ABSTRACT
Candida auris, a multidrug resistant pathogenic yeast, has spread worldwide and caused several outbreaks in healthcare settings. Here, we report the first case of C. auris candidemia in Taiwan in a patient with a two-month history of hospitalization in Vietnam. We performed further investigation on the isolate from the present case as well as the previously reported C. auris isolate identified from a wound in 2018 in Taiwan, which was the first case reported in Taiwan. Both C. auris isolates were found to be susceptible to fluconazole, amphotericin B, and echinocandins. Additionally, mutations in ERG11 or FKS1 were not detected in either isolate. Microsatellite genotyping revealed that both isolates belonged to the South Asian clade. In recent years, C. auris has emerged as a global concern, and differences in clades and susceptibility patterns mandate further awareness and systematic surveillance.
Introduction
Candida auris is an emerging pathogenic yeast that is unique because of its multidrug resistance and easily transmittable potency in nosocomial environments. C. auris was first identified from external ear discharge of a Japanese patient in 2009 [Citation1], and has been documented in over 40 countries across six continents [Citation2]. C. auris has resulted in several outbreaks in hospitals and other healthcare facilities [Citation3]. It can cause invasive infections and can colonize skin and mucosa. Bloodstream infections are the most common invasive infections and are associated with high mortality rates, ranging from 30 to 60% [Citation4,Citation5]. Most isolates of C. auris are multidrug-resistant and in particular exhibit high resistance to fluconazole. Although echinocandins are the recommended regimen for treating C. auris infections [Citation6], isolates resistant to all three classes of antifungal agents (azoles, polyenes, and echinocandins) have been reported [Citation7].
C. auris isolates can be divided into four major clades: South Asia (I), East Asia (II), South Africa (III), and South America (IV) [Citation8]. A potential fifth clade was reported in Iran in 2019 [Citation9]. The genomic epidemiology of C. auris has been described on a global scale but little is known about its genomic epidemiology in Southeast Asia [Citation10–12]. C. auris was not identified in a multicenter survey of 5064 clinical isolates of Candida species collected in Taiwan between 1999 and 2016, which were analyzed by DNA sequencing [Citation13]. The first reported C. auris isolate in Taiwan was isolated from a patient with a superficial wound infection in 2018; however, no clade information was available [Citation14]. Here, we report the first case of invasive C. auris causing candidemia in a patient with a two month history of hospitalization in Vietnam. Furthermore, we investigated the epidemiological relatedness and microbiological characteristics of the two C. auris isolates reported in Taiwan, the previously reported isolate [Citation14], and the isolate from the present case.
Materials and methods
Laboratory investigations
Two C. auris isolates, one from Kaohsiung Medical University Hospital (KMUH; Kaohsiung, Taiwan) and the other from Chi Mei Medical Center (CMMC; Tainan, Taiwan), which was previously reported [Citation14], were enrolled for analysis in the present study. The two isolates were identified by matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF MS) (Bruker Daltonik GmbH, Bremen, Germany) initially. Species identification was further confirmed by PCR following by sequencing using ITS1/ITS4 primers for the variable internal transcribed spacers (ITS1 and ITS2 regions) and 5.8SrDNA gene; and NL1/NL4 primers used to detect the D1/D2 region of the 28S ribosomal DNA(rDNA) [Citation15,Citation16], followed by GenBank basic local alignment search tool (BLAST) pairwise sequence alignment (http://www.ncbi.nlm.nih.gov/BLAST/Blast.cgi). The ITS and D1/D2 region sequences of two clinical isolates from the present study and C. auris isolates from different countries, retrieved from GenBank were included in the phylogenetic tree along with closely related Candida species, such as C. haemulonii complex and C. pseudohaemulonii. Antifungal susceptibility testing was performed using the broth microdilution method following the guidelines of the Clinical and Laboratory Standards Institute (CLSI) M27-S4 [Citation17] and using the Sensititre YeastOne panel (Trek Diagnostic System, Cleveland, OH, USA). Because there are currently no established susceptibility breakpoints for C. auris, the minimum inhibitory concentrations (MICs) of the isolates were interpreted according to the MIC breakpoints suggested by the US Centers for Disease Control and Prevention (CDC) [Citation18]. ERG11 and FKS1 gene amplification and sequencing were performed for the two isolates [Citation16]. The sequences were analyzed using Mutation Surveyor with GenBank. The ERG11 and FKS1 sequences of representative strains of each genotype in this study were deposited in GenBank (). Primers for amplification the ITS, D1/D2 region, ERG11 and FKS1 were listed in Appendix ().
