ABSTRACT
Acinetobacter baumannii is an important nosocomial pathogen. Upon colonizing a host, A. baumannii are subjected to selective pressure by immune defenses as they adapt to the host environment. However, the mechanism of this pathoadaptation is unknown. Here, we established an in vitro system to evolve A. baumannii driven by the continuous selective pressure exerted by epithelial cells, and we used a combination of experimental evolution, phenotypic characterization and multi-omics analysis to address the underlying mechanism. When continuously exposed to selective pressure by pulmonary epithelial cells, A. baumannii showed ptk mutation-mediated mucoid conversion (reduced adhesion and increased anti-phagocytic ability) by enhancement of capsular exopolysaccharide chain length; rsmG mutation-mediated deficiency of 7-methylguanosine modification in the 524th nucleotide of 16S rRNA, which increased ribosome translation efficiency; and rnaseI mutation-mediated changes in outer membrane permeability and efflux pump expression. Together, these mutations altered susceptibility to a variety of antimicrobial agents, including the novel antibiotic cefiderocol, by regulating siderophore and siderophore-receptor biosynthesis. In conclusion, pulmonary epithelial cells modulate A. baumannii pathoadaptation, implicating the host–microbe interaction in the survival and persistence of A. baumannii.
Introduction
Upon colonizing a host, bacterial pathogens rapidly adapt to survive under multiple selective pressures. Invading bacteria must overcome host defenses, including nutrient limitation and immune response [Citation1]. Bacterial pathogens have evolved numerous strategies to this end [Citation2], for example, by acquiring new gene by horizontal gene transfer or by undergoing pathoadaptive mutations [Citation3]. Pathogen mutations during persistent host infection are likely to be pathoadaptive [Citation4,Citation5].
Acinetobacter baumannii, an opportunistic pathogen, is increasing in incidence, and is prevalent in hospital environments, particularly in intensive care units and in patients with pneumonia and bacteremia [Citation6]. Translocation of A. baumannii from the lungs to the bloodstream is associated with severe complications (e.g. sepsis), particularly in immunosuppressed patients or those on mechanical ventilation, leading to a high mortality rate [Citation7]. Virulence factors such as OmpA, lipopolysaccharides, and capsule [Citation8,Citation9] mediate host colonization and damage. Especially, capsular exopolysaccharide and associated mucoid phenotype have emerged as a universal virulence factor owing to several observations [Citation10]. Previous evidence supports a role for capsule structures in contributing to defenses against antibiotics such as antimicrobial peptides [Citation11–13]. Because of the variability of the Acinetobacter genome, adaptive evolution under host stress can also promote persistence [Citation14].
The lung is the main colonization and infection site of A. baumannii, and the airway epithelium is the first line of defense against invasion by pathogenic microorganisms [Citation15,Citation16]. Epithelial cells trigger the host inflammatory responset [Citation17]. Although epithelial cells are nonspecific phagocytes, they are important in the pathogenesis of A. baumannii [Citation18]. A. baumannii changes at multiple levels as it interacts with a host, for example, undergoing transcriptome remodelling and gene mutation [Citation19–21]. Experimental evolution in controlled environments that mimic host selective pressures, combined with advanced DNA sequencing techniques, enables the investigation of adaptive evolution. To the best of our knowledge, the host cell-mediated adaptive evolution of A. baumannii has not been investigated using an experimental evolution model.
We established an in vitro system to evolve A. baumannii under continuous selective pressure from epithelial cells. Epithelial cell-adapted clones displayed not only the ability to evade adherence and phagocytosis due to a mucoid phenotype but also increased resistance to several antibiotics.
Materials and methods
Bacterial strains, cells, plasmids, and infection procedure
The wild-type (WT) A. baumannii strain ATCC17978 was used, and ASD9 and ACD9 were obtained from a laboratory evolution experiment (Table S1). One-step chromosomal gene mutation based on recombination-mediated genetic engineering (pMo130-Hyg) was performed to establish mutant strains; the pYMAb2-Hyg shuttle vector was used to establish gene-complemented strains (Table S1). Strains were cultured in Mueller-Hinton (MH) Broth (Oxoid; Thermo Scientific, Waltham, MA); gene-complemented strains were cultured in MH Broth supplemented with hygromycin (100 μg/mL). All strains were grown at 37°C.
