ABSTRACT
In vaccinees who were infected with SARS-CoV in 2003, we observed greater antibody responses against spike and nucleoprotein of both SARS-CoV-2 and SARS-CoV after a single dosage of inactivated SARS-CoV-2 vaccine. After receiving the second vaccination, antibodies against RBD of SARS-CoV-2 Wuhan, Beta, Delta, and recently emerged Omicron are significantly higher in SARS-CoV experienced vaccinees than in SARS-CoV naïve vaccinees. Neutralizing activities measured by authentic viruses and pseudoviruses of SARS-CoV, SARS-CoV-2 Wuhan, Beta, and Delta are greater in SARS-CoV experienced vaccinees. In contrast, only weak neutralizing activities against SARS-CoV-2 and variants were detected in SARS-CoV naïve vaccinees. By 6 months after the second vaccination, neutralizing activities were maintained at a relatively higher level in SARS-CoV experienced vaccinees but were undetectable in SARS-CoV naïve vaccinees. These findings suggested a great possibility of developing a universal vaccine by heterologous vaccination using spike antigens from different SARS-related coronaviruses.
The 2019 novel coronavirus disease (COVID-19) outbreak triggered a worldwide pandemic and caused a global health crisis [Citation1–3]. The rapid development and massive immunization of COVID-19 vaccinees significantly prevent infection and severe diseases caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and its variants [Citation4–6].
The SARS-CoV and SARS-CoV-2 are SARS-related coronavirus that belong to the subgenus sarbecovirus of betacoronavirus genus. These two SARS-related coronaviruses are placed in two antigenetically distinct phylogenetic clades [Citation1,Citation2]. The spike (S) protein of SARS-CoV and SARSCoV-2 shares 77.4% amino acid sequence similarity, with RBD shares 73.1% and the S2 region sharing the highest similarity at 87.3%. The nucleoprotein (N) shares even up to 90.9% amino acid sequence similarity (Figure S1). The SARS-CoV is responsible for the infection of more than 8,000 patients and spread to 26 countries on five continents during the 2002–2003 outbreak [Citation7]. Even at 17 years after infection, serum samples from SARS-CoV survivors still possessed binding antibodies to S protein of both SARS-CoV and SARS-CoV-2 and a low level of neutralizing activity against SARS-CoV, but no SARS-CoV-2 could be detected [Citation8]. Likewise, convalescent serum samples from SARSCoV-2 infected people can neutralize SARS-CoV-2 but not SARS-CoV [Citation8,Citation9]. Nevertheless, a few monoclonal antibodies (mAbs) with naturalizing activities against SARS-CoV and SARS-CoV-2 could be isolated from SARS-CoV or SARS-CoV-2 convalescents [Citation10–14]. These mAbs target spike protein's receptor-binding domain (RBD) [Citation15]. Recently, several reports revealed that a single dose of mRNA vaccine in SARS-CoV-2 convalescent people could induce a higher antibody response to the level similar to people who received two doses of mRNA vaccinees [Citation16–18]. In this study, we evaluated the magnitude and breadth of antibody response after one and two dosages of inactivated SARS-CoV-2 vaccine in a small cohort of people who have history of SARS-CoV infection during 2003 outbreak (SARS-CoV experienced) and people who have no history of SARS-CoV infection (SARS-CoV naïve).
