ABSTRACT
Convergence of mcr and carbapenemase genes has been sporadically detected in Enterobacter cloacae complex (ECC) with an upward trend. However, the state of the epidemic and underlying mechanism of such convergence has been poorly understood. In this study, the co-occurrence of MCR and carbapenemases was systematically analyzed in 230 clinical ECC isolates collected between 2000 and 2018 together with a global dataset consisting of 3,559 ECC genomes compiled from GenBank. We identified 48 mcr-9/mcr-10-positive isolates (MCR-ECC) (20.9%) in our collection, and a comparable ratio of MCR-ECC (720/3559, 20.2%) was detected in the global dataset. A high prevalence of carbapenemase-producing MCR-ECC (MCR-CREC) was further identified in the MCR-ECC of both datasets (16/48, 33.3%; 388/720, 53.9%), demonstrating a frequent convergence of mcr-9/10 and carbapenemase genes in ECC worldwide. An epidemic IncHI2/2A plasmid with a highly conserved backbone was identified and largely contributed to the dissemination of mcr-9 in ECC worldwide. A highly conserved IncX3-type NDM-1-carrying plasmid and IncN-type IMP-4-carrying plasmid were additionally detected in MCR-CREC isolated in China. Our surveillance data showed that MCR-CREC emerged (in 2013) much later than MCR-ECC (in 2000), indicating that MCR-CREC could be derived from MCR-ECC by additional captures of carbapenemase-encoding plasmids. Tests of plasmid stability and incompatibility showed that the mcr-9/mcr-10-encoding plasmids with the NDM-1-encoding plasmids stably remained in ECC but incompatible in Escherichia coli, suggesting that the convergence was host-dependent. The findings extend our concern on the convergence of resistance to the last resort antibiotics and highlight the necessity of continued surveillance in the future.
Introduction
Clinical infections caused by multidrug-resistant (MDR) bacteria with an increasing trend have become a critical threat to the public health network [Citation1]. In particular, the dramatic increase of carbapenem resistance worldwide has greatly compromised the efficacy of carbapenems and prompted renewed attention to the importance of the last-line antibiotics colistin (polymyxin E). However, with the heavy consumption of colistin in veterinary medicine and clinical settings, colistin-resistant isolates have emerged globally [Citation2–4], causing a therapeutic challenge in the clinical setting.
Colistin resistance was thought to be intrinsic in the past until the report of the first mobilized colistin resistance (mcr) gene mcr-1 in 2015 [Citation5]. Since then, an addition of nine mcr genes has been described and was assigned new numerical designations, namely mcr-2 ∼ mcr-10 [Citation6–14]. Plasmid-mediated mcr genes have been widely spread in various bacterial species, the vast majority belonging to Escherichia coli, followed by other Enterobacteriaceae (e.g. Klebsiella pneumoniae, Enterobacter spp., and Salmonella enterica), and Aeromonas hydrophila [Citation9, Citation11, Citation15–17]. Currently, mcr-1 is the most widely disseminated member of the mcr family, followed by mcr-9, mcr-3, and mcr-5 [Citation18]. Up to now, all mcr genes except for mcr-6 are frequently harboured on conjugative plasmids or ColE-type plasmids mobilizable with helper plasmids [Citation9, Citation19]. The mcr-1-harboring plasmids have been extensively studied and assigned to a large number of incompatibility groups, of which IncHI2, IncX4, and IncI2 were the predominant ones responsible for the dissemination of the mcr-1 gene among E. coli isolates [Citation20]. Most of the mcr-9-carrying plasmids were typed to be IncHI2 [Citation17], while the type of mcr-5-encoding plasmids was highly diverse [Citation8, Citation21].
E. coli and K. pneumoniae are the major reservoirs for most of the mcr genes, while mcr-9 and mcr-10 have been frequently reported in Enterobacter cloacae complex (ECC) [Citation14, Citation16, Citation22, Citation23]. ECC is an important nosocomial pathogen capable of causing a wide variety of infections, such as pneumonia, urinary tract infections, and septicemia [Citation24]. Molecular typing methods have identified more than 20 species/phylogenetic clusters in the complex [Citation25]. Epidemiological studies show that Enterobacter hormaechei is the predominant species among ECC in the clinical setting, contributing to increased morbidity and mortality in hospitalized patients, particularly when infected by carbapenem-resistant isolates [Citation24, Citation26].
Recently, the co-occurrence of carbapenemases and MCR in a single strain has been reported sporadically [Citation27, Citation28], especially the combination of genes encoding metallo-beta-lactamases and MCR-9. Up to date, such genotype (i.e. coexistence of carbapenemase genes and mcr-9) has been found in Enterobacter spp., Klebsiella spp., Escherichia spp., and Citrobacter spp. [Citation17, Citation22, Citation29–31]. An MDR E. hormaechei isolate co-harboring blaNDM-1 and mcr-9 genes was identified in a retrospective screen of carbapenemase- and MCR-producing Enterobacterales recovered from a tertiary hospital in Hangzhou, China [Citation30]. Two carbapenemase genes (blaIMP-4 and blaNDM-1) were simultaneously detected in an mcr-9-positive E. hormaechei strain [Citation32]. In particular, a high incidence of carbapenemases, especially metallo-beta-lactamases, in mcr-9-positive genomes was detected in Enterobacter spp. collected from a tertiary hospital recently [Citation33]. These data consistently suggest that Enterobacter spp. could be an important reservoir for carbapenemase genes and mcr-9. However, the underlying mechanism involved has been poorly understood. Herein, we aimed to systematically study the co-existence of mcr and carbapenemase genes in ECC isolates collected between 2000 and 2018 in a tertiary hospital of China together with a global dataset retrieved from GenBank. The plasmidom encoding mcr and carbapenemase genes were dissected, and their incompatibility and stability were tested to explore the co-occurrence mechanism.
Materials and methods
Bacterial isolates, mcr-positive-ECC (MCR-ECC) bacteria identification, and clinical data collection
ECC isolates were collected at a tertiary hospital of China between 2000 and 2018, and have been species-typed previously [Citation34]. The presence of mcr genes including mcr-1 ∼ mcr-10 were assessed by using primers reported previously [Citation16]. The mcr positive isolates were determined as MCR-ECC. Metadata including patients’ gender and age, dates of specimen collection, and specimen types were recorded.
