ABSTRACT
Kidney samples from 300 bat cadavers from the Czech and Slovak Republics were tested for Leptospira DNA using PCR and sequencing of three genes (lipL32, flab, and 16S ribosomal RNA). Overall detection rate was 4.7% and two bat species (Myotis myotis and Nyctalus noctula) were PCR-positive for at least one gene. Detected Leptospira sequences were similar to L. interrogans and L. borgpetersenii, and included a potentially novel species related to L. weilii.
European bats are carriers or reservoirs of several zoonotic bacterial pathogens, including Leptospira spp. [Citation1]. Using different approaches, Leptospira spp. have been detected in more than 50 bat species from different geographical regions, but mainly from the tropics and subtropics [Citation2]. In Europe, the possible role of bats in the epidemiology of leptospirosis remains to be clarified, as the occurrence of Leptospira spp. in bats is poorly known. A survey of wildlife conducted by Fennestad and Borg-Petersen [Citation3] in Denmark showed that four bat species had leptospires visible by dark-field microscopy in urine and/or kidney tissue suspensions: Myotis daubentonii, Pipistrellus pipistrellus, Nyctalus noctula, and Eptesicus serotinus. They also observed regular excretion of leptospires in bat urine visible by microscopy in three Nyctalus noctula kept alive in captivity for up to 20 weeks. Bai et al. [Citation4] detected Leptospira DNA via PCR in 25 bat kidney samples from two species, Myotis blythii and Miniopterus schreibersii, in the Republic of Georgia. Recently, Leptospira spp. DNA was detected in urine samples of four bat species from the Czech Republic and Poland: Barbastella barbastellus, Myotis bechsteinii, Myotis myotis, and Myotis nattereri [Citation5].
To study the occurrence of Leptospira in Central European bats in greater detail, 300 carcasses of 13 bat species were collected by passive surveillance (due to strict protection of bats that prohibit invasive sampling) from 2009 to 2019 at different sites in the Czech Republic and the Slovak Republic (see Appendix 1 for details). For the animals investigated in this study, carcasses of deceased bats found in the Czech Republic were kindly provided by bat researchers and animal rehabilitation centres. Bat species were identified by experienced bat biologists based on morphological traits and available keys [Citation6]. Team members were authorized to handle wild bats according to the Czech Certificate of Competency (No. CZ01341; §17, Act No. 246/1992 Coll.) and all sampling complied with Czech Law No. 114/1992 on Nature and Landscape Protection. Bat samples were frozen at −80°C after collection and this material was processed immediately. Bat kidneys were dissected, homogenized, and DNA extraction was performed using the QIAamp DNA Mini Kit (Qiagen, USA) according to the manufacturer's protocols. Conventional PCR protocols targeting the outer membrane lipoprotein (lipL32) gene [Citation4], the 16S ribosomal RNA (rRNA) gene [Citation7], and the flagellin B (flaB) [Citation7] were used to detect Leptospira DNA. The lipL32 gene is only present in pathogenic and intermediate Leptospira species (lineages P1 and P2 in the classification scheme from Vincent et al. [Citation8]), but flaB and 16S rRNA are present in all Leptospira lineages. Bidirectional sequencing was performed using an ABI PRISM 3100 Genetic Analyzer (Applied Biosystems, USA). The raw DNA sequences were edited and aligned using the Seqman module in Lasergene v6 (DNASTAR, USA) and manually checked. The BLAST algorithm (http://www.ncbi.nlm.nih.gov/blast) was used to confirm that sequences represented Leptospira DNA. A database was compiled consisting of lipL32 and flaB sequences from bats, canonical Leptospira species, and newly isolated species from environmental samples [Citation8], then phylogenetic trees were inferred using the maximum likelihood method (see Appendix 2 for details).
In total, 13/290 (4.5%) bats from the Czech Republic and 1/10 (10%) bats from the Slovak Republic were positive for Leptospira DNA via amplification and sequencing of one or both the lipL32 and flaB genes (4.7% overall). Attempts to amplify the 16S rRNA gene were unsuccessful with the primers and protocol used [Citation7]. As for bat species, Leptospira DNA was amplified in 3/69 Nyctalus noctula specimens (4.3%) and 11/187 Myotis myotis specimens (5.9%). The detection rate did not differ between these two species (chi-square test of proportions, p = 0.633) and sexes (chi-square test of proportions, Nyctalus noctula p = 0.090 and Myotis myotis p = 0.632), respectively. The remaining 11 species tested were all negative (see Appendix 1). The majority of positive lipL32 sequences (n = 12/14, 11 from M. myotis and one from N. noctula) were identical to pathogenic Leptospira interrogans (GenBank accession number MT4823) previously detected in urine from Myotis myotis in the Czech Republic [Citation5]. Two identical sequences (samples N8 and N45 from N. noctula, GenBank accession number OM307661) were phylogenetically distinct from other Leptospira species detected in bats and instead clustered with L. weilii, L. mayottensis, and L. alexanderi (); the novel sequences from bats shared 93.1%, 92.2%, and 92.7% sequence identity with these three species, respectively. Leptospira weilii was first described from the blood of a human patient in Australia [Citation9], L. mayottensis was isolated from blood of leptospirosis patients on the island of Mayotte [Citation10], and L. alexanderi was isolated from humans in China [Citation11]. The flaB gene was successfully amplified from sample N8 only (Nyctalus noctula from Czech Republic), and the sequence showed multiple peaks in the electropherogram, suggesting co-infection with multiple Leptospira species. Using the Mixed Sequences Reader tool [Citation12], two separate sequences (major and minor) were identified. The N8 major sequence (GenBank accession number ON552553) shared 94.7% sequence identity with L. borgpetersenii and the N8 minor sequence (GenBank accession number ON552554) shared 93.4% sequence identity with L. interrogans (B).
