are provided in Supplementary Table 6. b: The patient underwent CT examinations on March 20, 2019; September 20, 2019; May 27, 2020; July 8, 2020; July 21, 2020; August 17, 2020; December 21, 2020; and August 24, 2021. Each CT examination was compared with the previous examination to assess whether the lesions had improved. The CT examinations performed on September 20, 2019, and July 21, 2020, are not shown in this article, and the others can be found in . c: Data are not shown. d: AST results showed that GZ2020T is susceptible to linezolid and possibly to moxifloxacin.
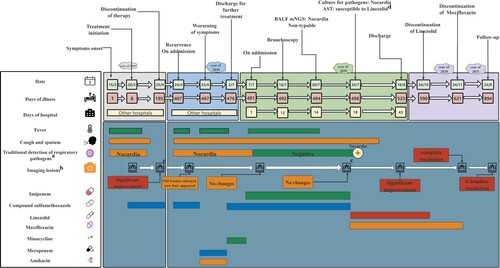
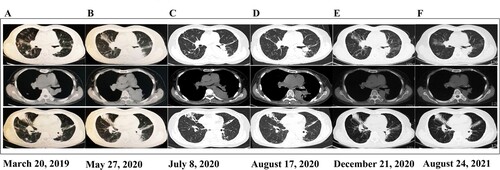
Figure 2. Imaging changes over time The upper section is the lung window, the middle section is the mediastinal window, and the lower section shows bronchiectasis lesions. A (March 20, 2019) shows the first chest CT obtained during the infection. Lesions were mainly observed in the upper lobe of the right lung, and bronchiectasis manifestation was observed, whereas in the lower lobe of the left lung, few significant lesions were seen. Subsequently, the patient was treated with trimethoprim-sulfamethoxazole (SMZ/TMP 400/80 mg; 0.96 g, po, q8 h) for 6 months. Her symptoms improved, and discontinuation of the medication was allowed. B (May 27, 2020) shows the first CT with recurrence of symptoms 1 year later—old lesions persisted in the right lung, and new-onset solid foci were observed in the left lung. She was then given SMZ/TMP (0.96 g, po, q8 h), meropenem (0.5 g, iv, q8 h) plus amikacin (0.4 g, iv, qd) and minocycline (100 mg, po, q12 h) plus SMZ/TMP (0.96 g, po, q8 h), but her symptoms were not completely resolved. C (July 8, 2020) shows the first chest CT scan obtained in our hospital, which showed no change compared with the last image. D (August 17, 2020) shows a chest CT scan after 24 days of linezolid (0.6 g, iv, q12 h) plus moxifloxacin (0.4 g, po, qd) treatment, which showed significant lesion absorption compared with the last image. E (December 21, 2020) shows a follow-up chest CT scan obtained after 4 months of linezolid (0.6 g, po, q12 h) plus moxifloxacin (0.4 g, po, qd) treatment and another 1 month of moxifloxacin monotherapy (0.4 g, qd, po), which resulted in complete absorption of the lesion. F (August 24, 2021) shows a follow-up chest CT scan obtained after drug withdrawal for more than 8 months, which showed no change compared with the last image. A, B, C, D, E and F indicate that precise antibiotic strategies were effective and necessary for treating the drug-resistant pathogen infection, and no exacerbation of bronchiectasis can be seen in these CT images.
She had a history of bile reflux gastritis (in April 2019) but had been cured. She had no history of other underlying diseases, smoking or allergies. Physical examination revealed the following parameters: temperature, 39.5 °C; blood pressure, 95/56 mmHg; pulse, 132/min; respiratory rate, 20/min; and oxygen saturation, 98% while breathing ambient air. Laboratory tests (Supplementary Table 5) revealed a high white blood cell count of 17000 (normal range: 4000–10000) cells per cubic millimeter and a high neutrophil ratio of 85.9% (normal range: 40%−70%). The serum level of procalcitonin (PCT) was elevated at 0.2 ng/mL, and the erythrocyte sedimentation rate (ESR) was increased to 64 mm/h. However, pathogenetic examinations for viruses, fungi, Mycobacterium tuberculosis, and other bacteria (Supplementary Table 6) did not indicate the presence of any pathogen in sputum and blood specimens. Chest CT scans revealed bronchiectasis in both lungs and a large area of consolidation in the left lower lung (C). Based on available evidence, the possibility of infection was high. Given the patient’s previous history of Nocardia infection and antibiotic treatment, imipenem and cilastatin sodium (1 g, iv, q6 h) plus SMZ/TMP (1.44 g, po, q8 h) were administered.