Antimicrobial susceptibility testing
The minimum inhibitory concentrations (MICs) of 14 antibiotics were evaluated according to the guidelines of the Clinical and Laboratory Standards Institute (CLSI) (M100-S30, 2020). MICs of colistin and tigecycline were determined by the broth dilution method, and the results were interpreted according to European Committee on Antimicrobial Susceptibility Testing (EUCAST) (version 10.0) criteria (https://eucast.org/clinical_breakpoints/). The other 12 antibiotics were determined using the agar dilution method. E. coli ATCC 25922 and Pseudomonas aeruginosa ATCC 27853 were used as quality control standards. MCR-ECC isolates resistant to imipenem and/or meropenem (MIC ≥ 4 mg/L) were determined as carbapenemase-producing MCR-ECC (MCR-CREC).
Whole genome sequencing (WGS) and data analysis
Genomic DNA was extracted from all of MCR-ECC isolates using a Gentra Puregene Yeat/Bact. Kit (Qiagen, San Francisco/Bay area, CA, USA) and subjected to WGS in an Illumina novaseq 6000 system (Illumina, San Diego, United States) to obtain 150-bp paired-end reads. Raw reads were trimmed using Trimmomatic [Citation35] and then assembled using SPAdes v3.12.0 [Citation36]. MCR-CREC isolates were further sequenced by a Nanopore PromethION platform (Nanopore, Oxford, UK) following a 10-Kbp library protocol. The hybrid assembly of both short Illumina reads and long PromethION reads was performed using Unicycler v0.4.8 [Citation37]. The assembled contigs were circularized by Circlator [Citation38] and plasmid circularity was confirmed by PCR () followed by Sanger sequencing. The assembled genomes were annotated using Prokka v1.17 [Citation39] and were analyzed using Centre for Genomic Epidemiology (CGE, https://cge.cbs.dtu.dk/services/) for detecting the presence of antimicrobial resistance (AMR) genes (ResFinder v3.2) [Citation40] and plasmid replicon typing (PlasmidFinder v2.1) [Citation41]. The pair-wise average nucleotide identity (ANI) between the queries and type/protype genomes of Enterobacter species was determined by FastANI (https://github.com/ParBLiSS/FastANI), with a cut-off of 95% [Citation42]. PubMLST (https://pubmlst.org/) was used for multilocus sequence typing (MLST) analysis, and ParSNP v1.2 [Citation43] was used to align the core genomes. The synteny analysis was performed and visualized by using Easy Figure 2.2.2 [Citation44]. Comparative analysis of plasmids was determined using the BLAST Ring Image Generator (BRIG) [Citation45]. To evaluate the presence of a plasmid in draft genomes, contigs were blasted against the reference using Blastn v2.9.0 [Citation46] to calculate the coverage.
Table 1. Primers used in this study.
A total of 3,559 genomes of ECC were retrieved from the NCBI RefSeq database as of October 1 2021, and were subjected for the identification of mcr-9/mcr-10 positive isolates using Abricate v1.0.1 (https://github.com/tseemann/abricate) and ResFinder v3.2 [Citation40].
Conjugation assay
In order to determine whether colistin and carbapenem resistance was transferable, conjugation assays were carried out in LB broth with using E. coli EC600 as the recipient according to a method previously described [Citation16]. Briefly, logarithmic-phase cultures of donor and recipient cells were added to fresh LB broth and incubated overnight without shaking. Transconjugants were selected on LB agar plates containing 600 mg/L rifampicin plus 2 mg/L meropenem/imipenem and/or 2 mg/L colistin. The presence of carbapenemase and/or mcr genes in transconjugants was confirmed using PCR and Sanger sequencing, and the species of transconjugants was confirmed using MALDI-TOF Mass Spectrometry (Hexin Instrument Co., Ltd, Guangzhou, China).
Plasmid stability assay
Plasmid stability assay was performed as previously described with slight modifications [Citation47]. The donor strains were grown in antibiotic-free LB broth. Cultures grew at 37°C in a shaking bath, followed by 1:1000 dilution in fresh antibiotic-free LB broth, and passaged for 10 successive days. Cultures of day 10 were serially diluted and plated on antibiotic-free LB agar plates, and incubated overnight. Fifty colonies were randomly picked from plates for each isolate. The presence of carbapenemase and mcr genes was validated by colony-PCR.
Plasmid incompatibility assay
Plasmid incompatibility assay was performed as previously described [Citation47]. Briefly, the transconjugants bearing both carbapenemase-encoding and MCR-encoding plasmids were cultured in antibiotic-free LB broth and were grown at 37°C in a shaking bath. Overnight cultures were serially diluted and plated on antibiotic-free LB agar. Ninety-four colonies of each culture were selected for the detection of carbapenemase and mcr genes by PCR. Plasmids were considered incompatible when more than 80% of colonies lost either or both of carbapenemase-encoding and MCR-encoding plasmids.
Accession numbers
The genome sequences have been deposited into GenBank. The detail accession numbers are listed in Table S1.
Results
Prevalence of MCR-ECC among ECC population
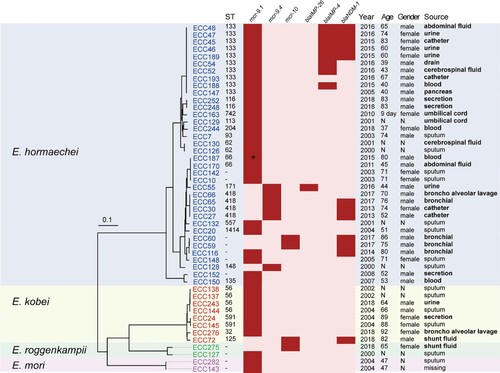
We collected 230 non-repetitive clinical ECC isolates at a tertiary hospital between 2000 and 2018. The isolates were previously assigned to eight species: E. hormaechei (n = 164), Enterobacter kobei (n = 26), Enterobacter roggenkampii (n = 17), Enterobacter cloacae (n = 13), Enterobacter asburiae (n = 2), Enterobacter ludwigii (n = 3), Enterobacter bugandensis (n = 2), Enterobacter mori (n = 2), and Enterobacter cancerogenus (n = 1) [Citation34]. Most of the isolates were obtained from sputum (20.9%, 48/230), followed by urine (14.8%, 34/230), secretion (12.2%, 28/230), and blood (10.4%, 24/230) (Table S2). Forty-eight isolates were PCR positive for mcr genes (20.9%; 48/230) [Citation48], of which 44 and four were positive for mcr-9 (19.1%) and mcr-10 (1.7%), respectively. The 48 MCR-ECC isolates belonged to four species: E. hormaechei (75.0%; 36/48), E. kobei (16.6%; 8/48), E. roggenkampii (4.2%; 2/48), and E. mori (4.2%; 2/48). They were recovered from various specimens according to the traceable metadata, and sputum accounted for the highest proportion (29.8%; 14/47), followed by urine (10.6%; 5/47). Of note, at least 28 isolates (59.6%; 28/47) were recovered from the sterile-site specimens () of patients. Of the patients positive for MCR-ECC with traceable metadata, 63.2% (24/38) were men, and 65.0% (26/40) were aged ≥ 60 years old (). The first mcr-9-producing ECC isolate was detected in 2000, which is much earlier than the emergence of mcr-10-producing isolates (in 2017).