Results
We first used ELISA to measure IgG titres to S, RBD, and N of SARS-CoV and SARS-CoV-2 (Wuhan strain). At day 28 (D28) after the first vaccination in SARS-CoV experienced vaccinees, there was a significant induction of IgG titres against not only SARS-CoV-2 S, RBD, and N but also SARS-CoV S, RBD, and N. In contrast, there were no induction of IgG against S, RBD, and N of either SARS-CoV-2 or SARS-CoV in SARS-CoV naïve vaccinees. At 28 days (D56) after the second vaccination, the IgG against S, RBD, and N proteins of either SARS-CoV-2 or SARS-CoV were elevated but were lower in SARS-CoV naïve vaccinees (). The median (half-maximal effective concentration) EC50 IgG titres against SARS-CoV-2 S, RBD, and N were 2269.9, 1896.7, and 2679.2 in SARS-CoV experienced vaccinees, but only 449.8, 545.8 and 696.6 in SARS-CoV naïve vaccinees (Table S2). The IgG antibodies against SARS-CoV RBD and SARS-CoV-2 RBD in SARS-CoV experienced vaccinees were 8.3-fold and 3.5-fold higher than that in SARS-CoV naïve vaccinees, respectively. The IgG antibodies against SARS-CoV N and SARS-CoV-2 N were 10.1fold and 3.9-fold higher in SARS-CoV experienced vaccinees than SARS-CoV naïve vaccinees (Table S2). These results suggested the recall response of memory B cells to SARS-CoV upon stimulation of SARS-CoV-2. We also found that the IgG antibodies against SARS-CoV-2 S2 were 7.0-fold higher in SARS-CoV experienced vaccinees than in SARS-CoV naïve vaccinees after the first vaccination. However, the difference narrowed to 2.5-fold higher after the second vaccination (Table S2, Figure S2A). In addition, we measured IgG antibodies against RBD-Beta and RBD-Delta variants after the second vaccination ((A,B)). The IgG antibodies against RBD-Beta and RBD-Delta were 6.5-fold and 5.3-fold higher 28 days after the second vaccination, and were 6.7-fold and 6.7-fold higher 3 months after the second vaccination in SARS-CoV experienced vaccinees than SARS-CoV naïve vaccinees (Table S3, (A,B)). During the preparation of this manuscript, the emergence of the SARS-CoV-2 Omicron variant raised the concern of immune evasion from established immunity. We compared the serum IgG that binds to RBD-Omicron 3 months after the second vaccination. RBD-Omicron IgG titres in SARS-CoV experienced vaccinees with median EC50 at 345.6 (268.3 - 662.6) than SARS-CoV naïve vaccinees with median EC50 at 47.0 (37.6 - 51.2). Although RBD-Omicron IgG in SARS-CoV experienced vaccinees had a 4.6-fold decrease compared to RBD-Wuhan IgG, the EC50 of RBD-Omicron IgG was 7.4-fold higher in SARS-CoV experienced vaccinees than SARS-CoV naïve vaccinees who decreased to near background level ((B), Table S3). These results indicated a significant expansion of B cells that can produce cross-reactive antibodies against SARS-CoV and SARS-CoV2 variants by heterologous vaccination with antigens from SARS-related coronaviruses.
Figure 1. The N, S and RBD protein-specific IgG antibody response in SARS-CoV experienced and SARS-CoV naïve vaccinees. Serum samples were collected at 28 days after the first dosage (D28) and 28 days after the second dosage (D56). (A-C) serum IgG binding antibodies against SARS-CoV-2 S, RBD, and N proteins were measured by ELISA; (D-F) serum IgG binding antibodies against SARS-CoV S, RBD, and N proteins were measured by ELISA. The half-maximal titre (EC50) was calculated using four-parameter logistic curve fit. Red squares and lines represent SARS-CoV experienced vaccinees. Blue circles and lines represent SARS-CoV naïve vaccinees. The pink and blue lines represent the median values of the SARS-CoV experienced vaccinees and the SARS-CoV naïve vaccinees, respectively. The dashed black lines represent the positive cutoff value. Statistical significance was determined using one-way ANOVA-test. * denotes p < 0.05, ** denotes p < 0.01, *** denotes p < 0.001, **** denotes p < 0.0001, ns, non-significant.