The A549 human pulmonary adenocarcinoma cell line (#CCL185; ATCC, Manassas, VA) and HBE human bronchial epithelial cell line (#CRL2741; ATCC, Manassas, VA) were cultivated in Dulbecco’s modified Eagle’s medium (Cat. No: 10-013-CV; Corning Inc., Corning, NY) supplemented with 10% fetal bovine serum (35-081-CV; Corning Inc., Corning, NY) and 1% penicillin/streptomycin (Cat. No: 30-002-CI; Corning Inc., Corning, NY) and incubated at 37°C in an atmosphere of 5% CO2. THP-1 human monocytes were cultivated in Roswell Park Memorial Institute-1640 medium (10-040-CV; Corning Inc., Corning, NY) with 10% fetal bovine serum and 1% penicillin/streptomycin. THP-1 cells were differentiated into macrophage-like cells (THP-1 Mø) by incubation in the presence of phorbol myristate acetate (Cat. No: HY-18739; MCE, Monmouth Junction, NJ) at 200 ng/mL for 48 h. For cell infection, fetal bovine serum-free medium without penicillin/streptomycin containing bacteria at a multiplicity of infection (MOI) of 100 was added to cell plates and incubated at 37°C in a 5% CO2 atmosphere.
Laboratory evolution experiment
A single colony of ATCC17978 was cultured in 2 mL MH broth overnight at 37°C. Fetal bovine serum-free medium without penicillin/streptomycin containing bacteria at an MOI of 1:1 (106 A. baumannii to 106 cells) was added to cell flasks and incubated for 24 h at 37°C in an atmosphere of 5% CO2, which was considered a passage. Bacteria were subsequently bottlenecked to start the next passage. As a control, equal numbers of bacteria evolved under identical conditions in the absence of cells. All populations evolved for 9 days, and populations on days 3, 6, and 9 were collected. The populations that evolved with and without cells were designated A1/A2/A3 and H1/H2/H3, and A4/A5/A6 and H4/H5/H6, respectively.
Genomic DNA sequencing and analysis
The genomic DNA of ACD9 and HCD9 was extracted using a QIAamp DNA Minikit (Qiagen Valencia, CA) following the manufacturer’s protocol. Libraries were prepared using the TruePrepTM DNA Library Prep Kit V2 for Illumina (Vazyme, Nanjing, China). Using a single “transposase” enzymatic reaction, sample DNA is simultaneously fragmented and tagged with adapters, an optimized, limited-cycle PCR protocol amplifies tagged DNA and adds sequencing indexes. Individual libraries were assessed on the QIAxcel Advanced Automatic nucleic acid analyzer and then were quantitated through qPCR by the use of KAPA SYBR FAST qPCR Kits (Kapabio systems, Boston, US). The bacterial genome was sequenced on an Illumina HiSeq platform (Illumina, San Diego, CA, USA) as described previously [Citation22]. Breseq v. 0.33.0 was used for comparative genome analyses [Citation23], referencing the original genome of ATCC17978 (GenBank accession NZ_CP018664.1). Mutations were confirmed by PCR and Sanger sequencing.
Cloning, expression, and recombinant RsmG protein purification
The full-length coding sequences of A. baumannii ATCC17978 rnaseI (AUO97_RS04110) and rsmG (AUO97_RS15310) were amplified using DNA from A. baumannii ATCC17978 and cloned into the pYMAb2 vectors to generate pYMAb2-rnaseI and pYMAb2-rsmG. Plasmids were verified by DNA sequencing and are listed in Table S2.
Bacterial expression and purification of His-tagged RsmG (RsmG-WT, RsmGΔT, and RsmG(TTG)2→3) were performed as described previously [Citation24]. Briefly, the rsmG sequence (AUO97_RS15310) was amplified using DNA from A. baumannii ATCC17978/ACD9/HCD9, cloned into the pET28A vector to generate the pET28A-rsmG-WT/ pET28A-rsmGΔT/ pET28A-rsmG(TTG)2→3 constructs, and transformed into competent BL21 (DE3) cells. Isopropyl-β-d-thiogalactoside was added to the bacterial suspension to a final concentration of 1 mM to induce target protein production. Details of protein purification are provided in the Supplementary Methods.
Minimum inhibitory concentration determination
The minimum inhibitory concentration (MIC) of the following antibiotics was determined by the broth microdilution method with cation-adjusted MH broth (Oxoid, Hampshire, UK): cefoperazone/sulbactam (1:1, CFU/SU), ceftazidime (CAZ), cefepime (FEP), cefiderocol (CFDC), imipenem (IPM), meropenem (MEM), colistin (COL), amikacin (AMK), and gentamicin (GEN). MIC values were interpreted according to the Clinical and Laboratory Standards Institute guidelines (CLSI) [Citation25].