Due to recurrent fever over the disease course, several malignancies and autoimmune diseases were ruled out before arriving at a definitive diagnosis. The patients tested negative for all autoimmune antibodies, but the levels of lung cancer markers were high (NSE: 21.48 ng/mL, CA125: 37.23 U/mL). Bronchoscopy was performed on July 18, which revealed bronchiectasis manifestations, and BALF and lung tissue were collected for pathogen detection. Subsequently, a metagenomic next-generation sequencing (mNGS) of BALF on July 20 showed Nocardia as the only pathogen (total reads: 50876 with Nocardia genus (49850 reads) accounting for the majority), but the species was unknown (Supplementary Table 7). Furthermore, the lung tissue and BALF culture results obtained on July 24 suggested the presence of Nocardia spp. (Supplementary Table 6). The histopathologic analysis results indicated inflammatory lung lesions. However, despite treatment from the time of admission (July 7) to July 23, the symptoms persisted. A repeat chest CT scan showed no obvious change in the lung lesion. Finally, the AST results on July 24 indicated that the pathogen was definitely susceptible to linezolid and possibly susceptible to moxifloxacin ( and Supplementary and ); subsequently, a combination of linezolid (0.6 g, iv, q12 h) plus moxifloxacin (0.4 g, po, qd) was administered. During 24 days of treatment, the patient’s symptoms showed gradual improvement, and a repeat chest CT scan revealed that the lesions were significantly absorbed on August 17 (D). The patient was in satisfactory condition and discharged (August 18, 2020). As an anti-infection strategy, linezolid (0.6 g, po, q12 h) plus moxifloxacin (0.4 g, po, qd) was administered for another 3 months, moxifloxacin monotherapy (0.4 g, qd, po) was administered for more than 1 month, and the patient then opted to discontinue treatment for financial reasons. A follow-up chest CT scan (December 21, 2020, E) revealed that the lesions had completely resolved, and this improvement in the clinical and microbial conditions has persisted steadily to date (August 24, 2021, F), without any recurrence of pneumonia or exacerbation of bronchiectasis.
Figure 3. Drug resistance profile of GZ2020T. a: The data were obtained from reference 8 (Clinical and Laboratory Standards Institute (CLSI), M24-A2, ISBN 1-56238-746-4). b: Breakpoints defined by the CLSI for Nocardia spp. were applied. c: The results for tobramycin, clarithromycin and moxifloxacin were interpreted according to disc diffusion tests with the CLSI breakpoints for Staphylococcus spp. as a reference, which showed that GZ2020T was resistant to tobramycin and clarithromycin and possibly susceptible to moxifloxacin. d: Although the result should be interpreted as “intermediate” according to the minimal inhibitory concentration (MIC) method, the poor clinical efficiency in the course of treatment and the markedly unsatisfactory outcomes from the disc diffusion tests instilled uncertainty. K-B method: Kirby-Bauer method; ND: no data.
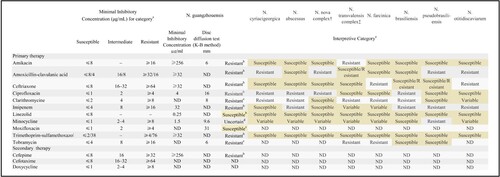
Figure 4. Phylogenetic tree analyses of GZ2020T. Species with a yellow background represent common Nocardia spp., and the strain with a green background represents GZ2020T. Different colored branches represent different groups.