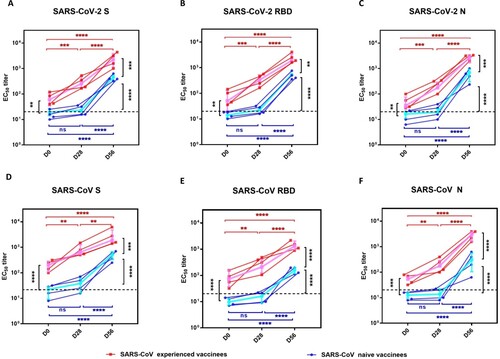
Figure 2. Neutralizing antibody responses against SARS-CoV-2 and SARS-CoV in SARS-CoV experienced and SARS-CoV naïve vaccinees. After the first dosage, serum samples were collected at day 28, day 56, day120, and day210. A second dosage was given at day 28 after the first dosage. (A) Serum IgG binding antibodies against RBDs of Wuhan, Beta, and Delta on day 56 after the first dosage. (B) Serum IgG binding antibodies against RBDs of Wuhan, Beta, Delta, and Omicron on day 120 after the first dosage. (C) Serum neutralizing antibody titres to live SARS-CoV-2. Wuhan and Beta on day 28 after the first dosage(D28) and day 28 after the second dosage(D56). (D) Serum neutralizing antibody titres to pseudotype SARS-CoV on day 28 after the second dosage(D56). The half-maximal neutralizing titre (NT50) was measured by a pseudovirus-based neutralizing assay and calculated by Reed Muench. (E) Serum neutralizing antibody titres to VSVpseudotype SARS-CoV-2 Delta on day 28 after the second dosage(D56). (F) A CLIA-based surrogate virus neutralization test (sVNT) to measure the RBD-targeted neutralizing antibodies that compete with the binding hACE2. Red squares and lines represent SARS-CoV experienced vaccinees. Blue circles and lines represent SARS-CoV naïve vaccinees. The pink and blue lines represent the median values of the SARS-CoV experienced vaccinees and the SARS-CoV naïve vaccinees, respectively. The dashed black lines represent the positive cutoff value. Statistical significance was determined using one-way ANOVA-test. * denotes p < 0.05, ** denotes p < 0.01, ***denotes p < 0.001, **** denotes p < 0.0001, ns = non-significant.
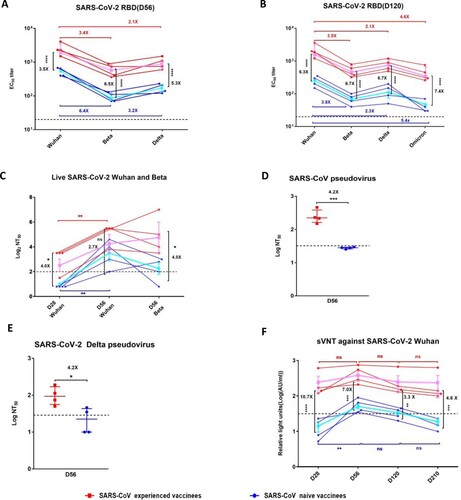
We next measured virus neutralization by a cytopathic plaque reduction assay using live SARSCoV-2 Wuhan (strain 2020XN4276). Live virus neutralization (>1:4) against SARS-CoV-2 Wuhan was detectable in SARS-CoV experienced vaccinees after the first dosage, which further increased after the second dosage ((C)). In contrast, in SARS-CoV naïve vaccinees, virus neutralization was not detectable after one dosage and became detectable after the second vaccination but was much lower ((C)). Live virus neutralization against authentic SARSCoV-2 Wuhan was 2.7-fold higher in SARS-CoV experienced vaccinees than SARS-CoV naïve vaccinees. Compared to virus neutralization against authentic SARS-CoV-2 Wuhan, there was no significant reduction of neutralizing activity against authentic SARS-CoV-2 Beta in SARS-CoV experienced vaccinees SARS-CoV-1 naïve vaccinees had a significant reduction of neutralization activity against SARS-CoV-2 Beta ((C), Table S4). Since the authentic SARS-CoV was not available, we used a lentivirus based pseudovirus neutralization assay to determine the neutralizing activity against SARS-CoV. Neutralizing activity against SARS-CoV was 4.2-fold higher in SARS-CoV experienced vaccinees ((D)). We also used a VSV based pseudovirus neutralization assay to determine the neutralizing activity against SARS-CoV-2 Delta for the serum samples collected on D56. Neutralization against Delta variant was 4.2-fold higher in SARS-CoV experienced vaccinees than in SARS-CoV naïve vaccinees (1:96 VS 1:23) ((E)).