Quantitative biofilm assay
Biofilm formation on polystyrene was measured via crystal violet staining [Citation26]. Each strain was cultivated overnight at 37°C, and the cultures were diluted 1:100 into 150 µL MH broth in 96-well plates and incubated at 37°C for 24 h. The cultures were washed three times with PBS and stained with 0.1% crystal violet, washed with PBS, and 200 µL 30% acetic acid was added to each well. The optical density at 550 nm (OD550) of each well was quantified using a microplate reader.
Bacterial adherence and invasion assay
For bacterial adherence assay, A. baumannii was added to a monolayer of epithelial cells at an MOI of 100 and incubated in a 5% CO2 atmosphere at 37°C for 1 h. For bacterial invasion assay, A. baumannii was added to THP-1 Mø cells at an MOI of 100 and incubated in a 5% CO2 atmosphere at 37°C for 1 h. The cells were washed three times with PBS and a fresh culture medium containing 100 μg/mL gentamicin was added, followed by incubation for 2 h. The cells were washed three times with PBS and lysed with 0.2% Triton X-100 for 20 min, and the lysate was plated on MH agar after gradient dilution. Details are provided in the Supplementary Methods.
Membrane permeability assay
Membrane permeability was measured using To-Pro-3 (Invitrogen, Thermo Fisher Scientific) and DiOC2(3) (3, 3′-diethyloxacarbocyanineiodide) as described previously [Citation27]. Membrane permeability was expressed as the ratio of To-Pro-3 fluorescence to green fluorescence or the frequency of To-Pro-3+ cells. Details are provided in the Supplementary Methods.
Bacterial capsule detection, surface polysaccharide extraction, and Alcian blue staining
Bacterial capsules were stained with Congo red and observed under a microscope (Olympus, Tokyo, Japan). Surface polysaccharides were extracted using the hot aqueous-phenol method [Citation27]. Polysaccharide extracts were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and stained with Alcian blue (Sangon Biotech, Co., Ltd.).
Direct RNA sequencing and EpiNano-based m7G-modified site prediction
m7G-modified sites were predicted as described previously [Citation28]. Briefly, A. baumannii total RNA samples were used to prepare RNA libraries for direct RNA sequencing; the sequence data were analyzed using EpiNano. Modification scores at each site were obtained for ATCC17978 and the rsmG mutant strains (rsmGΔT and rsmG(TTG)2→3). Details are provided in the Supplementary Methods.
In vitro m7G methyltransferase activity by LC-MS/MS
RNA probes derived from 16S rRNA of A. baumannii ATCC17978 were purchased from Beijing Tsingke Biotechnology Co., Ltd. In vitro methyltransferase activity assay was performed as described previously [Citation29]. Briefly, a 50 μL reaction mixture containing the RNA probe, recombinant protein (RsmG-WT, RsmGΔT, and RsmG(TTG)2→3), and SAM was incubated at 37°C for 3 h, and the reaction products were digested by nuclease P1 and alkaline phosphatase for QQQ LC-MS/MS analysis [Citation30]. Total m7G and G contents were quantified based on standard curves (Figure S2 A–D) to calculate the m7G/G ratio. Details are provided in the Supplementary Methods.
In vitro translation assay
Cell-free translation of eGFP mRNA was performed as described previously [Citation31]. Briefly, a homemade Escherichia coli in vitro transcription-translation PURE system was used to assess 70S ribosome activity. PURE reaction mix (15 μL), which translates the eGFP gene, was added to 70S ribosome (rsmG-WT, rsmGΔT, and rsmG(TTG)2→3). Protein synthesis was performed by incubation at 37°C (lid temperature 42°C), and eGFP fluorescence intensity was read at 0.5, 1, 1.5, and 2 h on a GloMax 20/20 luminometer (Promega).
Growth assay
Growth rates were determined in microplates using a Bioscreen C Microplate Reader (Oy Growth Curves Ab Ltd., Finland). The strains were cultured overnight at 37°C, and the cultures were diluted 1:100 into 200 µL MH broth with or without the ferrous iron chelator, 2,2′-dipyridyl (DIP), and incubated at 37°C with shaking. Absorbance at 600 nm was measured at 5 min intervals. Each strain was assayed in three independent cultures and three separate experiments. Growth was determined by calculating the area under the growth curve [Citation32].
Transcriptome analysis of A. baumannii rsmG and rnaseI mutants
To investigate the role of rnaseI and rsmG in antibiotic resistance, RNA-seq of A. baumannii ATCC17978, rnaseIΔA, rsmGΔT, and rsmG(TTG)2→3 was performed as described previously [Citation33]. Briefly, A. baumannii ATCC17978, rnaseIΔA, rsmGΔT, and rsmG(TTG)2→3 total RNA were subjected to RNA-sequencing, and genes differentially expressed in the rsmG and rnaseI mutants were screened. Details are provided in the Supplementary Methods.