Figure 1. Phylogenetic analysis and characteristics of the 48 MCR-ECC isolates collected in this study. A core-genome phylogenetic tree including the 48 MCR-ECC isolates was constructed and mid-point rooted. The isolates clustered into four clades corresponding to four species (blue: E. hormaechei, yellow: E. kobei, green: E. roggenkampii, purple: E. mori). The ST of each isolate is shown, and the novel ST is indicated by a dash. The absence (in pink) or presence (in red) of colistin and carbapenem resistance genes is diaplayed by a heatmap, and a truncated mutant of mcr-9 was marked with an asterisk. Clinical information of the isolates and patients are listed. The sterile-site specimens were in bold.
To understand whether mcr-9/mcr-10 are prevalent in ECC population worldwide, we further identified the two genes in a collection of 3,559 ECC genomes retrieved from the NCBI RefSeq database. We detected 720 genomes positive to mcr-9 and/or mcr-10, of which 657 (18.5%) and 70 (2.0%) were positive to mcr-9 and mcr-10, respectively. Seven of the 720 genomes co-carried both genes. The prevalence ratio was comparable with that of our collection. The MCR-ECC isolates were assigned to 10 species: E. hormaechei (72.5%, 522/720), E. roggenkampii (7.5%, 54/720), E. kobei (6.7%, 48/720), E. asburiae (4.7%, 34/720), E. cloacae (4.2%, 30/720), E. quasihormaechei (3.2%, 23/720), E. ludwigii (0.6%, 4/720), E. chengduensis (0.3%, 2/720), E. sichuanensis (0.3%, 2/720) and E. cancerogenus (0.1%, 1/720) (Table S3). The results demonstrate that mcr-9 and mcr-10 were prevalent in ECC, where E. hormaechei was the predominant species.
Emergence of MCR-CREC was later than that of MCR-ECC
The 48 MCR-ECC isolates harboured 7∼27 antibiotic resistance genes (ARGs) (median 23) (Table S1). Of these, 44 isolates carried mcr-9.1 and its variants, including mcr-9.1 (n = 37), mcr-9.2 (n = 6), and an unknown variant showing 88.7% coverage and 100% nucleotide-acid identity in comparison to the sequence of mcr-9.1 (). Close inspection of the unknown variant identified an IS15-like insertion sequence, resulting in a 1437-bp fragment left, thus was designated as Δmcr-9.1 ( and Table S1). The mcr-10 gene was confirmed in four isolates (ECC59, ECC60, ECC72, and ECC275). Carbapenemase genes were detected in 16 isolates (13 mcr-9- and three mcr-10-positive isolates), including blaNDM-1 (n = 12), blaIMP-4 (n = 8), and blaIMP-26 (n = 1) ( and Table S1). Five isolates (ECC45, ECC46, ECC47, ECC48, and ECC189) coproduced NDM-1 and IMP-4. The 16 MCR-CREC isolates were assigned to E. hormaechei (n = 15) and E. kobei (n = 1).
Among the 48 MCR-ECC isolates, 26 were resistant to colistin (54.2%), 15 to tigecycline (31.2%), and 16 to carbapenems (imipenem and/or meropenem). At least four isolates were resistant to two of the three classes (colistin + tigecycline: 4; colistin + carbapenems: 5; tigecycline + carbapenems: 8), and two were resistant to all. The MICs of the 48 MCR-ECC isolates are summarized in and Table S1. Of note, all MCR-CREC isolates were detected in 2013∼2018, which is much later than the emergence of MCR-ECC (in 2000). We, therefore, suppose that MCR-CREC might be derived from MCR-ECC by capturing the carbapenemase genes in our collection.
Table 2. Susceptibility profiles and MICs for 48 MCR-ECC strains.
Among the 720 MCR-ECC genomes retrieved from GenBank, carbapenemase genes were detected in 388 genomes (53.9%), mainly including blaIMP-4 (19.8%; 77/388), blaVIM-1 (18.0%; 70/388), blaVIM-4 (14.2%; 55/388), blaKPC-3 (11.1%; 43/388) and blaNDM-1 (10.3%; 40/388) (Table S3). Most of the genomes were assigned to E. hormaechei (77.8%; 302/388), demonstrating that the convergence of carbapenemase and mcr-9/10 genes was highly frequent in E. hormaechei.
Prevalent clones shared by MCR-ECC and MCR-CREC
The population structure of the 48 MCR-ECC isolates was highly diverse, the top three STs were ST133, ST418, and ST56 () [Citation48]. The 16 MCR-CREC were assigned to ST133 (n = 8), ST418 (n = 3), ST171 (n = 1), ST125 (n = 1) and novel STs (n = 3) ().
A total of 152 STs were identified among the 720 MCR-ECC genomes retrieved from GenBank. ST133 and ST114 were the predominant clone (4.9% for each; 35/720), followed by ST171 (4.4%; 32/720) and ST93 (3.9%; 28/720) (Table S3). Sixty-two isolates were assigned to be novel STs. Ninety-eight STs were identified in the 388 MCR-CREC genomes, of which ST171 was the predominant clone (7.0%; 27/388), followed by ST93 (5.9%; 23/388), ST873 (5.4%; 21/388) and ST78 (4.9%; 19/388) (Table S3). Collectively, these data show that some prevalent clones were shared by MCR-ECC and MCR-CREC.
An epidemic incHI2/2A plasmid highly contributes to the dissemination of mcr-9 in ECC worldwide
To identify how the carbapenemase and MCR genes were captured by MCR-CREC, we randomly chose 11 of 16 MCR-CREC isolates (eight positive to mcr-9, and three to mcr-10) for long-read sequencing to dissect the structure of plasmids. The sequencing results and assembly qualities are summarized in Table S4. The plasmidome of the 11 isolates can be classified into six groups according to Inc typing results, i.e. ECC59/ECC60 (IncHI2A, IncHI2, IncFIA, and IncX3), ECC27/ECC65 (IncFIB, IncHI2A, IncHI2, IncX3, IncFII, and IncFIB), ECC45/ECC46/ECC47/ECC48 (IncHI2, IncHI2A, IncX3, and IncN), ECC30 (IncHI2A, IncHI2, IncX3, and IncFIB), ECC72 (IncX3, IncFII, and IncFIB), and ECC116 (IncHI2A, IncHI2, IncFIB, IncR, and IncX3) (Table S4). The mcr and carbapenemase genes were located on different plasmids in each isolate, which were demonstrated by circularizing all plasmids carrying mcr and/or carbapenemase genes (see details in method) (Table S4).