In addition, we used a surrogate virus neutralization test (sVNT) based on chemiluminescent immunoassay (CLIA) to evaluate the neutralizing antibodies that block the binding of SARS-CoV-2 RBD with cellular receptor ACE2. After the first vaccination, sVNT titre increased to 126.47 AU/ml in SARS-CoV experienced vaccinees but was undetectable in SARS-CoV naïve vaccinees ((F)). sVNT titre was 7.0-fold higher in SARS-CoV experienced vaccinees after the second dosage. At 6 months after the second vaccination, sVNT titre in SARS-CoV experienced vaccinees decreased 3.6-fold from 312.61 AU/ml to 87.30 AU/ml, but was still much higher than that in SARS-CoV naïve vaccinees, which decreased to below detection limit ((F), Table S5). Interestingly, although the median S-specific IgM level in SARS-CoV naïve vaccinees was low, the median Sspecific IgM in SARS-CoV naïve vaccinees appeared to be higher in SARS-CoV experienced vaccinees after the first and second dosage (Figure S2C–E, Table S5).
One limitation of this study is that the cohort is relatively small, because it is difficult to recruit vaccinees with a history of SARS-CoV infection. To support the validity of the study, we also tested another 271 healthy cohort at day 28 after the first dosage of COVID-19 vaccine and compared with our cohort in the present study using CLIA to measure sVNT, RBD-specific IgG, and S-specific IgM (Figure S2B, S2C, S2E). Although there is a range of distribution of these antibody levels among 271 donors, the levels of sVNT, RBD-specific IgG, and S-specific IgM of SARS-CoV experienced vaccinees are located at the high end, whereas those of SARS-CoV naïve vaccinees are located around the average value of all 271 samples tested, supporting our cohort’s validity. Notably, only 9 out of 271 (3.3%) of these SARS-CoV naïve individuals had RBD-specific IgG reached the median level of SARS-CoV experienced vaccinees. The median RBD-specific IgG level was 127.90 AU/ml in SARS-CoV experienced vaccinees, 18.3-fold higher than SARS-CoV naïve vaccinees.
Taken together, people with a history of SARS-CoV infection even after 17 years ago could elicit greater and broader IgG antibody response against both SARS-CoV and SARS-CoV-2 variants after vaccination with an inactivated SARS-CoV-2 vaccine.
Discussion
Our study revealed that people with a history of 2003 SARS-CoV infection could elicit greater S-, RBD-, and N-specific binding IgGs and neutralizing antibodies against SARS-CoV and SARS-CoV-2 Wuhan and variants after vaccination with an inactivated SARS-CoV-2 vaccine, even after one dosage. In contrast, SARS-CoV naïve vaccinees required at least two dosages to induce antibody responses, and the neutralizing antibodies were only against SARS-CoV-2 but not SARS-CoV.
One important finding of this study was that vaccination with an inactivated SARS-CoV-2 vaccine could elicit a broad spectrum of neutralizing antibodies against SARS-CoV, SARS-CoV-2 and variants in SARS-CoV experienced vaccinees. SARS-CoV and SARS-CoV-2 share amino acid sequence identity of 77.4% for S protein (Figure S1). In our study, neutralizing antibodies, as reflected by live virus neutralization showed no significant reduction against Beta variant compared to SARS-CoV-2 Wuhan in SARS-CoV experienced vaccinees. The recently emerged Omicron variant has the most significant antibody escape to vaccinated and convalescent sera. We also observed a significant reduction in IgG binding titre to RBD-Omicron as compared to RBD-Wuhan. Nevertheless, RBD-specific IgG titre was 7.4-fold higher in SARS-CoV experienced vaccinees, indicating the presence of broadly reactive antibodies against Omicron.