Microsatellite genotyping of C. auris isolates was performed using a recently developed short tandem repeat (STR) method [Citation19]. The copy numbers of the 12 markers were determined using GeneMapper software (Applied Biosystems, Waltham, MA, USA). Relatedness between isolates was analyzed using BioNumerics software version 7.6.3 (bioMérieux, Marcy-l'Étoile, France) via the unweighted pair group method with arithmetic averages (UPGMA) using the multistate categorical similarity coefficient.
Case presentation
A 64-year-old male patient with diabetes mellitus and prior history of ischaemic stroke was hospitalized at the University Medical Center in Ho Chi Minh City, Vietnam, in May 2021. The patient is a Taiwanese businessman who has lived in Vietnam for the past five years. Initially, he presented with disturbance of consciousness, and septic encephalopathy was suspected. While hospitalized, the patient went into cardiac arrest because of ventricular fibrillation, and he gained consciousness and spontaneous circulation was restored after resuscitation. Thereafter, he underwent renal replacement therapy because of acute kidney injury after cardiac arrest and was placed on mechanical ventilation in an intensive care unit (ICU). He received several antibacterial agents for nosocomial infections. Additionally, caspofungin was prescribed for yeast (no final identification) isolated from culture of bronchoalveolar lavage and penile ulcer pus. He was then transferred to a tertiary medical centre in Kaohsiung, Taiwan, on 10 July 2021. He had an indwelling central venous catheter (CVC) and a haemodialysis catheter. The CVC was removed on the 12th day after transfer. On the 21st day after transfer, the patient developed a fever and was hypotensive; broad-spectrum antibiotics and vasopressors (norepinephrine) were initiated. A preliminary report of blood culture yielded a yeast-like organism that was further identified as C. auris. The patient was started on anidulafungin (200 mg on day 1 and then 100 mg once daily) on the 26th day after transfer. Repeated blood cultures were negative on the 27th day after transfer. The treatment course for anidulafungin was 15 days. C. auris was not isolated from the swap samples of the skin, axilla, or groin of the patient. The patient was transferred to the respiratory care unit for weaning from ventilator. Surveillance cultures of patients in the ICU and environmental swabs were negative for C. auris; no additional infected or colonized cases of C. auris were identified at the centre.
Results
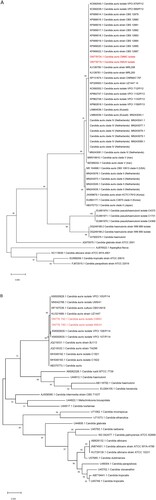
The two isolates, KMUH and CMMC, were identified as C. auris by MALDI-TOF MS with log (score) values of 1.84 and 1.82, respectively. Furthermore, conclusive identification was confirmed by nucleotide sequencing of the ITS and D1/D2 regions. The ITS and D1/D2 regions phylogenetic analyses showed that the two isolates in the present study were to be identical to many other clinical strains of C. auris from all over the world ((A,B)).
Figure 1. Phylogenetic tree generated by Maximum likelihood analysis using (A) ITS and (B) D1/D2 region of the Candida auris strains with closely related Candida species. The percentages of replicate trees in which the associated taxa clustered together in the bootstrap test (10,000 replicates) are indicated at the branches. The scale bar indicates the nucleotide substitutions per site. Two isolates in the present study are highlighted in red and two isolates are related to C. auris as it falls in the same clade.