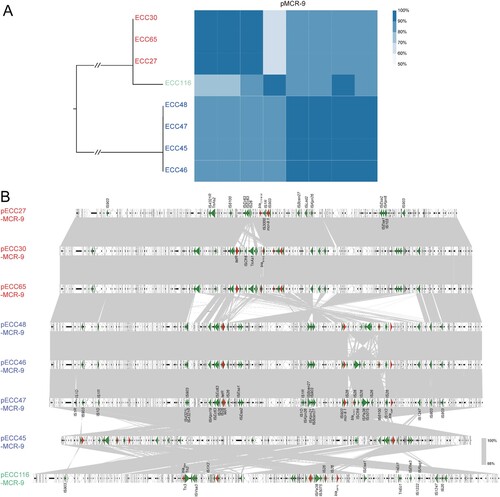
The mcr-9 gene detected on the eight long-read sequenced genomes (four ST133, three ST418, and one novel ST) located on IncHI2/2A plasmids with sizes ranging from 274,120 bp to 334,517 bp. Pairwise comparison of the eight circularized plasmids showed that the nucleotide similarity is accordant with the ST that plasmids carried by the same clone exhibited > 90% coverage and > 90% identity compared with each other, and pECC116-MCR-9 carried by the novel ST isolate shared < 70% coverage and < 90% coverage with the other plasmids (except for pECC45-MCR-9) (A). Synteny analysis showed that the diverse regions among the eight plasmids were mainly caused by mobile genetic elements (B), suggesting that they might have originated from a common ancestor with a conserved backbone, and had further co-evolved with the chromosome.
Figure 2. Comparative genomic analysis on eight complete sequenced mcr-9-harboring MCR-CRE isolates. (A) Core-genome phylogenetic tree accompanied by a similarity matrix of mcr-9 plasmids. The isolates are coloured according to the STs (blue: ST133; red: ST418; green: a novel ST). The pairwise similarity between plasmids is shown by a heatmap, and is defined as the coverage of homology regions for query plasmid (row-wise) and subject plasmid (column-wise). (B) Synteny analysis of eight circularized mcr-9 plasmids. Identical regions (i.e. 100% similarity) are highlighted by grey rectangles. Arrows with direction indicate the sense of transcription of genes. Δ represents truncated genes.
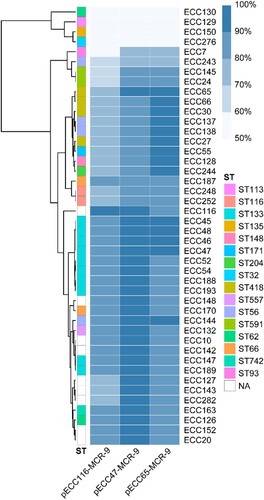
To further estimate the structure diversity of mcr-9 plasmids in our collection, the 44 mcr-9-harboring draft genomes were mapped to three representative circularized plasmids, i.e. pECC47-MCR-9, pECC65-MCR-9, and pECC116-MCR-9. According to the mapping coverage, the mcr-9 plasmids carried by the 44 isolates can be classified into at least six groups (), and 33 isolates (75%) shared a high similarity (≥ 90%) with pECC47-MCR-9 and/or pECC65-MCR-9. We also identified 309 of the 657 mcr-9-positive genomes (47.0%) retrieved from GenBank showing ≥ 90% coverage to pECC47-MCR-9 and/or pECC65-MCR-9, and these genomes were obtained from 22 countries across six continents. The 309 genomes were assigned to seven species, of which E. hormaechei was the predominant one (n = 254) (Table S5). The results indicate that mcr-9 dissemination among ECC was highly mediated by an IncHI2/2A epidemic plasmid.
Figure 3. Detection of pECC116-MCR-9, pECC47-MCR-9, and pECC65-MCR-9 in 44 mcr-9-carrying MCR-ECC. The percentage length of virulence plasmid sequences are obtained by mapping short reads of the 44 isolates to the three mcr-9 plasmids (pECC116-MCR-9, pECC47-MCR-9, and pECC65-MCR-9) used as references. The existence of plasmid is defined by that isolates having short reads mapped to ≥90% of the reference plasmid length. The isolates are clustered according to coverages using the Pearson method. STs of isolates are indicated, and NA represents the novel ST.
Highly conserved NDM-1- and IMP-4-carrying plasmids carried by MCR-CREC in China
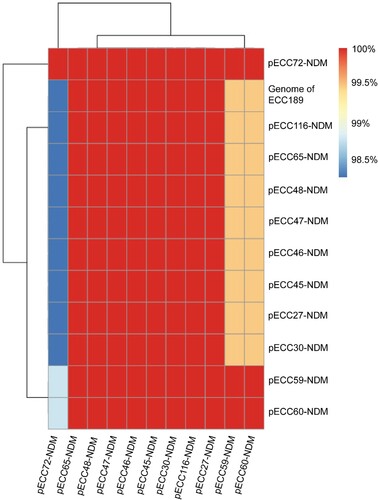
All of the 11 hybrid assembled genomes carried a blaNDM-1 gene, and an IncX3-type NDM-1-carrying circularized plasmid with the size of about 53 kb was identified in each genome, with limited sequence variations (> 98% coverage and > 90% nucleotide identity) (), suggesting the occurrence of cross-species horizontal transfer for the plasmid. The IncX3 plasmid additionally carried a bleomycin resistance gene bleMBL and a beta-lactam resistance gene blaSHV-12. Blasting the representative pECC27-NDM against NCBI nt database identified 34 genomes sharing > 90% coverage and identity, and all of them were carried by Enterobacterales, of which 32 were isolated from China (Table S6), indicating that the IncX3-type NDM-1-carrying plasmid was an epidemic plasmid in Enterobacterales in China.
Figure 4. Pairwise comparison of blaNDM-1-carrying plasmids. Twelve blaNDM-1-carrying plasmids are included in the analysis, of which 11 are circularized, and the other one (Genome of ECC189) is analyzed using the draft genome. The similarity between sequences is defined as the coverage of homology regions for query plasmid (row-wise) and subject sequences (column-wise).