Several recent reports demonstrated robust antibody responses after one dose of SARS-CoV-2 mRNA vaccine in SARS-CoV-2 convalescent individuals, which reached the level of two dosages in SARS-CoV-2 seronegative individuals [Citation16–18]. This robust antibody response after one dosage of mRNA vaccine can be considered as a homologous boost with the same spike antigen. Our study is unique that unlike the short-term homologous boost in SARS-CoV-2 convalescent or vaccinated people, we observed that even at 17 years after SARS-CoV infection, vaccination with one dosage of inactivated SARS-CoV-2 vaccine could elicit cross-reactive binding antibodies against S, RBD, and N proteins of both SARS-CoV and SARS-CoV-2. An earlier report showed that cross-clade pansarbecovirus RBD-targeted neutralizing antibodies, measured only by surrogate virus neutralization test (sVNT), were induced in SARS-CoV experienced vaccinees after two dosages of BNT162b2 mRNA vaccine [Citation19]. However, there were no reports regarding the binding antibodies to S, RBD, and N proteins in that study. BNT162b2 mRNA vaccine contains only S protein but no N protein, while an inactivated vaccine contains S, N, and other components of SARS-CoV-2. Furthermore, the extent of antibodies elicited by SARS-CoV experienced vaccinees after receiving the first dosage of mRNA vaccine was not reported in that paper. The duration of antibody titres after the second dosage was not investigated. The antibody potency against SARS-CoV-2 and variants remained unknown. Unlike BNT162b2 mRNA vaccine which delivers only S antigen, which has about 77% amino acid sequence identity between SARS-CoV and SARS-CoV-2, the inactivated SARS-CoV-2 vaccine contains integrated virus particles and thus includes N protein, which has over 90% sequence identity between SARS-CoV and SARS-CoV-2. We found that the increased magnitude of N-specific IgG was greater than that of RBD-specific IgG in SARS-CoV experienced vaccinees after one dosage. Although the N-specific antibodies do not directly contribute to virus neutralization, it provides an evidence that the memory B cells contribute to rapid antibody response due to previous exposure to antigens with a high degree of identity.
Intriguingly, we found no significant reduction of neutralizing activity against Beta variant compared to SARS-CoV2 Wuhan strain in SARS-CoV experienced vaccinees, whereas neutralizing activity against the Beta variant was significantly reduced to below detection level in SARS-CoV naïve vaccinees. It is important to note that only SARS-CoV experienced vaccinees, but not SARS-CoV naïve vaccinees, generated neutralizing antibodies against SARS-CoV after one dosage, which further increased after the second dosage. The different patterns following vaccination in SARS-CoV experienced and SARS-CoV naïve vaccinees indicated that the recall response of memory B cells is a significant source for the rapid elevation of antibodies. The Omicron variant has around 15 mutations on RBD of spike protein, contributing to significant immune evasion of vaccine-elicited humoral immunity. Our data showed that RBD-Omicron IgG titres in SARS-CoV experienced vaccinees were higher than SARS-CoV naïve vaccinees. This observation suggested that antibodies generated by priming immunization with SARS-CoV followed by SARS-CoV-2 can confer a more significant cross-reactive antibody response to Omicron. Our study demonstrated that a heterologous immunization can rapidly elicit recall and expansion of B cells that recognize antigens with similarities to a previously infected SARS-related coronavirus.