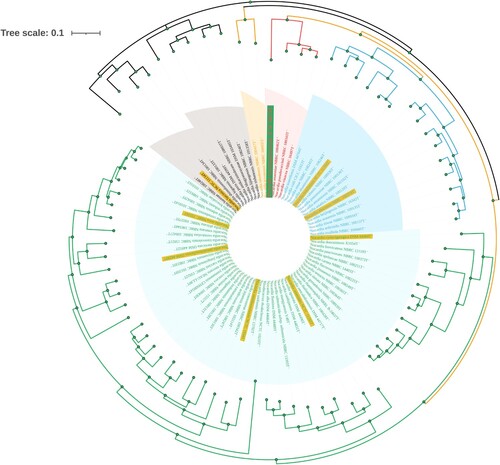
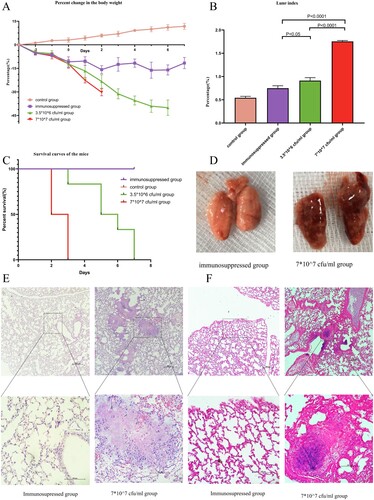
Figure 5. In vivo study of the bacterial pathogenicity. A: Percent change in body weight. B: Lung/body weight index of the groups. C Survival curves of the mice. Compared with the control and immunosuppressed groups, all mice in the infected groups died within 7 days, and the high-dose group (7*10^7/mL) showed 100% mortality at 72 h. D Gross pathology findings from the groups. Compared with the lungs of the control group, the lungs of the infected group were significantly congested and edematous. E (upper part: 100X magnification, lower part: 400X magnification) HE staining of samples from the immunosuppressed and infected groups (7*107 cfu/mL). Suppurative inflammatory changes, such as exudation, necrosis and infiltration of neutrophils, were observed in the infected group. F (upper part: 100X magnification, lower part: 400X magnification) Gram staining of samples from the immunosuppressed group (7*107 cfu/mL). Gram staining indicated rod-shaped bacterial aggregates in the lung of the infected group.
Methods
Specimen collection and bacterial culture
Blood, sputum, BALF and lung tissue samples were collected from the patient according to clinical practices [Citation9,Citation10]. In the laboratory, small aliquots of the samples were removed, plated onto Columbia blood agar (CBA) and incubated aerobically at 37 °C for 3 days. The bacteria were then prepared for staining and microscopic examination, including Gram staining, weak acid-fast staining, acid-fast staining and transmission electron microscopy.
Genome sequencing and phylogenetic analysis
DNA was extracted from strain GZ2020T using a nucleic acid extraction kit (Hangzhou Matridx Biotechnology). A 500-bp insert-size library was constructed with 0.2 μg of genomic DNA, and 136,235,859 single-end 75-bp clean reads were obtained using the NextSeq 500 platform. The sequencing data were assembled using Spades (v3.13.0), which resulted in 138 scaffolds of more than 1000 bp. The final genome sequence was annotated with Prokka (v1.14.6) and assessed with BUSCO (v4.1.4). Seventy-six previously published genome sequences of model Nocardia strains were obtained for phylogenetic tree construction, and the genomes and associated accession numbers are shown in Supplementary Table 1. For improved consistency, Rhodococcus rhodochrous NCTC 10210T (LT906450), which is in the same family as Nocardia, was selected as the outgroup. The single nucleotide polymorphisms (SNP) information for 76 Nocardia strains was obtained using MUMmer (version 3.1) software [Citation11,Citation12]. Subsequently, to elucidate the exact taxonomic position of these novel Nocardia spp., a maximum-likelihood phylogenetic tree based on SNPs was constructed using FastTree (version 2.1.10) software [Citation13]. The average nucleotide identity (ANI) [Citation14] between the genomes was calculated using the Python module pyani based on MUMmer (ANIm) algorithms, and digital DNA:DNA hybridization (dDDH) similarities were computed with the Genome-to-Genome Distance Calculator (version 3, https://ggdc.dsmz.de/ggdc.php#). Subsequently, basic functional gene analysis was performed to compare with the other 3 relatives using eggnog (http://eggnog5.embl.de/)
Physiological and chemotaxonomic analysis
All culture characteristics of strain GZ2020T were determined by growing the strain on CBA at 37 °C for 3 days. Gram staining was conducted with Hucker’s method [Citation15]. The morphological characteristics of the strain after growth on CBA at 37 °C for 3 days were observed by light microscopy (BX43, Olympus) and scanning electron microscopy (S-3000N, Hitachi). The growth of GZ2020T on CBA was tested over 3 days of incubation at different temperatures (4, 10, 20, 28, 30, 37, 40 and 45 °C), with different NaCl concentrations (0–15%, w/v, at intervals of 1%) and at different pH values (4.0–12.0, at intervals of 1.0 pH units). Catalase activity was evaluated by observing whether bubbles were produced after pouring 3% H2O2 (v/v) on the colonies. The hydrolysis of starch and Tween (20, 80) was tested as described previously [Citation16]. The utilization of single carbon sources was determined with a GEN III MicroPlate (Biolog) according to the manufacturers’ instructions. In vitro AST was performed using the E-test and disc diffusion test, as described previously for Nocardia species [Citation17–19]. Breakpoints defined by the CLSI were applied for the E-test, [Citation20,Citation21] and the results from disc diffusion tests were interpreted according to the CLSI breakpoints for Staphylococcus spp.[Citation18,Citation22] The interpretation of antimicrobial susceptibility was based on the result of an E-test, and the disc diffusion test was used as a supplement when the E-test was unavailable. For further chemotaxonomic analyses, GZ2020T was incubated in CBA for 3 days under aerobic conditions at 37 °C. The cells were harvested by centrifugation, washed twice with distilled water, recentrifuged and freeze-dried. The freeze-dried cell biomass was used to analyze polar lipids, quinones and cell sugars at the Guangdong Institute of Microbiology (Guangzhou, Guangdong, PR China), as described by Wu et al. [Citation23] and Zhuang et al. [Citation24].