An IncN-type blaIMP-4-encoding circularized plasmid with the size of about 43 kb was detected in the four hybrid assembled genomes carrying a blaIMP-4 gene (ECC45, ECC46, ECC47, and ECC48) with highly limited sequence variations (100% coverage and > 99% nucleotide-acid identity). The four isolates belong to ST133, suggestive of a clonal dissemination. pECC45-IMP-4 was almost identical to pIMP-SZ1502 (GenBank accession no. KU051707) carried by an E. coli strain (99.9% identity and 100% coverage) (Figure S1). A quinolone resistance gene qnrS1 co-located on the blaIMP-4-encoding plasmid. A pECC45-IMP-4-like plasmid was also detected in the remaining four IMP-4-positive genomes by mapping the reads to pECC45-IMP-4 (100% coverage and > 99% nucleotide-acid identity). These data indicate that the emergence of MCR-CREC was derived from MCR-ECC by capturing an IncX3-type NDM-1-carrying and/or an IncN-type IMP-4-encoding plasmid in China.
The structure of mcr-10-carrying plasmids is highly diverse
A 64,293-bp IncFIA-type circularized plasmid was detected in two of the three hybrid assembled mcr-10-positive genomes (ECC59 and ECC60), since their plasmids (pECC59-MCR-10 and pECC60-MCR-10) were almost identical (100% coverage and 99.99% nucleotide-acid identity), leaving four core-genome SNPs. They were different from a previously reported 71,775-bp IncFIA-type plasmid pMCR10_090065 (GenBank accession no. CP045065) with 25% coverage and 99.92% identity (Figure S2) [Citation14]. Blasting in the nt database showed that the IncFIA-type mcr-10 plasmid shared the highest similarity (51% coverage and 98.2% identity) with a 160,042-bp IncFIA-FII-type plasmid pRHBSTW-00675_2 (GenBank accession no. CP056767) carried by an E. cloacae strain isolated from the wastewater. A 137,379-bp IncFIB-FII-type mcr-10 circularized plasmid (pECC72-MCR-10) was identified in ECC72, and the most homologous plasmid identified in the nt database was a 240,613-bp IncFIB-FII-IncR-type plasmid pRHBSTW-00542_2 (GenBank accession no. CP056712) carried by an E. asburiae isolate, with 63% coverage and 99.44% identity.
The mcr-10 of ECC275 was detected on a 35,290-bp contig encoding an IncFIB-type replicon. Mapping the sequencing reads of ECC275 to the three circularized plasmids (pECC59-MCR-10, pECC60-MCR-10, and pECC72-MCR-10) resulted in a low coverage ranging between 24.44% and 53.08%. When mapping the 70 mcr-10-positive genomes retrieved from GenBank to the three circularized plasmids, only four showed ≥ 90% coverage to pECC72-MCR-10 (Table S7). These results indicate that the structure of mcr-10 plasmids might be highly diverse.
Plasmids encoding mcr-9/mcr-10 are stable with NDM-1-encoding plasmids in ECC but incompatible in E. coli.
Conjugation assays were performed to evaluate the transferability of plasmids encoding MCR and carbapenemases. The mcr plasmids carried by most of MCR-ECC isolates were self-transmissible to E. coli EC600, including 32 mcr-9 and four mcr-10 isolates, and all plasmids encoding carbapenemase-encoding genes can be transferred successfully. The carbapenemase and mcr genes were able to be transferred simultaneously to EC600 from all MCR-CREC isolates.
The stability of plasmids encoding carbapenemases and MCR was evaluated in the 16 MCR-CREC by a 10-day passage. Fifty colonies of each isolate were tested (see details in method). All of the mcr-, blaNDM-1-, and blaIMP-26-harboring plasmids displayed high stability, as shown by the retention rates of over 80% at the end of the passages. While blaIMP-4-harboring plasmids showed low retention rates (10%∼74%) in six strains (ECC45, ECC47, ECC48, ECC52, ECC54, and ECC188), and ECC45 lost its IMP-4 activity after 10 days with 10% retention rate, suggestive of the instability for blaIMP-4-harboring plasmids in ECC (Table S8).
Plasmid incompatibility was further assessed in the transconjugants of the 16 MCR-CREC isolates. Ninety-four colonies of each isolate were tested (see details in method). The plasmids encoding blaNDM-1 or blaIMP-26 remained in all colonies overnight culture, while those with mcr or blaIMP-4 were completely lost (Table S9). These results suggest that the mcr and carbapenemase-producing plasmids were incompatible in E. coli EC600. The observed plasmid incompatibility is consistent with the current epidemiological data that in E. coli mcr-9 and mcr-10 was rarely detected and blaNDM-1 was prevalent.
Genetic context of MCR and carbapenemase genes
At least five types of mcr-9 genetic contexts were identified in the 44 genomes (A), and a module IS903B (intact or fragmented)-mcr-9 was detected in all of them, implying the role of IS903B in the transmission of mcr-9. Type I genetic context was identical to that of pW17-1 (CP031102), and compared with the other types it exclusively encodes a two-component regulator qseB/C probably involved in mcr-9 expression, an ATPase gene, and an orf with unknown function at downstream of wbuC (encoding a cupin fold metalloprotein). Type II might be derived from type I by the deletion of orf-ATPase-qseB/C, which is almost identical to that detected on p17277A_477 (CP043927). Compared with type II, IS26-wbuC was replaced by IS3000Δ-IS1R in type III as detected in pMRVIM0813 (KP97507). Different from the three types, pcoS-pcoE-rcnA-rcnR locating at the mcr-9 upstream flanking was absent in type IV and V, and a gene encoding a hypothetical protein was detected in type IV instead. Note that, we were unable to determine the intact mcr-9 genetic environments in 34 draft genomes due to the short mcr-9-bearing contigs available for the comparison.
Figure 5. Synteny analysis for genetic environments of (A) mcr-9, (B) mcr-10, and (C) blaIMP-4. Identical regions are highlighted by dark grey rectangles (i.e. 100% similarity). Arrows with direction indicate the sense of transcription of genes, and Δ represents truncated sequences.
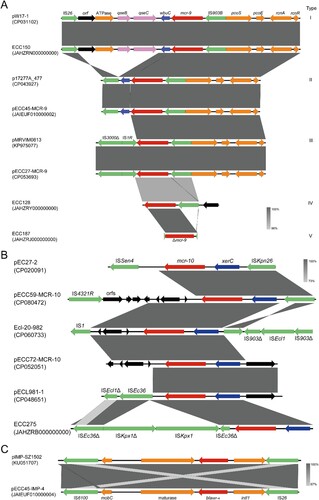
The genetic context of mcr-10 detected on pECC59-MCR-10 and pECC60- MCR-10 was identical (ISkpn26-xerC-mcr-10-orfs-IS4321R), while on pECC72-MCR-10 genes locating at upstream and downstream of xerC-mcr-10 were replaced by those encoding hypothetical proteins without any insert sequences. An ISEc36 interrupted by the insertion of an ISKpx1 and a truncated ISKpx1 were located downstream of mcr-10 in ECC275. The genetic contexts of mcr-10 identified in this study are different from those described previously (B) [Citation16].