Results of antifungal susceptibility testing are shown in . The two isolates, KMUH and CMMC, exhibited high MICs for fluconazole of 8 and 16 mg/L, respectively, and both MICs were interpreted as susceptible according to the tentative breakpoints suggested by CDC [Citation18]. Both isolates had low MICs for amphotericin B, echinocandins (anidulafungin and micafungin), and 5-flucytosine. The two isolates were screened for F126, Y132, K143, and S639, which confer resistance to azoles and echinocandins, respectively [Citation5]. Mutations in ERG11 or FKS1 were not detected in either isolate ((A,B)). STR genotypes and UPGMA dendrogram revealed that the two isolates were closely related to the South Asian clade (I) (). Despite being related to the South Asian clade, the CMMC isolate showed small variations (copy number 2) in the STR marker M3-IIc.
Figure 2. (A) Sequence alignments of ERG11 gene in C. auris isolates. (B) Sequence alignments of FKS1 hot spot 1 region in C. auris isolates. Mutations in ERG11 or FKS1 were not detected in either isolate.
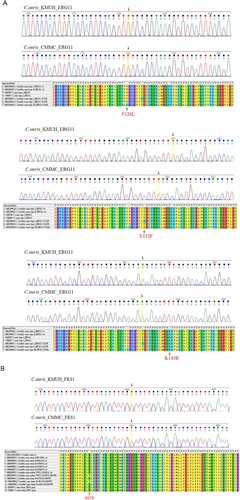
Figure 3. Short tandem repeat genotypes of 44 Candida auris isolates. Unweighted pair group method with arithmetic averages dendrogram of both isolates (KMUH and CMMC) and representative isolates from the South Asian clade and four clades are shown. Abbreviations: UK, United Kingdom; SA, South Africa.
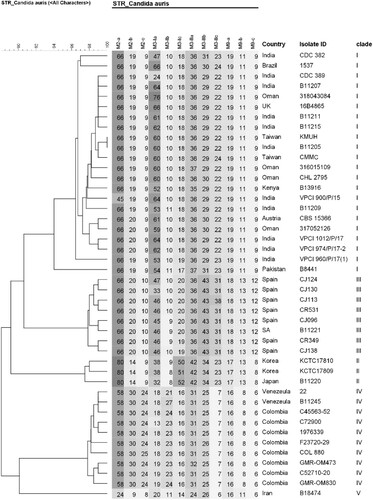
Table 1. Characteristics and antifungal susceptibility profiles of Candida auris isolates in Taiwan.
The sequence data for the ITS and D1/D2 regions, the ERG11 and FKS1 genes sequences were submitted to GenBank (ITS accession number: ON778716-ON778717 for KMUH isolate and ON778724-ON778725 for CMMC isolate; D1/D2 region accession number: ON778740-ON778741 for KMUH isolate and ON778742-ON778743 for CMMC isolate; ERG11 accession number: ON853792, ON853793 for KMUH and CMMC isolate, respectively; FKS1 accession number: ON853794, ON853795 for KMUH and CMMC isolate, respectively).
Discussion
Here, we present the first case of an invasive C. auris infection in Taiwan. The patient had multiple risk factors [Citation20] for susceptibility to C. auris infection including mechanical ventilation, prolonged ICU stay, central line indwelling, and prior antifungal exposure. The epidemiological details of the two C. auris cases in Taiwan were different; in our case, the patient had a recent history of prolonged hospitalization in Vietnam, whereas in the other seemed to be indigenous and the patient had no exposure to any foreign healthcare worker [Citation14].