Bacterial pathogenicity in vivo
Forty-eight BALB/C mice were divided into 4 groups: high-dose infection group (7.0 x107 cfu/mL, n = 12), low-dose infection group (3.5 x106 cfu/mL, n = 12), immunosuppressed group (n = 12), and control group (n = 12). The mice in the infection and immunosuppressed groups were intraperitoneally injected with 150 mg/kg cyclophosphamide on days −3, −1, and +1 relative to infection, as in another study [Citation24]. The experimental mice in the infection group were administered 50 µL of bacterial culture solution via nasal drip on day 0, and 50 µL of phosphate-buffered saline was administered to the mice in the immunosuppressed and control groups. On days 2 and 3, the mice were euthanized for lung pathologic examination assays and bacterial culture of lung tissue homogenates.
Results
Under gross observation, colonies cultured on CBA plates were light yellow and circular and measured 0.5–1.0 mm in diameter after 3 days at 37 °C (Supplementary Figure 1). Bacteriologic analysis revealed positive Gram staining and weak acid-fast staining, and negative acid-fast staining was also observed (Supplementary Figure 2). Analysis of the cell morphology of this bacterial strain under an optical microscope revealed that the strain was beaded and had a rod shape with branching filaments. Scanning electron microscopy (SEM) analysis revealed a filamentous network covered with an opaque biofilm matrix (Supplementary Figure 3).
The genome (CDS) of GZ2020T was found to have a length of 8,350,551 bp, with an average GC content of 66.89% (Supplementary Figure 6), which is similar to the genomes of other Nocardia spp. A maximum-likelihood phylogenetic tree based on SNPs was constructed using FastTree (version 2.1.10) software, and the results revealed that the closest known relatives were Nocardia anaemiae NBRC 100462T, Nocardia pseudovaccinii NBRC 100343T, and Nocardia vinacea NBRC 16497T, which form a distinct branch. However, considering the branch lengths, substantial genetic variation was observed between GZ2020T and these 3 strains. Moreover, common species of Nocardia were even more distinct from GZ2020T because these pathogens formed different branches in the phylogenetic tree, which belonged to different generations (; details on the genetic, physiological and chemical characteristics are provided in the Supplementary Material).
The heatmap from the identity alignment of GZ2020T with the other 3 pathogens showed ANI values < 95% and a similar dDDH result of less than 70% (Supplementary Figure 7). Physiological, chemotaxonomic, molecular biological methods and basic functional gene analysis indicated that this strain was somewhat different from known species (Supplementary Tables 2–4 and Supplementary Figure 8). Due to the gene analysis (ANI,dDDH and basic functional gene analysis) results and differences in physiological characteristics between GZ2020T and these species, we concluded that GZ2020T represented a novel Nocardia species and named it Nocardia guangzhouensis GZ2020T ( = GDMCC 4.187T = JCM 34519T). To further estimate its pathogenicity, in vivo virulence testing was performed.