A highly conserved genetic environment of blaNDM−1 was found in all isolates that an ISAba125 interrupted by the insertion of an IS5 lied upstream of blaNDM-1, and the fragment bleMBL-trpF-dsbD-cutA-groS-groL-orf lied downstream of blaNDM-1 (Figure S3). The blaIMP-4 was harboured by a class I integron in the four genomes with an identical structure (an IS26-truncated intI, blaIMP-4, a group II intron reverse transcriptase/maturase, a mobilization protein MobC, and an IS6100) (C).
Discussion
Carbapenems and colistin are the last resort antibiotics used for severe infections caused by multidrug-resistant bacteria. The worrisome fact is the emergence and spread of bacteria co-producing carbapenemases and mcr genes, largely limiting clinical treatment strategies and worsening outcomes. Understanding the occurrence and mechanism of such convergence of resistance to the last-resort antibiotics is imperative for making tailored strategies to control its further spread. In this study, we aimed to answer this question by systematically studying the co-existence of mcr and carbapenemase genes in ECC isolates collected between 2000 and 2018 in a tertiary hospital of China together with a global dataset retrieved from GenBank.
mcr-9 has been detected in multiple species of Enterobacterales mainly isolated from clinical samples worldwide, e.g. E. coli [Citation49], S. enterica [Citation13], ECC [Citation17], and K. pneumoniae [Citation50]. In contrast, mcr-10 was almost hosted in ECC and sporadically isolated from environment [Citation16], animal [Citation51] and clinical samples [Citation52]. Previous studies have shown that mcr-9 and mcr-10 were frequently carried in ECC by using single centre or endemic data [Citation14, Citation16, Citation22, Citation23]. To our knowledge, this is the first report demonstrating that mcr-9/mcr-10 were prevalent in ECC globally with the ratio of ca. 20%, and mcr-9 was much more prevalent than mcr-10 (18.5% vs 2.0%). The mcr-9 gene was firstly identified in a clinical S. enterica serotype Typhimurium strain isolated in 2010 [Citation13], and mcr-10 was in an Enterobacter roggenkampii clinical strain recovered in 2016 [Citation14]. In this study, the earliest mcr-9 and mcr-10 isolates were obtained in 2000 and 2018, respectively, implying that mcr-9 might have emerged much earlier than mcr-10. We further pinpoint that E. hormaechei was the major host of mcr-9. Accumulating epidemiological data suggest that E. hormaechei is the predominant species among ECC frequently obtaining multi-drug resistance and causing infections in the clinical setting [Citation26, Citation53, Citation54]. Therefore, the high prevalence of mcr-9 in E. hormaechei would further challenge the clinical treatment.
Different from the other mcr variants, the activities of mcr-9 and mcr-10 to colistin are much weaker in numerous isolates [Citation13, Citation29, Citation55]. The first reported isolate carrying mcr-9 or mcr-10 was susceptible to colistin with an MIC of 0.25-0.5 mg/L, despite both genes did confer colistin MICs of > 2.5 mg/L when overexpressed in the laboratory strains [Citation13, Citation14]. A recent study found that mcr-9 expression was inducible in the presence of colistin when located upstream of the two-component system qseB/qseC, and the mcr-9-harboring isolates were susceptible to colistin when lacked the qseB/qseC regulatory operon [Citation56]. In this study, qseB/qseC was detected at downstream of mcr-9 in seven isolates resistant to colistin, further supporting the role of qseB/qseC in the induction of mcr-9-mdiated colistin resistance. While others lacking this two-component system at downstream of mcr-9 also showed colistin resistance with even higher MIC values (≥ 32 mg/L), and no functional mutations (i.e. PhoPD56 and PhoQH277) were detected in their chromosomes, suggestive of unknown mechanism involved.
Our analysis showed that the dissemination of mcr-9 worldwide was mainly mediated by an IncHI2/2A epidemic plasmid. This is consistent with a previous study that IncHI2 was identified to be the dominant replicon type (90.1%; 64/71) of the mcr-9-carrying plasmids disseminating worldwide [Citation57]. Notably, IncHI2/2A type plasmids are highly associated with multidrug resistance genes, which raising a concern that the mcr-9-carrying epidemic plasmid could become multidrug resistant in the future. In contrast, a highly diversity was found for the mcr-10 plasmids that three different replicons (IncFIA, IncFIA-II, and IncFIB) were detected in the four mcr-10-carrying plasmids, and only one genome of the global dataset showed ≥ 90% coverage with one of the four plasmids. This is accordant with the finding of a recent study [Citation33], and may be one of the causes for the less prevalence of mcr-10 observed here.
Of greater concern, a high carbapenem resistance rate was detected in our MCR-ECC collection and also the global dataset, accounting for 33.3% (16/48) and 53.89% (388/720), respectively. These data demonstrate that the convergence of carbapenemase and mcr-9/mcr10 genes was highly frequent in ECC. The discrepancy of plasmid stability observed between blaIMP-4-harboring and blaIMP-26-harboring plasmids might be caused by their different hosts that the host of blaIMP-4-harboring plasmids belongs to ST133, while that of blaIMP-26-harboring plasmid belongs to ST171. Our surveillance data showed that MCR-CREC emerged later than MCR-ECC in China, and the two populations shared prevalent clones (ST133 and ST418), we thus suspect that MCR-CREC might have been derived from MCR-ECC recently through obtaining plasmids encoding carbapenemases in clinical settings. This can be further supported by that the two prevalent carbapenemases detected in our collection, i.e. NDM-1 and IMP-4, were carried by an IncX3-type and an IncN-type epidemic plasmid with very limited sequence variations, respectively. Additionally, our results underpin that such convergence could be dependent on the host preference of plasmids, since mcr-9/mcr-10-encoding plasmids can be remained in ECC stably but not in E. coli though the plasmids encoding carbapenemase were stable in both.
Of clinical and epidemiological concern, various combinations of resistance to the last resort antibiotics were detected in MCR-ECC, and two of them were even resistant to colistin, carbapenems, and tigecycline. Such convergence would largely worsen clinical outcomes in the future, although the tigecycline resistance determinants could not be defined in this study. Additionally, a majority of MCR-CREC belonged to epidemic clones, e.g. ST171, ST93, ST133, and ST418 [Citation48, Citation58, Citation59], and the plasmids encoding MCR or carbapenemases were able to remain stable in ECC and self-transmissible as shown here. Despite that it remains unclear whether the resistance convergence in these epidemic clones has established evolutionary advantages, continuous surveillance is imperative to prevent them from being high-risk clones.