RNA preparation, quantitative reverse transcription-PCR, and mRNA stability measurement
Total RNA was extracted, and RT–PCR was performed following the manufacturer’s instructions (Cat. No. RR037A; TaKaRa, Tokyo, Japan). TB Green Premix Ex Taq II (Cat. No. RR820A; TaKaRa, Tokyo, Japan) was used for signal detection, followed by real-time PCR analysis (Roche LightCycler480; Roche Molecular Systems, Basel, Switzerland) of gene expression levels; the primer sequences are listed in Table S3. For mRNA stability measurement, bacterial cultures were harvested at 0, 1, and 3 min after rifampicin addition (1 mg/mL), and total RNA samples were prepared and subjected to qRT-PCR. Details are provided in the Supplementary Methods.
Statistical analysis
Data are expressed as means ± standard deviations. Comparisons between two groups were performed using two-tailed unpaired Student’s t-tests. One-way analysis of variance was used for multiple-group comparisons. Two-way repeated measures analysis of variance was used to compare changes in a single variable over time in each group. Statistical analyses were performed using Prism v. 9.0 (GraphPad Software, San Diego, CA). P < 0.05 was considered to indicate statistical significance.
Results
The emergence of morphological and phenotypic diversity
We generated four evolved A. baumannii populations (all from the same ancestral colony of ATCC17978) that adapted to the antagonistic interaction with A549 and HBE cells. The populations (A1 to A3 and H1 to H3) evolved in a complete culture medium with A549 or HBE cells at an MOI of 1:1 (106 A. baumannii to 106 cells). Populations (A4 to A6 and H4 to H6) evolved under identical conditions in the absence of cells and were used as the controls. Bacteria from A1-A6 and H1-H6 were streaked on plates, and a randomly selected colony was cultured in liquid medium (A). These colonies are hereafter referred to as ASD3/ASD6/ASD9/ACD3/ACD6/ACD9 and HSD3/HSD6/HSD9/HCD3/HCD6/HCD9 (Table S4).
Figure 1. Evolution experiment and phenotypic characterization. (A) Pathogen–host cell co-culture-based laboratory evolution experiment. (B) Representative images of the morphological diversity of A. baumannii populations adapted to pulmonary epithelial cells (A549 and HBE). (C) Representative Congo red-stained images of capsules of colonies (ASD9/ACD9/HSD9/HCD9) derived from evolved strains. Scale bar, 10 μm (D) OD600 values of culture supernatants. (E) Biofilm formation is revealed by crystal violet staining. (F) Adherence to epithelial cells and internalization by macrophages. Data are from at least three independent experiments; error bars represent standard deviations. **P < 0.01 vs. blank group, ##P < 0.01, #P < 0.05.
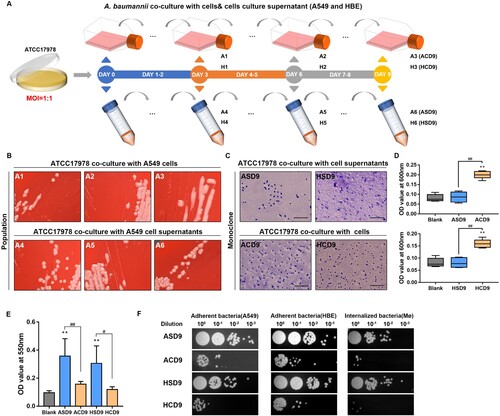
Adaptation of A. baumannii in the presence of epithelial cells was characterized by mucoid phenotype emergence. After 3 days, mucoid colonies were observed in populations co-cultured with A549 and HBE cells. This was not observed in the control groups ( and Figure S1). Congo red capsule staining showed a thickened capsule in colonies from evolved populations (ACD9 and HCD9) compared to those from control populations (ASD9 and HSD9). A sedimentation assay of microviscosity showed that the OD600 of the supernatant of evolved strains (ACD9 and HCD9) was higher than that of the control strains (ASD9 and HSD9). Crystal violet staining showed that the evolved strains had considerably weaker biofilm formation than the control strains. Regarding cell adherence and internalization, the evolved strains showed less adhesion to epithelial cells and less internalization into macrophages than the control strains. The evolved strains showed significantly different antibiotic susceptibilities, and their resistance to cephalosporins increased significantly ().