Compared with the immunosuppressed and control groups, all infected mice died within 7 days, and the high dose group (7*10^7 cfu/mL) showed 100% mortality at 72 h (C). Their body weight decreased progressively (A), and the lung index increased significantly (B). The lungs of the infected group were significantly congested and edematous (D), and HE staining revealed suppurative inflammatory changes, such as exudation, necrosis and infiltration of neutrophils (E). Gram staining indicated that rod-shaped bacteria aggregated in the lung (F). Lung homogenates from the infected group were plated on CBA plates and cultured for gene sequencing, which showed that species isolated from the mice and GZ2020T belonged to the same genus (Supplementary Figure 9).
Discussion
Herein, we report a confirmed case of recurrent pulmonary infection caused by a novel community-acquired Nocardia species in Guangzhou, China. The patient did not belong to the traditional immunosuppressed population but had bronchiectasis. This pathogen is characterized by its low susceptibility to multiple antibiotics, including routine therapeutic antibacterial agents, SMZ/TMP, amikacin and imipenem. In vivo experiments indicated that the main injury mechanism was peribronchial purulent inflammation with massive neutrophilic infiltration. The substantial potential health threat posed by GZ2020T deserves attention.
Bacteriological analysis revealed that GZ2020T exhibited positive Gram staining and weak acid-fast staining, and microscopic examination showed that the bacteria tended to be beaded and to have a rod shape with branching filaments (Supplementary Figure 2), which is consistent with the common characteristics of Nocardia [Citation1,Citation2,Citation6,Citation25]. However, further phylogenetic analysis and physiological examinations ( and Supplementary Tables 1–4) revealed that it belonged to the Nocardia species but was different from known strains, which indicated that GZ2020T is a novel pathogen. Notably, compared with other Nocardia spp., GZ2020T was isolated and propagated relatively easily on CBA plates, as demonstrated by the finding that bacterial colonies were visible to the naked eye within 72 h (Supplementary Figure 1 and 2A), which is the minimum time (48 h-72 h) needed before colonies of other Nocardia spp. are evident[Citation25], and colonies of some for Nocardia spp. sometimes need 2–14 days to appear[Citation26]. Most AR mechanisms are associated with a fitness cost, which typically manifests as a reduced bacterial growth rate in the absence of antibiotic pressure [Citation27]. Hence, the growth rate of GZ2020T indicates that this strain could be a naturally selective pathogen rather than a strain that evolved from a known species through antibiotic induction within a short time. However, the original pathogen source and method of spreading remain uncertain, and additional research is needed. Based on its gross appearance (Supplementary Figure 1), GZ2020T appeared drier and rougher than known species, which may also contribute to its low susceptibility to multiple antibiotics, as previous studies have revealed [Citation28,Citation29].
Immunosuppressed patients were previously the main human population susceptible to Nocardia infection [Citation2], but a history of bronchiectasis was also recently found to be a key risk factor [Citation5,Citation7,Citation30]. It has been reported that the proportion of patients with bronchiectasis and Nocardia infection significantly increased from 11% during 1996–2001 to 33% during 2008-2013, and the increasing number of Nocardia infections over time could be driven by the occurrence of bronchiectasis instead of the immunocompromised status of the population [Citation30]. Bronchiectasis is a predisposing condition for Nocardia infection, although the definite mechanism remains uncertain [Citation7,Citation30]. In addition, other CLDs, such as COPD, have been found to be related [Citation4,Citation5,Citation7]; thus, attention should be appropriately shifted from the immunosuppressed population toward patients with CLD, particularly those with bronchiectasis. Changes in susceptible populations could be implicated in the alteration of the pathogenic spectrum and the emergence of novel strains. Enhanced surveillance is necessary for this population.
The most concerning health issue associated with GZ2020T is its low susceptibility to first-line antibiotic treatment. The emergence and rapid spread of AR poses a severe threat to human health, and in 2019, AR could have been the third leading Global Burden of Diseases (GBD) Level 3 cause of death and was second only to ischemic heart disease and stroke [Citation31]. The scenario might be even worse by 2050 if no effective measures are taken [Citation32]. The majority of known Nocardia species are susceptible to first-line antibiotics, despite the emergence of some drug-resistant species[Citation2,Citation8]. However, GZ2020T is the first Nocardia species to exhibit little susceptibility to multiple antibiotics (), and the notable side effects of the most effective drug (linezolid) make its treatment challenging [Citation33]. Although the original pathogen source and method of spreading remain uncertain, our animal model and this case demonstrate the existence of a possible mode of transmission from the environment to humans. Because this species was found in the community [Citation34,Citation35], assessment of the drug resistance burden caused by GZ2020T could be difficult and complex. The current key to antibiotic resistance (AR) management lies in controlling the spread of resistant strains and genes [Citation36]. Thus, much vigilance should be maintained for this novel pathogen and its spread. We will also perform detailed epidemiological research and analyses of resistance genes, which may be beneficial for understanding the development of drug resistance.