We and others found that the serum in 2003 SARS-CoV convalescent individuals had relatively low but detectable titres against S and N protein of SARS-CoV even after 17 years [Citation8,Citation9]. One retrospective study revealed that anti-SARS-CoV IgG antibodies peaked in 2004 (one year after infection), declined rapidly from 2004-2006, and persisted for up 12 years [Citation20]. These long-existing antibodies in the circulation may be a concern that vaccinees who had previous SARS-CoV infection may elicit a biased antibody response when receiving COVID-19 vaccine and thus have reduced immunity against SARS-CoV-2. Notably, all 4 SARS-CoV experienced vaccinees reported no adverse events after the first or second vaccination dosage. A higher magnitude and broader spectrum of antibody response against both SARS-CoV-2 and SARS-CoV was achieved after the second dosage. Our study demonstrated a possibility to develop a heterologous prime-boost vaccine strategy for a universal vaccine that can protect against a broad spectrum of SARS-related coronaviruses. A recent report also showed that in mice, sequential immunization S/RBD vaccine of SARS-CoV, SARS-CoV-2, and MERS-CoV led to induction of cross-reactive antibodies and isolation of monoclonal antibodies with broadly neutralizing activities [Citation21].
An ideal COVID-19 vaccine should elicit potent and broad-spectrum neutralizing antibodies against current circulating SARS-CoV-2 and future variants. This study suggested that heterologous primeboost vaccination using different S antigens of SARS-related coronaviruses provides a strategy for developing a universal vaccine against sarbecoviruses.
Methods
Study subjects
Four healthy SARS-CoV experienced vaccinees and 4 healthy SARS-CoV naïve vaccinees (Table S1) who received an inactivated COVID-19 vaccine (BBIBP-CorV/Sinopharm, Beijing). All these 8 vaccinees are female healthcare workers in the First Affiliated Hospital of Guangzhou Medical University in Guangzhou, China. The other 271 healthcare workers were also recruited for control.
The vaccine was intramuscularly injected on day 0 and a second dosage was given on day 28. Blood samples were collected on day 0, day 28, day 56, day 120, and day 210 (six months after the second dosage). The study was approved by the Ethics Committee of the First Affiliated Hospital of Guangzhou Medical University (Ethics number 2021-31). Written informed consents were obtained from all participants. All donors were in good health condition and had no reports of serious adverse events related to the vaccination. The hospital, where the donors located, regularly conducted SARS-CoV-2 RT-qPCR test 1–2 times each week for the healthcare workers and inpatients. No positive cases were reported throughout the vaccination and sampling period.
Enzyme-linked immunosorbent assay (ELISA)
The ELISA assay was used to quantify the serum binding titres to the nucleoprotein (N), spike and RBD protein of SARS-CoV and SARS-CoV-2, and S2 protein of SARS-CoV-2 (Sinobiological, Beijing, SARS-CoV N Cat:40143-V08B, SARS-CoV S Cat:40634-V08B, SARS-CoV RBD Cat:40150-V08B2, SARS-CoV-2 N Cat:40588-V08B, SARS-CoV-2 S Cat:40589-V08B1, SARSCoV-2 S2 Cat:40590-V08B, SARS-CoV-2 RBD proteins of Wuhan strain, Cat:40592-V08B, Beta strain, Cat: 40592-V08H85-B, Delta strain, Cat: 40592-V08H115, Omicron strain, Cat:40592V08H121). The proteins were coated on 96-well plates with 50 ul per well of 1ug/ml protein solution in PBS overnight at 4°C. Plates were washed 6 times with washing buffer (1x PBS with 0.05% Tween-20) and incubated with 200 ul blocking buffer (1x PBS with 2% BSA and 0.05% Tween-20) for 1 h at room temperature. The inactivated serum was diluted in PBS starting dilution at 1:30 and incubated for 1 h at room temperature. Plates were washed 6 times with washing buffer and then incubated with anti-human IgG second antibody conjugated to horseradish peroxidase (HRP)
(Sigma) in blocking buffer at 1:5,000 dilution. Plates were developed by adding the HRP substrate, TMB (Biohao Biotechnology) for 10 min. The developing reaction was then stopped by adding 50 ul 1M H2SO4, and absorbance was measured at 450 nm with an ELISA microplate reader (BioTek). The half-maximal effective concentration (EC50) was determined using four-parameter nonlinear regression (GraphPad Prism v.8.0).