Four distinct clades of C. auris are associated with specific geographic distribution [Citation21–23]. In Asia, isolates from India and Pakistan mainly belong to the South Asian clade, whereas isolates from Japan and South Korea belong to the East Asian clade. Isolates from different regions of China have been reported to belong to the South Asian and South African clades [Citation24,Citation25]. Although several outbreaks of C. auris have been documented in South Asia [Citation5,Citation20], epidemiological reports for Southeast Asia are limited; cases have been only reported in Malaysia [Citation10,Citation26], Thailand [Citation11], and Singapore [Citation12,Citation27]. The geographical locations of cases reported in Southeast Asia and neighbouring countries are shown in . Tan et al. [Citation12] reported seven cases in Singapore, and most isolates belonged to the South Asian clade. Borman et al. [Citation32] analyzed C. auris isolates in the United Kingdom and compared them to strains from diverse geographic origins; the isolate reported from Malaysia (KU321688) belonged to the same lineage as India and Kuwait, which is consistent with the South Asian clade. Fifteen cases of C. auris colonization were reported in Hong Kong [Citation30], and all the isolates belonged to the South Asian clade. In the present study, using STR typing, it was confirmed that the two isolates from Taiwan belonged to the South Asian clade (). The performance of the STR-based genotyping technique is comparable to that of whole genome sequencing [Citation19]. STR genotyping is a rapid, reliable, and cost-effective assay, and has accurate discrimination power regarding the relatedness of isolates. Several studies have used this method for epidemiological and outbreak surveys [Citation33,Citation34]. The KMUH isolate had the STR genotype 17, while the CMMC isolate had small variations (copy number < 5) in the STR marker M3-IIc (). Given that persistent skin colonization of C. auris lasting 1–3 months, as well as environmental contamination lasting 2–3 months, have been described previously [Citation35], we cannot exclude the possibility that our patient was already colonized with C. auris in Vietnam, even though only one possible C. auris case has been reported from Vietnam [Citation36]. Further genomic epidemiology of C. auris is required in Taiwan and Vietnam.
Figure 4. Distribution of reported C. auris cases in Southeast Asia and neighbouring regions/countries. Case counts were based on an epidemiological report [Citation22] and studies from Japan [Citation28], South Korea [Citation29], Taiwan [Citation14] (including the present study), China [Citation25], Hong Kong [Citation30], Malaysia [Citation10], Singapore [Citation12], Thailand [Citation11], Bangladesh [Citation31], and India and Pakistan [Citation5].
![Figure 4. Distribution of reported C. auris cases in Southeast Asia and neighbouring regions/countries. Case counts were based on an epidemiological report [Citation22] and studies from Japan [Citation28], South Korea [Citation29], Taiwan [Citation14] (including the present study), China [Citation25], Hong Kong [Citation30], Malaysia [Citation10], Singapore [Citation12], Thailand [Citation11], Bangladesh [Citation31], and India and Pakistan [Citation5].](/cms/asset/92956dd8-b1a2-4f8f-ab2d-11ac5efb22af/temi_a_2100280_f0004_oc.jpg)
Conventional phenotypic methods commonly misidentify C. auris as other Candida species, such as Candida haemulonii, Candida famata, Candida catenulata, Candida sake, Rhodotorula glutinis, and Saccharomyces cerevisiae [Citation13]. Accurate identification of C. auris can be achieved using MALDI-TOF and molecular identification based on sequencing of ITS regions of rDNA and the D1/D2 domain of 28S rDNA [Citation37].
MALDI-TOF MS is an efficient and reliable diagnostic tool for C. auris, given that it presents with reference spectra in database or research use only library (RUO) [Citation13]. Additionally, MALDI-TOF has advantage of reduced turnaround time compared to conventional or molecular methods [Citation38]. rDNA sequencing of ITS and 28S D1/D2 region will accurately identify C. auris to the species level, but it is not routinely available in clinical laboratories due to higher technical and instrumental requirements [Citation38]. Our two isolates (KMUH and CMMC) were confirmed by MALDI-TOF MS and rDNA sequencing methods. Laboratories in Asia mainly rely on conventional phenotypic and morphological methods for identification of fungi, and very few laboratories rely on DNA sequencing (16.9%) or MALDI-TOF MS (12.3%) for isolate identification [Citation39]. Therefore, the true burden of C. auris infections in Asia remains largely unknown.
Most C. auris isolates are resistant to fluconazole [Citation5]. Three hot spot mutations (Y132F, K143R, and F126L) have been reported in ERG11 in fluconazole-resistant C. auris strains belonging to different genetic clades [Citation5]; however, these mutations were not detected in the present study in either isolate (). Most South Asian clade I isolates are resistant to fluconazole and demonstrate high MICs for polyenes [Citation40–42]. However, fluconazole-susceptible C. auris isolates belonging to the South Asian clade (I) with low MICs for azoles have been reported [Citation33,Citation43]. The two C. auris isolates in Taiwan had elevated MICs for fluconazole but were still susceptible to fluconazole. Majority of the South Asian clade isolates from India and Pakistan were resistant to fluconazole (97–100%) with varying resistance to polyenes (7.9–93.7%) [Citation44,Citation45], respectively; however, only one South Asian clade isolate in China [Citation25,Citation43] and two isolates in Taiwan ([Citation14] and the present report) were susceptible to all three classes of antifungal agents. Whether a subclade exhibiting low MICs for fluconazole within the South Asian clade has circulated in Taiwan and Vietnam requires further investigation.