Table 1. Antimicrobial susceptibility of ATCC17978 and in vitro evolved strains
Identification and investigation of genetic mutations associated with morphological and phenotypic diversity
To detect mutations, the genomes of ACD9 and HCD9 were subjected to whole genome sequencing (WGS). Three and two mutations were detected in ACD9 and HCD9, respectively, and confirmed by Sanger sequencing (). ptk (AUO97_RS06965) and rsmG (AUO97_RS15310) were mutated in ACD9 and HCD9, and rnaseI (AUO97_RS04110) was mutated only in ACD9. The mutation in ptk resulted in a substitution of valine for glutamic acid at amino acid 653 (ptk V653E). rsmG harboured a missense mutation (584/633nt) and glutamine insertion (175/633nt). The mutation in rnaseI (rnaseIΔA) resulted in premature termination of translation (rnaseIM122*).
Table 2. Mutations acquired by evolved clones identified through whole genome re-sequencing (WGS).
Next, the impact of each mutation on morphological and phenotypic diversity was investigated (). We introduced four mutations into the genome of the parental strain ATCC17978 to generate four mutant strains. The effect of each mutation was determined by evaluating mucoid phenotype and antibiotic susceptibility. Only ptk V653E showed a distinct mucoid phenotype, as indicated by a thickened capsule and increased microviscosity, decreased adhesion to epithelial cells and internalization into macrophages, and decreased biofilm formation. The rsmG and rnaseI mutations but not ptk mutations significantly altered antibiotic susceptibility (Table S5). ATCC17978 and the rsmG and rnaseI mutations (rsmGΔT, rsmG(TTG)2→3, and rnaseIΔA) were transformed with a plasmid encoding WT rsmG and rnaseI, which restored their susceptibility to cephalosporins (Table S6).
Figure 2. ptk mutation induces the mucoid phenotype and reduces adhesion and internalization. (A) Representative images of morphological diversity of ATCC17978 and its mutant strains (ptk V653E, rsmGΔT, rsmG(TTG)2→3, and rnaseIΔA). (B) Adherence to epithelial cells and internalization by macrophages of ATCC17978 and its mutant strains. (C) OD600 values of culture supernatants of ATCC17978 and its mutant strains. (D) Biofilm formation by ATCC17978 and its mutant strains as revealed by crystal violet staining. (E) Representative images of morphological diversity of the evolved and ptk-WT complemented strains. (F) Representative Congo red-stained images of the capsules of evolved strains and ptk-WT complemented strains. Scale bar, 10 μm. (G) Adherence to epithelial cells and internalization by macrophages of evolved strains and ptk-WT complemented strains. (H) Capsular exopolysaccharide stained with Alcian blue (sup supernatant, arrows HMW exopolysaccharides). (I–K) IL-6, IL-8, and TNF-α mRNA levels in A549 cells (I), HBE cells (J), and THP-1 derived macrophages (K) at 3 h after infection. **P < 0.01 vs. ATCC17978 group.
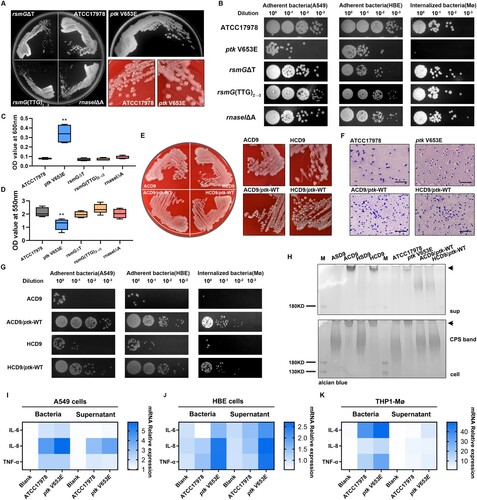
Enhancement of capsular exopolysaccharide chain length in ptk mutation mediates mucus phenotype and reduces bacterial adhesion and internalization
Next, we replaced the mutated ptk in ACD9 and HCD9 with the WT ptk, thus generating the revertant mutants ACD9/ptk-WT and HCD9/ptk-WT (). Compared to their parental strains, the two revertant mutants lacked a mucoid phenotype and had a thinner capsule and increased epithelial cell adhesion and macrophage internalization. SDS-PAGE and Alcian blue staining showed that the lysates of ACD9, HCD9, and ptk V653E had a very high-molecular-weight (HMW) polysaccharide that replaced the capsular polysaccharide band of the WT. This polysaccharide was also present in the culture supernatant. The polysaccharide bands of the two revertant mutants were similar to those of the WT. Therefore, ptk mutation increased the capsule polymer chain length. To investigate the effect of the extended polymer chain on host cells, we determined the mRNA levels of proinflammatory cytokines, including IL-6, IL-8, and TNF-α in A549, HBE, and THP-1 Mø. The IL-6, IL-8, and TNF-α transcript levels were significantly upregulated in the ptk V653E-infected groups, compared to the control and WT-infected groups.