The chemiluminescent immunoassay (CLIA)-based binding and surrogate virus
Neutralization test (sVNT)
ACCORDING TO THE MANUFACTURER'S INSTRUCTIONS, the CLIA assays were performed on AutoLumo A1000 (Autobio Diagnostics, China). In brief, for the IgG/IgM antibodies in the serum samples, the recombinant spike protein coated microparticles and diluted serum was incubated, and the mixture generated a solid phase, and HRP-conjugated anti-human IgG/IgM was added, and then the chemiluminescent substrate was added, and the complex catalyzes substrate, resulting a chemiluminescent reaction. The resulting chemiluminescent reaction is measured as Relative Luminescent Units (RLU). The RLU is proportional to the amount of the according to IgG/IgM in the serum. For the surrogate virus neutralization test (sVNT), the serum mixed with the HRP-labeled RBD protein neutralizes the combination of RBD protein and hACE2 coated on the microparticles. Then the chemiluminescent substrate is added, and the complex catalyzes substrate, resulting in a chemiluminescent reaction. The resulting chemiluminescent reaction is measured as Relative Luminescent Units (RLU). The RLU is inversely proportional to the amount of SARS-coV-2 neutralizing antibodies of the serum. All the sVNT assays were performed on AutoLumo A1000 following the instruction.
Authentic SARS-CoV-2 virus neutralization assay
All microneutralization assays with authentic SARS-CoV-2 virus were performed in a BSL-3 facility.
Heat-inactivated sera were 4-fold serially diluted starting at 1:4 with DMEM supplemented with 2% FBS and 1% penicillin and streptomycin, and mixed with the equal volumes of 100 half tissue culture infective doses (100 TCID50) SARS-CoV-2 of 2020XN4276 strain, or a beta lineage GDPCCnCoV84 strain at 37 °C for 2 h. Afterwards, the sera-virus mixture was added to Vero-E6 cells in 96well cell culture plates, then incubated for an additional 96 h at 37 °C in a 5% CO2 incubator and then observed the virus-induced cytopathic effect (CPE) at 40X magnification. All diluted serum samples were tested in duplicate. The neutralization antibody titres of all the sera were defined as the reciprocal of the serum dilution that neutralizes 50% of virus infection at 4 days post-infection.
Pseudovirus neutralization assay
The pseudovirus neutralization assay was performed to evaluate the sera neutralization ability to SARS-CoV. In brief, 1.5 X 104 HEK293T-hACE2 cells were seeded into 96-well culture plates and incubated overnight at 37 °C in 5% CO2 incubator. The sera were three-fold serially diluted with complete DMEM medium starting at 1:30 and preincubated with an equal volume of 650 TCID50 SARS-CoV pseudovirus at 37 °C for 2 h. The mixture was subsequently incubated with HEK293T-hACE2 cells for 72 h. The cells were lysed with lysis buffer for luciferase activity. All diluted serum samples were tested in three repeats. For the VSV pseudovirus-based neutralization assay, a pseudovirus bearing SARS-CoV-2 delta of S proteins was incubated with three-fold serially diluted sera with complete DMEM medium starting at 1:30 for 1 h at 37 °C. The mixture was inoculated in 3 × 104 Huh7 cells (JCRB, Cat#0403) seeded in 96-well solid white flat-bottom plates, and then incubated for 24 h at 37 °C with 5% CO2. The cells were lysed with lysis buffer for luciferase activity. Neutralizing activity of serum is defined as when the luciferase activity was reduced to 50% of that from the virus-only wells.
Supplemental Material
Download PDF (1.4 MB)Acknowledgements
L.C., X.N. and B.S designed and initiated the project. B. S., P. Z., X.N, H.L., J.M. recruited the patients. H.L., Q. W., Y. D., D. L., H. Y., Y. C., P. H., P. Z., S. H., X. L. conducted the experiments, L. C., X. N., H.L., Q. W., X. Z., Y. Z., Y.Y contributed to data analysis, L. C., X. N., H. L. wrote the manuscript, L.C., X.N., B. S., C. K. contributed to revise and improve the manuscript.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Wu F, Zhao S, Yu B, et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265–269.