Travel restrictions due to the ongoing coronavirus disease (COVID-19) pandemic were expected to reduce the transmission of C. auris between countries. However, superinfection cases and outbreaks of C. auris in COVID-19 care facilities have been reported in several countries [Citation46–49], and were associated with high mortality rates, up to 60% [Citation46]. Owing to long-term survival of C. auris on inanimate surfaces, contact precautions are crucial for the prevention of C. auris transmission in healthcare settings [Citation8,Citation13]. Fortunately, in the present case, the patient was cared for in a single room with contact precautions since being transferred to our hospital according to the COVID-19 quarantine policy, and there was no indication of hospital transmission. In addition to COVID-19 screening, active surveillance for C. auris should be considered for high-risk patients [Citation3,Citation13].
Conclusions
In summary, we presented the first case of C. auris candidemia in Taiwan. STR genotyping revealed that two isolates from Taiwan, the previously reported isolate and the isolate from the present case, belonged to the South Asian clade. The prevalence of C. auris in Southeast Asia may be underestimated, and development of quality mycology laboratories and further surveillance are required.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki, and reviewed and approved by the Institutional Review Board of Kaohsiung Medical University Hospital (IRB no. KMUHIRB-E(I)-20210303).
Informed Consent Statement
Informed consent was obtained from the patient’s next of kin.
Acknowledgments
The authors thank Hung-Jen Tang for kindly providing the Candida auris isolate from Chi Mei Medical Center, Tainan, Taiwan.
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- Satoh K, Makimura K, Hasumi Y, et al. Candida auris sp. nov., a novel ascomycetous yeast isolated from the external ear canal of an inpatient in a Japanese hospital. Microbiol Immunol. 2009;53:41-44.
- Centers for Disease Control and Prevention: General Information about Candida auris: Tracking Candida auris [cited 2021 Dec 27]. Available from: https://www.cdc.gov/fungal/candida-auris/tracking-c-auris.html. 2021.
- Ahmad S, Alfouzan W. Candida auris: epidemiology, diagnosis, pathogenesis, antifungal susceptibility, and infection control measures to combat the spread of infections in healthcare facilities. Microorganisms. 2021;9:807.
- Osei Sekyere J. Candida auris: A systematic review and meta-analysis of current updates on an emerging multidrug-resistant pathogen. Microbiologyopen. 2018;7:e00578.
- Lockhart SR, Etienne KA, Vallabhaneni S, et al. Simultaneous emergence of multidrug-resistant candida auris on 3 continents confirmed by whole-genome sequencing and epidemiological analyses. Clin Infect Dis. 2017;64:134–140.
- Farmakiotis D, Kontoyiannis DP. Epidemiology of antifungal resistance in human pathogenic yeasts: current viewpoint and practical recommendations for management. Int J Antimicrob Agents. 2017;50:318–324.
- Ostrowsky B, Adams E, et al. Candida auris isolates resistant to three classes of antifungal medications - New York, 2019. MMWR Morb Mortal Wkly Rep. 2020;69:6–9.
- Du H, Bing J, Hu T, et al. Candida auris: epidemiology, biology, antifungal resistance, and virulence. PLoS Pathog. 2020;16:e1008921.
- Chow NA, de Groot T, Badali H, et al. Potential fifth clade of Candida auris, Iran, 2018. Emerg Infect Dis. 2019;25:1780–1781.
- Mohd Tap R, Lim TC, Kamarudin NA, et al. A fatal case of Candida auris and Candida tropicalis candidemia in neutropenic patient. Mycopathologia. 2018;183:559–564.