Community-acquired GZ2020T could make the management of Nocardia more challenging in patients with bronchiectasis. Drug-resistant bacterial infections pose an increased risk of pathogen persistence and clinical deterioration [Citation37]. Poorly controlled or recurrent chronic infections in patients with bronchiectasis could contribute to worsening of the quality of life, strengthening of bronchiectasis, deterioration of lung function [Citation38], and increased mortality [Citation39,Citation40]. Therefore, a precise anti-infective strategy is necessary to rapidly eliminate pathogens and prevent the exacerbation of bronchiectasis. In this case, initial empiric antibiotic treatment was not sufficient to obtain a favorable effect. Subsequently, the administration of treatment guided by AST results gradually improved the symptoms and imaging results, and no clinical relapse or exacerbation of bronchiectasis has been observed to date (), which indicates that the precise antibiotic strategies based on AST results were effective and necessary. Similarly, this pathogen could also increase the risk of immunocompromised patients. In addition, more than half of these Nocardia pneumonia patients could show a combination of extrapulmonary disease, and the brain is the most common site of metastasis [Citation37]. When this pathogen breaks the blood–brain barrier, high mortality could be observed (even reaching 90% in cases of delayed treatment) [Citation37]. Therefore, the emergence of GZ2020T is of concern because this novel, multidrug-resistant pathogen could easily lead to late diagnosis and treatment and thus causes high mortality if central nervous system infections occur.
In conclusion, this case highlights the substantial health threat posed by novel, highly drug-resistant Nocardia spp. to patients with CLD and immunocompromised individuals, particularly those with bronchiectasis. Vigilance of its spread in the population and the transmission of AR genes in the environment should be maintained. This report also serves as a reminder that despite the achievements made to date, much more effort should be paid to the issue of AR.
Declaration
The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention (CDC). The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper. Informed consent and authorization were obtained from the patient, and the file is attached as Supplementary Material (Additional documents 1 and 2). All experiments involving Nocardia and other microorganisms in this article were carried out under standard biosecurity and institutional safety standards. All experiments were conducted in biosafety level 2 laboratories and strictly performed in accordance with laboratory biosafety regulations (procedurehttp://www.nhc.gov.cn/wjw/gfxwj/201304/64601962954745c1929e814462d0746c.shtml). Furthermore, this study was approved by the Ethics Committee of the First Affiliated Hospital of Guangzhou Medical University (ethical approval number: 2019-26).
GZ2020T was identified and is preserved in the Guangdong Microbial Culture Collection Center (https://www.gdmcc.net/main.do?method=load&css=1&englist=) (GDMCC 4.187T) and Microbe Division, RIKEN BioResource Research Center, JAPAN COLLECTION OF MICROORGANISMS (https://jcm.brc.riken.jp/en/) (JCM 34519T) (Additional documents 3 and 4).
The raw sequence data for GZ2020T identified in this study have been deposited under accession number GWHBEDI00000000 in the Genome Warehouse at the National Genomics Data Center, Beijing Institute of Genomics, Chinese Academy of Sciences/China National Center for Bioinformation, which is publicly accessible at https://ngdc.cncb.ac.cn/gwh. All other data are available from the authors upon reasonable request. Genome Warehouse: A Public Repository Housing Genome-scale Data. Genomics Proteomics Bioinformatics 2021. https://doi.org/10.1016/j.gpb.2021. 04.001; and Database Resources of the National Genomics Data Center, China National Center for Bioinformation in 2021. Nucleic Acids Res 2021, 49(D1): D18–D28.
The raw sequence data for GZ2020T identified in this study have also been deposited in the NCBI database (accession number JAIRBR000000000).
The type strain, Nocardia guangzhouensis GZ2020T ( = GDMCC 4.187T = JCM 34519T), was isolated from the lung tissue and BALF specimens of a patient at the First Affiliated Hospital of Guangzhou Medical University in Guangzhou, Guangdong, China.