- Zhou P, Yang XL, Wang XG, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273.
- Guan WJ, Ni ZY, Hu Y, et al. Clinical characteristics of Coronavirus disease 2019 in China. N Engl J Med. 2020;382(18):1708–1720.
- Wibmer CK, Ayres F, Hermanus T, et al. SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma. Nat Med. 2021; 27(4):622–625.
- Baden LR, El Sahly HM, Essink B, et al. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N Engl J Med. 202;384(5):403–416.
- Jara A, Undurraga EA, Gonzalez C, et al. Effectiveness of an inactivated SARS-CoV-2 vaccine in Chile. N Engl J Med. 2021;385(10):875–884.
- Peiris JS, Yuen KY, Osterhaus AD, et al. The severe acute respiratory syndrome. N Engl J Med. 2003;349(25):2431–2441.
- Anderson DE, Tan CW, Chia WN, et al. Lack of cross-neutralization by SARS patient sera towards SARS-CoV-2. Emerg Microbes Infect. 2020;9(1):900–902.
- Lv H, Wu NC, Tsang OT, et al. Cross-reactive antibody response between SARS-CoV-2 and SARS-CoV infections. Cell Rep. 2020;31(9):107725.
- Pinto D, Park YJ, Beltramello M, et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature. 2020;583(7815):290–295.
- Wec AZ, Wrapp D, Herbert AS, et al. Broad neutralization of SARS-related viruses by human monoclonal antibodies. Science. 2020;369(6504):731–736.
- Liu H, Wu NC, Yuan M, et al. Cross-Neutralization of a SARS-CoV-2 antibody to a functionally conserved site is mediated by avidity. Immunity. 2020;53(6):12721280, e5.
- Liu H, Yuan M, Huang D, et al. A combination of cross-neutralizing antibodies synergizes to prevent SARS-CoV-2 and SARS-CoV pseudovirus infection. Cell Host Microbe. 2021;29(5):806–818. e6.
- Pinto D, Sauer MM, Czudnochowski N, et al. Broad betacoronavirus neutralization by a stem helix-specific human antibody. Science. 2021 Sep 3;373(6559):1109–1116.
- Corti D, Purcell LA, Snell G, et al. Tackling COVID-19 with neutralizing monoclonal antibodies. Cell. 2021;184(12):3086–3108.
- Ebinger JE, Fert-Bober J, Printsev I, et al. Antibody responses to the BNT162b2 mRNA vaccine in individuals previously infected with SARS-CoV-2. Nat Med. 2021;27(6):981–984.
- Krammer F, Srivastava K, Alshammary H, et al. Antibody responses in seropositive persons after a single dose of SARS-CoV-2 mRNA vaccine. N Engl J Med. 2021;384(14):1372–1374.
- Saadat S, Rikhtegaran Tehrani Z, Logue J, et al. Binding and neutralization antibody titers after a single vaccine dose in health care workers previously infected With SARSCoV-2. JAMA. 2021;325(14):1467–1469.
- Tan CW, Chia WN, Young BE, et al. Pan-Sarbecovirus neutralizing antibodies in BNT162b2-immunized SARS-CoV-1 survivors. N Engl J Med. 2021;385(15):14011406.
- Xiaoqin Guo ZG, Duan C, Chen Z, et al. Long-term persistence of IgG antibodies in SARS-CoV infected healthcare workers. medRxiv; 2020.
- Onodera T, Kita S, Adachi Y, et al. A SARS-CoV-2 antibody broadly neutralizes SARSrelated coronaviruses and variants by coordinated recognition of a virus-vulnerable site. Immunity. 2021;54(10):2385–2398. e10.