In summary, our study revealed a high prevalence of mcr-9 and carbapenemase genes co-existing in ECC, and the convergence was driven by epidemic plasmids. The data suggest that MCR-CRE could be derived from MCR-ECC, and multiple epidemic clones have mediated the dissemination of MCR-CRE worldwide, highlighting that effective measures should be taken to control its further spread.
Supplemental Material
Download Zip (4.3 MB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Giamarellou H, Poulakou G. Multidrug-Resistant gram-negative infections. Drugs. 2009;69:1879–1901.
- van Duin D, Doi Y. Commentary: Outbreak of colistin-resistant, carbapenemase-producing klebsiella pneumoniae: Are We at the End of the road? J Clin Microbiol. 2015;53:3116–3117.
- D'Onofrio V, Conzemius R, Varda-Brkic D, et al. Epidemiology of colistin-resistant, carbapenemase-producing enterobacteriaceae and acinetobacter baumannii in Croatia. Infect Genet Evol. 2020;81:104263.
- Kieffer N, Ahmed MO, Elramalli AK, et al. Colistin-resistant carbapenemase-producing isolates among klebsiella spp. and acinetobacter baumannii in Tripoli, Libya. J Glob Antimicrob Resist. 2018;13:37–39.
- Liu Y-Y, Wang Y, Walsh TR, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. 2016;16:161–168.
- Xavier BB, Lammens C, Ruhal R, et al. Identification of a novel plasmid-mediated colistin-resistance gene, mcr-2, in Escherichia coli, Belgium, June 2016. Eurosurveillance. 2016;21:pii = 30280.
- AbuOun M, Stubberfield EJ, Duggett NA, et al. mcr-1 and mcr-2 (mcr-6.1) variant genes identified in moraxella species isolated from pigs in Great Britain from 2014 to 2015. J Antimicrob Chemother. 2017;72:2745–2749.
- Borowiak M, Fischer J, Hammerl JA, et al. Identification of a novel transposon-associated phosphoethanolamine transferase gene, mcr-5, conferring colistin resistance in d-tartrate fermenting salmonella enterica subsp. enterica serovar paratyphi B. J Antimicrob Chemother. 2017;72:3317–3324.
- Carattoli A, Villa L, Feudi C, et al. Novel plasmid-mediated colistin resistance mcr-4 gene in salmonella and Escherichia coli, Italy 2013, Spain and Belgium, 2015 to 2016. Eurosurveillance. 2017;22:pii = 30589.
- Yin W, Li H, Shen Y, et al. Effects of a bio-invasion of the pacific oyster, crassostrea gigas (thunberg, 1793) in five shallow water habitats in scandinavia. mBio. 2017;8:543–00517.
- Wang X, Wang Y, Zhou Y, et al. Emergence of a novel mobile colistin resistance gene, mcr-8, in NDM-producing klebsiella pneumoniae. Emerg Microbes Infec. 2018;7:122.
- Yang YQ, Li YX, Lei CW, et al. Novel plasmid-mediated colistin resistance gene mcr-7.1 in klebsiella pneumoniae. J Antimicrob Chemother. 2018;73:1791–1795.
- Carroll LM, Gaballa A, Guldimann C, et al. Identification of novel mobilized colistin resistance gene mcr-9 in a multidrug-resistant, colistin-susceptible salmonella enterica serotype typhimurium isolate. mBio. 2019;10:e00853–e00819.
- Wang C, Feng Y, Liu L, et al. Identification of novel mobile colistin resistance gene mcr-10. Emerg Microbes Infect. 2020;9:508–516.
- Ma S, Sun C, Hulth A, et al. Mobile colistin resistance gene mcr-5 in porcine aeromonas hydrophila. J Antimicrob Chemother. 2018;73:1777–1780.
- Xu T, Zhang C, Ji Y, et al. Identification of mcr-10 carried by self-transmissible plasmids and chromosome in Enterobacter roggenkampii strains isolated from hospital sewage water. Environ Pollut. 2021;268:115706.
- Macesic N, Blakeway LV, Stewart JD, et al. Silent spread of mobile colistin resistance gene mcr-9.1 on IncHI2 ‘superplasmids’ in clinical carbapenem-resistant enterobacterales. Clin Microbiol Infect. 2021;27:1856.1856.e7–1856.e13.
- Ling Z, Yin W, Shen Z, et al. Epidemiology of mobile colistin resistance genes mcr-1 to mcr-9. J Antimicrob Chemother. 2020;75:3087–3095.
- Alba P, Leekitcharoenphon P, Franco A, et al. Molecular epidemiology of mcr-encoded colistin resistance in enterobacteriaceae from food-producing animals in Italy revealed through the EU harmonized antimicrobial resistance monitoring. Front Microbiol. 2018;9:1217.
- Jiang Y, Zhang Y, Lu J, et al. Clinical relevance and plasmid dynamics of mcr-1-positive Escherichia coli in China: a multicentre case-control and molecular epidemiological study. The Lancet Microbe. 2020;1:e24–e33.
- Nesporova K, Jamborova I, Valcek A, et al. Various conjugative plasmids carrying the mcr-5 gene in Escherichia coli isolates from healthy chickens in Paraguay. J Antimicrob Chemother. 2019;74:3394–3397.
- Chavda KD, Westblade LF, Satlin MJ, et al. First report of blaVIM-4- and mcr-9-coharboring enterobacter species isolated from a pediatric patient. mSphere. 2019;4:e00629–e00619.
- Yuan Y, Li Y, Wang G, et al. Coproduction of MCR-9 and NDM-1 by colistin-resistant Enterobacter hormaechei isolated from bloodstream infection. Infect Drug Resist. 2019;Volume 12:2979–2985.
- Davin-Regli A, Lavigne JP, Pages JM. Enterobacter spp.: Update on Taxonomy, Clinical Aspects, and Emerging Antimicrobial ResistanceEnterobacter spp.: update on taxonomy, clinical aspects, and emerging antimicrobial resistance. Clin Microbiol Rev. 2019;32:e00002–19.
- Sutton GG, Brinkac LM, Clarke TH, et al. Enterobacter hormaechei subsp. hoffmannii subsp. nov., Enterobacter hormaechei subsp. xiangfangensis comb. nov., Enterobacter roggenkampii sp. nov., and Enterobacter muelleri is a later heterotypic synonym of Enterobacter asburiae based on computational analysis of sequenced Enterobacter genomes. F1000Res. 2018;7:521.