The m7G524 deficiency caused by rsmG mutation induces antibiotic resistance
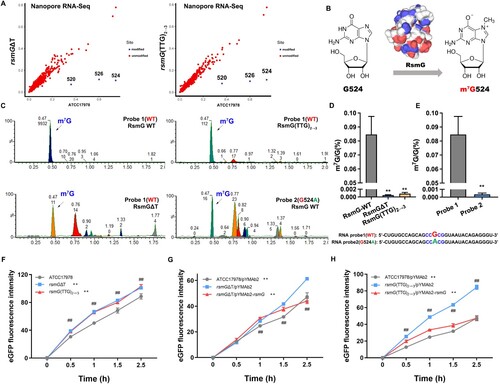
The m7G modification of 16S rRNA in the presence or absence of rsmG mutation was detected by Oxford Nanopore Technologies (ONT), a third-generation sequencing technology. m7G modification of G524 and G526 (replacement of guanosine with adenosine at nucleotide 520 in the 16S rRNA of A. baumannii) in 16S rRNA was missing in the rsmG mutant (A). RsmG mediates m7G modification of nucleotide 527 in the 16S rRNA of E. coli (corresponds to G524 of A. baumannii) (B). Consistent with previous studies, strains harbouring the rsmG mutation exhibited elevated MICs to streptomycin (Table S7). Next, an in vitro methyltransferase activity assay was performed to determine m7G activity and to verify modification sites for synthetic RNA probes with or without potential modification sites (obtained from ONT). Total m7G and G contents were measured via LC-MS/MS and quantified based on standard curves (C). The RsmG mutant protein (RsmGΔT, RsmG(TTG)2→3) showed no methyltransferase activity with RNA probe 1 (WT), whereas WT RsmG exhibited methyltransferase activity (D). Therefore, rsmG mutation results in the loss of RsmG methylation activity. RNA probes 2 and 4 (524G to 524A in the consensus sequence) resulted in inefficient methylation compared to RNA probes 1 and 3 (524G) (E and Figure S2). In addition, the m7G modification was downregulated by 526G to 526A (RNA Probe 3) but was significantly higher than that of RNA probes 2 and 4 (Figure S2F). The secondary structure of RNA Probe 3 (526G to 526A) changed significantly compared to RNA Probe 1(WT), potentially affecting methylation and indicating that RsmG has site specificity (Figure S1B). These results led us to hypothesize that rsmG mutations increase protein synthesis. Therefore, we measured the in vitro translational activity of ribosomes from the WT and rsmG mutant strains, using eGFP mRNA as a template. Ribosomes from the rsmG mutants (rsmGΔT, rsmG(TTG)2→3) exhibited higher eGFP synthetic activity than the WT (F). Ribosomes from rsmG-complemented strains (rsmGΔT/pYMAb2-rsmG and rsmG(TTG)2→3/pYMAb2-rsmG) showed decreased eGFP synthetic activity compared to the corresponding rsmG mutant strains (rsmGΔT/pYMAb2 and rsmG(TTG)2→3/pYMAb2) (G and H). Therefore, rsmG mutations resulted in m7G524 deficiency, which increased translation efficiency and acted as a global regulator.
Figure 3. m7G524 deficiency caused by rsmG mutation increases ribosomal translation efficiency. (A) Prediction of m7G modification site in rsmG mutant strains by third-generation sequencing. (B) RsmG mediated m7G modification of nucleotide 524 in the 16S rRNA of A. baumannii. (C) In vitro methyltransferase activity using 16S rRNA probes and RsmG protein, with detection of m7G and G by LC-MS/MS. (D and E) m7G/G ratio. (F–H) 70S ribosome activity of rsmG mutant (F) and complemented strains (G and H) by in vitro translation assay. **P < 0.01 vs. RsmG-WT (Probe 1, ATCC17978, rsmGΔT/pYMAb2 or rsmG(TTG)2→3/pYMAb2) groups, ##P < 0.01.