- Chayakulkeeree M, Ungulkraiwit P, Chongtrakool P, et al. The first case of Candida auris fungemia in Thailand. in: Gangneux JP, Lortholary O, Cornely OA et al. 9th trends in medical mycology held on 11-14 October 2019, nice, France, organized under the auspices of EORTC-IDG and ECMM. J Fungi. 2019;5:95.
- Tan YE, Teo JQ, Rahman NBA, et al. Candida auris in Singapore: genomic epidemiology, antifungal drug resistance, and identification using the updated 8.01 VITEK®2 system. Int J Antimicrob Agents. 2019;54:709–715.
- Lu PL, Liu WL, Lo HJ, et al. Are we ready for the global emergence of multidrug-resistant Candida auris in Taiwan? J Formos Med Assoc. 2018;117:462–470.
- Tang HJ, Lai CC, Lai FJ, et al. Emergence of multidrug-resistant Candida auris in Taiwan. Int J Antimicrob Agents. 2019;53:705–706.
- Leaw SN, Chang HC, Sun HF, et al. Identification of medically important yeast species by sequence analysis of the internal transcribed spacer regions. J Clin Microbiol. 2006;44:693–699.
- Chowdhary A, Prakash A, Sharma C, et al. A multicentre study of antifungal susceptibility patterns among 350 Candida auris isolates (2009-17) in India: role of the ERG11 and FKS1 genes in azole and echinocandin resistance. J Antimicrob Chemother. 2018;73:891–899.
- Clinical and Laboratory Standards Institute. Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeasts: Fourth Informational Supplement M27-S4; CLSI: Wayne, PA, USA. 2012.
- Centers for Disease Control and Prevention: Candida auris: Laboratorians and Health Professionals: Antifungal Susceptibility Testing. [cited 2021 Dec 27]. Available from: https://www.cdc.gov/fungal/candida-auris/c-auris-antifungal.html. 2020.
- de Groot T, Puts Y, Berrio I, et al. Development of Candida auris short tandem repeat typing and its application to a global collection of isolates. mBio. 2020;11:e02971–19.
- Rudramurthy SM, Chakrabarti A, Paul RA, et al. Candida auris candidaemia in Indian ICUs: analysis of risk factors. J Antimicrob Chemother. 2017;72:1794–1801.
- Rhodes J, Fisher MC. Global epidemiology of emerging Candida auris. Curr Opin Microbiol. 2019;52:84–89.
- Chen J, Tian S, Han X, et al. Is the superbug fungus really so scary? A systematic review and meta-analysis of global epidemiology and mortality of Candida auris. BMC Infect Dis. 2020;20:827.
- Desoubeaux G, Coste AT, Imbert C, et al. Overview about Candida auris: what's up 12 years after its first description? J Mycol Med. 2022;32:101248.
- Tian S, Bing J, Chu Y, et al. Genomic epidemiology of Candida auris in a general hospital in Shenyang, China: a three-year surveillance study. Emerg Microbes Infect. 2021;10:1088–1096.
- Bing J, Wang S, Xu H, et al. A case of Candida auris candidemia in Xiamen, China, and a comparative analysis of clinical isolates in China. Mycology. 2022;13:68–75.
- Ding CH, Situ SF, Steven A, et al. The pitfall of utilizing a commercial biochemical yeast identification kit to detect Candida auris. Ann Clin Lab Sci. 2019;49:546–549.
- Tan YE, Tan AL. Arrival of Candida auris fungus in Singapore: report of the first 3 cases. Ann Acad Med Singapore. 2018;47:260–262.
- Sekizuka T, Iguchi S, Umeyama T, et al. Clade II Candida auris possess genomic structural variations related to an ancestral strain. PLoS One. 2019;14:e0223433.
- Kwon YJ, Shin JH, Byun SA, et al. Candida auris clinical isolates from South Korea: identification, antifungal susceptibility, and genotyping. J Clin Microbiol. 2019;57:e01624–18.
- Tse H, Tsang AKL, Chu YW, et al. Draft genome sequences of 19 clinical isolates of Candida auris from Hong Kong. Microbiol Resour Announc. 2021;10:e00308–20.