Figure 1. Phylogenetic relationships between Leptospira lipL32 sequences (A) and flaB sequences (B). Separate groups, including new sequences detected in Central European bats, are indicated by distinct symbols. The maximum likelihood trees were inferred using a TIM3+F+I+G4 model for lipL32 and a TVMe+I+G4 model for flaB in IQ-TREE v2.1.1. Numbers next to nodes indicate the percent bootstrap support after 1000 replicates. Branch lengths are in units of substitutions per site.
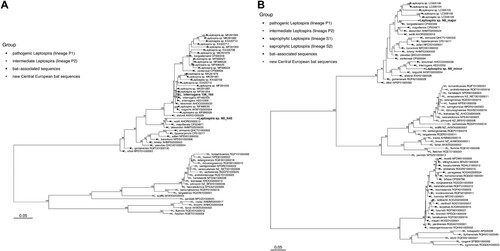
The presence of Leptospira spp. DNA was investigated in kidney tissues of bats from the Czech and Slovak Republics. The overall detection rate (4.7%) is within the range reported from other countries (2–35%; [Citation1]), although the detection rate can be highly variable even in the same species over time, suggesting a possible seasonal pattern of infection in bats that could be influenced by many factors such as sex, roosting behaviour, parturition, lactation, or weaning [Citation13]. While bats have been shown to be hosts of pathogenic Leptospira spp. strains, particularly in the tropics and subtropics, the role of European bats in maintaining and shedding Leptospira infection, and the potential public health risks from human exposure to bat leptospires remains poorly understood. In this study, we detected three lineages within the pathogenic clade of Leptospira (), one related to L. borgpetersenii, one related to L. interrogans, and another more distantly related to L. weilii, in kidney samples from bat cadavers. While the phylogenetic placement of these lineages suggests their pathogenic potential for humans and/or other mammals, this assessment is preliminary pending further clinical and ecological data. Clarifying the epizootiology of Leptospira in European bats would help to assess potential risks to public health and identify mitigation measures.
Authorship
VS, PS and IR conceived and designed the study; JZ, TB, JP collected material; PS, MN, JS, LD, RK, SŠ, JM and VS performed the laboratory analyses; JZ, JP, VS, PS, RK, CM, and IR analyzed the data and drafted the manuscript, to which all authors contributed with critical comments.
Data and materials availability
All data needed to evaluate the conclusions are present in the paper.
Supplemental Material
Download Zip (33.9 KB)Acknowledgements
We also thank to wildlife rescuers and rehabilitators from centres in Plzeň and Jinačovice, Dagmar Zieglerova from the Czech Union for Nature Conservation; Nyctalus Praha and Jiri Safar from the Nature Conservation Agency of the Czech Republic for providing bat cadavers.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Mühldorfer K. Bats and bacterial pathogens: a review. Zoonoses Public Health. 2013;60:93–103.
- Dietrich M, Mühldorfer K, Tortosa P, et al. Leptospira and bats: story of an emerging friendship. PLoS Pathog. 2015a;11:e1005176.
- Fennestad KL, Borg-Petersen C. Leptospirosis in Danish wild mammals. J Wildl Dis. 1972;8:343–351.
- Bai Y, Urushadze L, Osikowicz L, et al. Molecular survey of bacterial zoonotic agents in bats from the country of Georgia (Caucasus). PLoS One. 2017;12:e0171175.
- Seidlova V, Nemcova M, Pikula J, et al. Urinary shedding of leptospires in palearctic bats. Transbound Emerg Dis. 2021;68:3089–3095.
- Dietz CH, Kiefer A. Bats of Britain and Europe. Bloomsbury Publishing; London. 2018; 400 pp.
- Wajjwalku W, Sukmak M, Amavisit P, et al. Molecular characterization of flaB for Leptospira identification. Southeast Asian J Trop Med Public Health. 2015;46(2):262–267.
- Vincent AT, Schiettekatte O, Goarant C, et al. Revisiting the taxonomy and evolution of pathogenicity of the genus Leptospira through the prism of genomics. PLoS Negl Trop Dis. 2019;13:e0007270.
- Yasuda PH, Steigerwalt AG, Sulzer KR, et al. Deoxyribonucleic acid relatedness between serogroups and serovars in the family Leptospiraceae with proposals for seven new Leptospira species. Int J Syst Evol Microbiol. 1987;37:407–415.
- Bourhy P, Collet L, Brisse S, et al. Leptospira mayottensis sp. nov., a pathogenic species of the genus Leptospira isolated from humans. Int J Syst Evol Microbiol. 2014;64:4061–4067.
- Brenner DJ, Kaufmann AF, Sulzer KR, et al. Further determination of DNA relatedness between serogroups and serovars in the family Leptospiraceae with a proposal for Leptospira alexanderi sp. nov. and four new Leptospira genomospecies. Int J Syst Bacteriol. 1999;49:839–858.
- Chang CT, Tsai CN, Tang CY, et al. Mixed sequence reader: a program for analyzing DNA sequences with heterozygous base calling. Sci World J. 2012;2012:365104.
- Dietrich M, Wilkinson DA, Benlali A, et al. Leptospira and paramyxovirus infection dynamics in a bat maternity enlightens pathogen maintenance in wildlife. Environ Microbiol. 2015b;17:4280–4289.