Author contributions
Article design and writing: Zhengtu Li, Yongming Li, Shaoqiang Li, Nanshan Zhong, Feng Ye; patient management and treatment: Zhengtu Li, Shaoqiang Li, Yangqing Zhan, Nanshan Zhong, Feng Ye; clinical sample collection and detection: Ying Mai, Yongming Li, Zhun Li, Jing Cheng, Danhong Su. In vivo experiment: Yongming Li, Zhengtu Li, and Zhun Li.
All authors contributed to the acquisition, analysis, or interpretation of the data and reviewed and approved the final version of the manuscript.
Supplemental Material
Download PDF (1.9 MB)Acknowledgments
This work was funded by the ZHONGNANSHAN MEDICAL FOUNDATION OF GUANGDONG PROVINCE (ZNSA-2020003). We thank the staff members of the hospital for their effort in collecting the information used in this study; other staff members at the Clinical Microbiology Laboratory for the microbial cultivation; staff members at the Clinical Imaging Department for chest CT image processing; and staff members at the Clinicopathological Center for histopathological section detection. We also thank Jun Wang, Yi Zheng, and Boshi Yang from Hangzhou Matridx Biotechnology Co., Ltd. (Hangzhou, China), and Pengcheng Du and Le Yu from Beijing YuanShengKangTai (ProtoDNA) Genetech Co., Ltd. (Beijing, China) for the sequencing of GZ2020T with mNGS and Nanopore technology. Furthermore, we would like to thank the AJE team for providing language editing.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Potential conflicts of interest
No potential conflict of interest is reported by the author(s).
Additional information
Funding
References
- Beaman BL, Beaman L. Nocardia species: host-parasite relationships. Clin Microbiol Rev 1994;7(2):213–264.
- Dennis L, Kasper ASF. Harrison's TM Infectious Diseases. McGraw-Hill Education; 2017. (Filice GA, editor. Nocardiosis).
- Mamelak AN, Obana WG, Flaherty JF, et al. Nocardial brain abscess. Neurosurgery. 1994;35(4):622–631.
- Martínez Tomás R, Menéndez Villanueva R, Reyes Calzada S, et al. Pulmonary nocardiosis: risk factors and outcomes. Respirology. 2007;12(3):394–400.
- Ambrosioni J, Lew D, Garbino J. Nocardiosis: updated clinical review and experience at a tertiary center. Infection. 2010;38(2):89–97.
- Chaussade H, Lebeaux D, Gras G, et al. Nocardia arthritis. Medicine (Baltimore). 2015;94(42):e1671.
- Kandi V. Human Nocardia infections: A review of pulmonary nocardiosis. Cureus. 2015 Aug 15;7(8):e304.
- Woods GL, Brown-Elliott BA, Conville PS, et al. Susceptibility Testing of Mycobacteria, nocardiae, and Other Aerobic actinomycetes. 2nd ed. Wayne (PA): Clinical and Laboratory Standards Institute; 2011.
- Metlay JP, Waterer GW, Long AC, et al. Diagnosis and treatment of adults with community-acquired pneumonia. An official clinical practice guideline of the American thoracic society and infectious diseases society of America. Am J Respir Crit Care Med 2019;200(7):e45–e67.
- Wilson JW. Nocardiosis: updates and clinical overview. Mayo Clin Proc 2012;87(4):403–407.
- Delcher AL, Phillippy A, Carlton J, et al. Fast algorithms for large-scale genome alignment and comparison. Nucleic Acids Res 2002;30(11):2478–2483.
- Kurtz S, Phillippy A, Delcher AL, et al. Versatile and open software for comparing large genomes. Genome Biol 2004;5(2):R12.
- Price MN, Dehal PS, Arkin AP. Fasttree 2 – approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5(3):e9490.
- Yoon SH, Ha SM, Lim J, et al. A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek. 2017;110(10):1281–1286.
- Gerhardt P, Murray R, Costilow RN, et al. Manual of Methods for General Bacteriology. 1981.
- Smibert R, Krieg N, Gerhardt P, et al. Methods for general and molecular bacteriology. American Society for Microbiology, Washington, DC. 1994:607-654.
- CLSI/NCCLS. Susceptibility testing of mycobacteria, nocardiae, and other aerobic actinomycetes. National Committee for Clinical Laboratory Standards Wayne, PA; 2003.
- Perçin D, Sümerkan B, Inci R. [Comparative evaluation of e-test and disk diffusion methods for susceptibility testing of Nocardia species]. Mikrobiyol Bul. 2011 Apr;45(2):274–279.