- Dutta S, Rahman MH, Hossain KS, et al. Detection of Candida auris and its antifungal susceptibility: first report from Bangladesh. IMC Journal of Medical Science. 2020;13:18–22.
- Borman AM, Szekely A, Johnson EM. Isolates of the emerging pathogen Candida auris present in the UK have several geographic origins. Med Mycol. 2017;55:563–567.
- de Almeida JN Jr., Francisco EC, Hagen F, et al. Emergence of Candida auris in Brazil in a COVID-19 intensive care unit. J Fungi (Basel). 2021;7:220.
- Steinmann J, Schrauzer T, Kirchhoff L, et al. Two Candida auris cases in Germany with no recent contact to foreign healthcare-epidemiological and microbiological investigations. J Fungi (Basel). 2021;7:380.
- Jeffery-Smith A, Taori SK, Schelenz S, et al. Candida auris: a review of the literature. Clin Microbiol Rev. 2017;31:e00029–17.
- Xie O, Streitberg R, Hughes C, et al. Candida duobushaemulonii sepsis and Candida auris co-isolation following hospitalisation in Vietnam. Pathology. 2020;52:590–591.
- Centers for Disease Control and Prevention: Candida auris: Laboratorians and Health Professionals [cited 2021 Dec 27]. Available from: https://www.cdc.gov/fungal/candida-auris/identification.html. 2020.
- Mahmoudi S, Agha Kuchak Afshari S, Aghaei Gharehbolagh S, et al. Methods for identification of Candida auris, the yeast of global public health concern: A review. J Mycol Med. 2019;29:174–179.
- Chindamporn A, Chakrabarti A, Li R, et al. Survey of laboratory practices for diagnosis of fungal infection in seven Asian countries: An Asia Fungal Working Group (AFWG) initiative. Med Mycol. 2018;56:416–425.
- Szekely A, Borman AM, Johnson EM. Candida auris isolates of the southern Asian and South African lineages exhibit different phenotypic and antifungal susceptibility profiles in vitro. J Clin Microbiol. 2019;57:e02055–18.
- Chow NA, Munoz JF, Gade L, et al. Tracing the evolutionary history and global expansion of Candida auris using population genomic analyses. mBio. 2020;11:e03364–19.
- Maphanga TG, Naicker SD, Kwenda S, et al. In vitro antifungal resistance of Candida auris isolates from bloodstream infections, South Africa. Antimicrob Agents Chemother. 2021;65:e0051721.
- Wang X, Bing J, Zheng Q, et al. The first isolate of Candida auris in China: clinical and biological aspects. Emerg Microbes Infect. 2018;7:93.
- Shastri PS, Shankarnarayan SA, Oberoi J, et al. Candida auris candidaemia in an intensive care unit - prospective observational study to evaluate epidemiology, risk factors, and outcome. J Crit Care. 2020;57:42–48.
- Sayeed MA, Farooqi J, Jabeen K, et al. Clinical spectrum and factors impacting outcome of Candida auris: a single center study from Pakistan. BMC Infect Dis. 2019;19(384.
- Chowdhary A, Tarai B, Singh A, et al. Multidrug-Resistant Candida auris infections in critically ill coronavirus disease patients, India, April-July 2020. Emerg Infect Dis. 2020;26:2694–2696.
- Prestel C, Anderson E, Forsberg K, et al. Candida auris outbreak in a COVID-19 speciality care unit – Florida, July-August 2020. MMWR Morb Mortal Wkly Rep. 2021;70:56–57.
- Villanueva-Lozano H, Trevino-Rangel RJ, Gonzalez GM, et al. Outbreak of Candida auris infection in a COVID-19 hospital in Mexico. Clin Microbiol Infect. 2021;27:813–816.
- Magnasco L, Mikulska M, Giacobbe DR, et al. Spread of carbapenem-resistant gram-negatives and Candida auris during the COVID-19 pandemic in critically ill patients: one step back in antimicrobial stewardship? Microorganisms. 2021;9:95.
Appendix
Table A1. GenBank accession numbers, ERG11 and FKS1 genotypes, and clade information of representative strains included in this study.
Table A2. Sequences of primers used in the study.