The rnaseI mutation affects outer membrane permeability and efflux pump expression
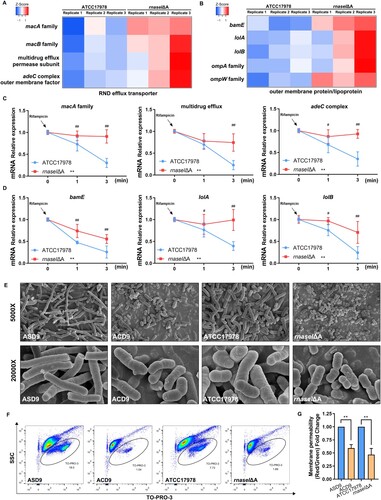
Transcriptomic analysis was performed to explore the mechanism of rnaseI mutation-mediated antibiotic resistance. The transcriptome of the WT and rnaseIΔA strains revealed upregulation of four genes encoding RND efflux transporters, which mediate antibiotic transport, and upregulation of five genes involved in outer membrane formation (A and B). We hypothesized that modulation of nucleotide homeostasis mediates some of the transcriptional and phenotypic changes observed in rnaseIΔA, therefore, we performed mRNA stability assays. The mRNA stability of RND efflux transporter-related genes and outer membrane protein/lipoprotein synthesis-related genes in rnaseIΔA were significantly higher than that of the WT after transcriptional repression by rifampicin (C and D). Next, we examined the effect of carbonyl cyanide 3-chlorophenylhydrazone (CCCP; efflux pump inhibitor) on MIC values. CCCP reduced the MIC values of rnaseIΔA (Table S8). Scanning electron microscopy (SEM) showed that the outer membrane morphologies of the ACD9 and rnaseIΔA strains were significantly changed compared to the ASD9 and ATCC17978 WT strains. Evolved and mutant cells were of heterogeneous shapes rather than the homogenous shape of the parental isolate (E). In addition, the outer membrane permeability was reduced in ACD9 compared to ASD9, and in the rnaseIΔA strain compared to the WT strain, as measured according to the positive cell frequency (F) and fluorescence intensity (G). Therefore, overexpression of RND efflux transporters and changes in outer membrane morphology and permeability are implicated in the antibiotic resistance of the rnaseIΔA strain.
Figure 4. rnaseI mutation affects outer membrane permeability and efflux pump expression. (A) Heatmap of RND efflux transporter gene expression. (B) Heatmap of outer membrane formation gene expression. (C and D) mRNA stability of RND efflux transporter-related genes (C) and outer membrane protein/lipoprotein synthesis-related genes (D) in rnaseIΔA compared to ATCC17978. (E) Representative images of the morphology of the evolved and rnaseI WT/ΔA strains by scanning electron microscopy (SEM). (F and G) Outer membrane permeability was measured via flow cytometry – the percentage of TO-PRO-3 positive cells (F) and adjusted fluorescence intensity (G). **P < 0.01, *P < 0.05 vs. ATCC17978 or ASD9 group, ##P < 0.01, #P < 0.05.
The rsmG and rnaseI mutation impairs iron acquisition, which may mediate resistance to cefiderocol
The rsmG and rnaseI mutants (rsmGΔT, rsmG(TTG)2→3, and rnaseIΔA) showed downregulation of bauB, bauE, bauF, basB, basC, basD, basE, basF, basG, and basJ (A). These genes are linked to siderophore and siderophore-receptor biosynthesis. We hypothesized that downregulated expression of these genes may suppress iron acquisition and utilization. In the completed medium, the growth of the rsmG and rnaseI mutants was not inhibited compared to the WT (B and H), whereas the rsmG and rnaseI mutants displayed significant growth restriction in an iron-limited medium (containing the ferrous iron chelator 2,2′-dipyridyl; DIP) (C, D, and H). In the presence of 100 μM DIP, only the rsmGΔT/pYMAb2-rsmG strain partially recovered from growth restriction (F, and I-K). In the presence of 200 μM DIP, the growth of the rnaseI and rsmG-complemented strains partially recovered (G, and I-K). Mutations in these genes impaired bacterial utilization of iron, possibly explaining the reduction in cefiderocol susceptibility.
Figure 5. rsmG and rnaseI mutations impair bacterial iron acquisition and utilization. (A) Siderophore and siderophore receptor gene expression data in rsmG and rnaseI mutants. (B–G) Growth curves of the mutants (B–D) and their complemented strains (E–G) in the presence or absence of the ferrous iron chelator 2,2′-dipyridyl (DIP). (H–K) Areas under the growth curves (AUC) of the mutants (H) and their complemented (I–K) strains in the presence or absence of DIP. **P < 0.01, *P < 0.05 vs. ATCC17978 or ATCC17978/pYMAb2 group, ##P < 0.01, #P < 0.05.