- Wallace RJJ, Septimus EJ, Musher DM, et al. Disk diffusion susceptibility testing of Nocardia species. J Infect Dis. 1977;135(4):568–576.
- Glupczynski Y, Berhin C, Janssens M, et al. Determination of antimicrobial susceptibility patterns of Nocardia spp. from clinical specimens by etest. Clin Microbiol Infect. 2006;12(9):905–912.
- Woods GL, Brown-Elliott BA, Conville PS, et al. CLSI standards: guidelines for health Care excellence. Susceptibility Testing of Mycobacteria, nocardiae, and Other Aerobic actinomycetes. Wayne (PA): Clinical and Laboratory Standards Institute; 2011.
- The Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing. 32 nd ed. CLSI supplement M100. Wayne, PA: Clinical and Laboratory Standards Institute. 2016.
- Wu W, Feng Y, Zong Z. Enterobacter sichuanensis sp. nov., recovered from human urine. Int J Syst Evol Microbiol 2018;68(12):3922–3927.
- Zhuang K, Liu Y, Dai Y, et al. Nocardia huaxiensis sp. nov., an actinomycete isolated from human skin. Int J Syst Evol Microbiol 2021;71(8):004970.
- Goodfellow M. The family nocardiaceae. In: A Balows, HGTM Dworkin, W Harden, KH Schleifer, editor. The prokaryotes, 2nd ed. New York, N.Y.: Springer-Verlag; 1992. p. 1188–1213.
- Berd D. Laboratory identification of clinically important aerobic actinomycetes. Appl Microbiol. 1973;25(4):665–681.
- Andersson DI, Hughes D. Antibiotic resistance and its cost: is it possible to reverse resistance? Nat Rev Microbiol. 2010;8(4):260–271.
- Drenkard E, Ausubel FM. Pseudomonas biofilm formation and antibiotic resistance are linked to phenotypic variation. Nature. 2002;416(6882):740–743.
- Cangelosi GA, Do JS, Freeman R, et al. The Two-component regulatory systemmtrABIs required for morphotypic multidrug resistance inmycobacterium avium. Antimicrob Agents Chemother 2006;50(2):461–468.
- Woodworth M, Saullo J, Lantos P, et al. Increasingnocardia incidence associated with bronchiectasis at a tertiary care center. Ann Am Thorac Soc. 2017;14(3):347–354.
- Collaborators AR. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. lancet (London, England). 2014;399(10325):629–655.
- On J. Review on Antimicrobial Resistance Antimicrobial Resistance: Tackling a crisis for the health and wealth of nations 2014 [cited 2021 July 7]. Available from: https://amr-review.org/sites/default/files/AMR%20Review%20Paper%20-%20Tackling%20a%20crisis%20for%20the%20health%20and%20wealth%20of%20nations_1.pdf.
- Zhang X, Falagas ME, Vardakas KZ, et al. Systematic review and meta-analysis of the efficacy and safety of therapy with linezolid containing regimens in the treatment of multidrug-resistant and extensively drug-resistant tuberculosis. J Thorac Dis. 2015 Apr;7(4):603–615.
- Jernigan JA, Hatfield KM, Wolford H, et al. Multidrug-Resistant bacterial infections in U.S. hospitalized patients, 2012-2017. N Engl J Med. 2020 Apr 2;382(14):1309–1319.
- Fang FC, Schooley RT. Antimicrobial resistance — The glass Is half full. N Engl J Med. 2020;382(14):1363–1365.
- Collignon P, Beggs JJ, Walsh TR, et al. Anthropological and socioeconomic factors contributing to global antimicrobial resistance: a univariate and multivariable analysis. The Lancet Planetary Health. 2018;2(9):e398–e405.
- Organization WH. Global action plan on antimicrobial resistance. 2015.
- Wilson CB, Jones PW, O'Leary CJ, et al. Effect of sputum bacteriology on the quality of life of patients with bronchiectasis. Eur Respir J. 1997;10(8):1754–1760.
- Martínez-García M, de Gracia J, Vendrell Relat M, et al. Multidimensional approach to non-cystic fibrosis bronchiectasis: the FACED score. Eur Respir J. 2014;43(5):1357–1367.
- Chalmers JD, Goeminne P, Aliberti S, et al. The bronchiectasis severity index. An international derivation and validation study. Am J Respir Crit Care Med. 2014;189(5):576–585.