- Chavda KD, Chen L, Fouts DE, et al. Enterobacter spp.: New Insights into Phylogeny, Population Structure, and Resistance MechanismsComprehensive genome analysis of carbapenemase-producing Enterobacter spp.: new insights into phylogeny, population structure, and resistance mechanisms. mBio. 2016;7:e02093–e02016.
- Mediavilla JR, Patrawalla A, Chen L, et al. Colistin- and carbapenem-resistant Escherichia coli harboring mcr-1 and blaNDM-5, causing a complicated Urinary Tract Infection in a patient from the United States. mBio. 2016;7:e01191–e01116.
- Yao X, Doi Y, Zeng L, et al. Carbapenem-resistant and colistin-resistant Escherichia coli co-producing NDM-9 and MCR-1. Lancet Infect Dis. 2016;16:288–289.
- Khalifa HO, Soliman AM, Saito T, et al. First report of blaVIM-1-, blaNDM-1-, and mcr-9-co-harboring foodborne klebsiella pneumoniae. Antimicrob Agents Chemother. 2020;64:e00882–e00820.
- Sun L, Zhao X, Wang L, et al. Coexistence of mcr-9 and blaNDM-1 in a multidrug-resistant Enterobacter hormaechei strain recovered from a bloodstream infection in China. J Glob Antimicrob Resist. 2021;24:440–442.
- Simoni S, Mingoia M, Brenciani A, et al. First inchi2 plasmid carrying mcr-9.1, blaVIM-1, and double copies of blaKPC-3 in a multidrug-resistant Escherichia coli human isolate. mSphere. 2021;6:e0030221.
- Chen W, Hu Z, Wang S, et al. Letter to the Editor: Characterization of a clinical Enterobacter hormaechei strain belonging to epidemic clone ST418 co-carrying blaNDM-1, blaIMP-4 and mcr-9.1. Microb Drug Resist. 2022;28:180–184.
- Liao W, Cui Y, Quan J, et al. High prevalence of colistin resistance and mcr-9/10 genes in Enterobacter spp. in a tertiary hospital over a decade. Int J Antimicrob Agents. 2022;106573.
- Ji Y, Wang P, Xu T, et al. Development of a one-step multiplex PCR assay for differential detection of four species (Enterobacter cloacae, Enterobacter hormaechei, Enterobacter roggenkampii, and Enterobacter kobei) belonging to Enterobacter cloacae complex with clinical significance. Front Cell Infect Mi. 2021;11:677089.
- Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120.
- Bankevich A, Nurk S, Antipov D, et al. Spades: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–477.
- Foong J, Girdea M, Stavropoulos J, et al. Prioritizing clinically relevant copy number variation from genetic interactions and gene function data. PLoS One. 2015;10:e0139656.
- Hunt M, Silva ND, Otto TD, et al. Circlator: automated circularization of genome assemblies using long sequencing reads. Genome Biol. 2015;16:294.
- Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069.
- Zankari E, Hasman H, Cosentino S, et al. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother. 2012;67:2640–2644.
- Carattoli A, Zankari E, García-Fernández A, et al. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother. 2014;58:3895–3903.
- Richter M, Rossello-Mora R. Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci U S A. 2009;106:19126–19131.
- Treangen TJ, Ondov BD, Koren S, et al. The harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol. 2014;15:524.
- Sullivan MJ, Petty NK, Beatson SA. Easyfig: a genome comparison visualizer. Bioinformatics. 2011;27:1009–1010.
- Alikhan NF, Petty NK, Ben Zakour NL, et al. BLAST ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 2011; 12: 402.
- Camacho C, Coulouris G, Avagyan V, et al. Blast+: architecture and applications. BMC Bioinformatics. 2009;10:421.
- Gao H, Liu Y, Wang R, et al. The transferability and evolution of NDM-1 and KPC-2 co-producing klebsiella pneumoniae from clinical settings. EBioMedicine. 2020;51:102599.
- Xu T, Xue C-X, Huang J, et al. Emergence of an epidemic hypervirulent clone of Enterobacter hormaechei coproducing MCR-9 and carbapenemases. The Lancet Microbe. 2022;3:e474–e475.
- Nakamura A, Nakamura T, Niki M, et al. Genomic characterization of ESBL- and carbapenemase-positive enterobacteriaceae co-harboring mcr-9 in Japan. Front Microbiol. 2021;12:665432.
- Khedher MB, Baron SA, Riziki T, et al. Massive analysis of 64,628 bacterial genomes to decipher water reservoir and origin of mobile colistin resistance genes: is there another role for these enzymes? Sci Rep. 2020;10:5970.
- Sato T, Usui M, Harada K, et al. Complete genome sequence of an mcr-10-possessing Enterobacter roggenkampii strain isolated from a dog in Japan. Microbiol Resour Announc. 2021;10:e0042621.
- Guan J, Li L, Zheng LA-O, et al. First report of the colistin resistance gene mcr-10.1 carried by Inc(pA1763-KPC) plasmid pSL12517-mcr10.1 in Enterobacter cloacae in Sierra Leone. Microbiol Spectr. 2022;e0112722. Online ahead of print.
- Mezzatesta ML, Gona F, Stefani S. Enterobacter cloacae complex: clinical impact and emerging antibiotic resistance. Future Microbiol. 2012;7:887–902.
- Zhou K, Yu W, Cao X, et al. Characterization of the population structure, drug resistance mechanisms and plasmids of the community-associated Enterobacter cloacae complex in China. J Antimicrob Chemother. 2018;73:66–76.
- Tyson GH, Li C, Hsu CH, et al. The mcr-9 gene of Salmonella and E. coli is not associated with colistin resistance in the United States. Antimicrob Agents Chemother. 2020;64:e00573–e00520.
- Borjesson S, Greko C, Myrenas M, et al. A link between the newly described colistin resistance gene mcr-9 and clinical enterobacteriaceae isolates carrying blaSHV-12 from horses in Sweden. J Glob Antimicrob Resist. 2020;20:285–289.
- Li Y, Dai X, Zeng J, et al. Characterization of the global distribution and diversified plasmid reservoirs of the colistin resistance gene mcr-9. Sci Rep. 2020;10:8113.
- Chen J, Tian S, Nian H, et al. Carbapenem-resistant Enterobacter cloacae complex in a tertiary hospital in northeast China, 2010-2019. BMC Infect Dis 2021;21:611.
- Jin C, Zhang J, Wang Q, et al. Molecular characterization of carbapenem-resistant Enterobacter cloacae in 11 Chinese cities. Front Microbiol. 2018;9:1597.