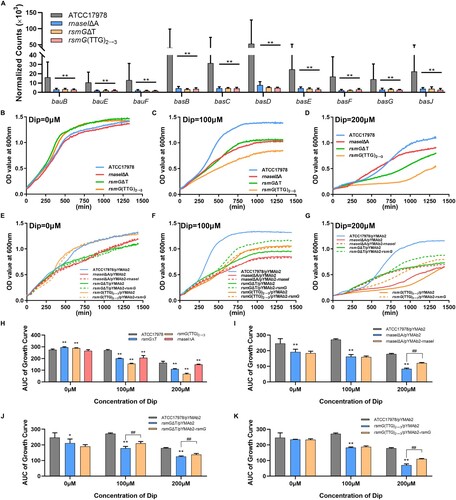
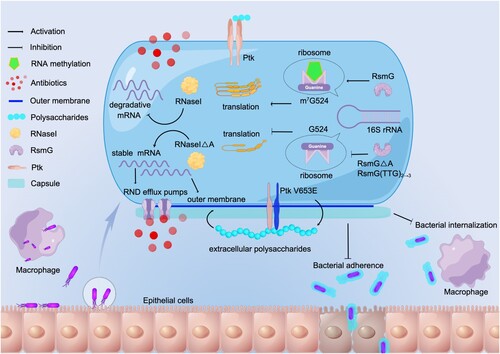
Figure 6. Proposed mechanism of A. baumannii pathoadaptation to epithelial cells. In A. baumannii, mutation of ptk mediates mucoid conversion and reduces adhesion and internalization. Mutations of rsmG and rnaseI involve in antibiotic resistance, and m7G524 deficiency in 16S rRNA caused by rsmG mutation increases ribosomal translation efficiency and may act as global regulators, and rnaseI mutation affects outer membrane permeability and efflux pump expression by modulating RNA metabolism. (By Figdraw, www.figdraw.com).
Discussion
Bacterial evolution from a commensal to a pathogen may occur by the acquisition of new gene via horizontal gene transfer (gain of function) or accumulation of pathoadaptive mutations (change in function) [Citation34,Citation35]. For example, macrophage-adapted E. coli exhibit increased intracellular survival and delayed phagosome maturation due to a single IS1 insertion upstream of yrfF [Citation3]. The loss of function of mucA in Pseudomonas aeruginosa results in an increased ability to escape phagocytosis and pulmonary clearance [Citation36]. The lung is the main colonization and infection site of A. baumannii, an opportunistic pathogen mainly in patients on mechanical ventilation [Citation7]. The airway epithelium is the first line of defense against invasion by pathogenic microorganisms, and epithelial cells trigger the host inflammatory response [Citation17]. Therefore, the acquisition of adaptive mutations is essential for A. baumannii persistence. It is shown that A. baumannii can very easily acquire and lose genes [Citation37]. Several pathogenic bacteria have evolved to survive and indeed take advantage of the host environment [Citation38]. In this study, we induced the evolution of A. baumannii in vitro with two pulmonary epithelial cell lines. Epithelial cell-adapted A. baumannii exhibited a mucoid phenotype and enhanced antibiotic resistance, and several gene mutations and deletions (Table S9) were found. The mucoid phenotype was characterized by reduced adhesion to epithelial cells and internalization by macrophages. The antibiotic resistance adaptation was associated with the overproduction of efflux pumps and reduced outer-membrane permeability ().
In a previous study, we found that clinical isolates from sputum showed enhanced resistance and morphological changes after within-host evolution [Citation39]. In that study, we identified mutations in ptk that caused mucoid conversion and decreased adherence and internalization, promoting dissemination. Macrophage- and neutrophil-adapted A. baumannii also undergo mucoid conversion and ptk mutation (unpublished data). The K locus, which encompasses ptk, is responsible for capsule biosynthesis, and ptk mutants produce very HMW polysaccharides [Citation40]. Our data confirm that ptk mutations trigger the production of HMW exopolysaccharides, which have a pro-inflammatory effect on host cells. Capsular exopolysaccharide is important for soft tissue infection, defense against serum factor-mediated killing, and biofilm formation [Citation41]. Therefore, the acquisition of a mucoid phenotype promotes the dissemination and persistence of A. baumannii in the host.
Antibiotic resistance contributes to the persistence of A. baumannii in the host [Citation39] and is an adaptive response to stress [Citation21]. In this study, epithelial cell-adapted A. baumannii showed enhanced resistance to ceftazidime, cefoperazone/sulbactam, cefepime, and cefiderocol, as a result of mutations to rsmG and rnaseI (linked to RNA modification and metabolism). rsmG, which encodes S-adenosylmethionine (SAM)-dependent 16S rRNA methyltransferase, is involved in 7-methylguanosine modification (m7G) of the 530 loops of 16S rRNA in a variety of Gram-negative bacteria [Citation42,Citation43]. In this study, mutations in rsmG resulted in the loss of the m7G modification of 16S rRNA in A. baumannii, and the modification site was G524 of 16S rRNA. We excluded G526 as a potential methylation modification site because the RNA probe retained the m7G modification after the change from 526G to 526A. Therefore, the reduction of the m7G/G ratio was likely related to the altered secondary structure of RNA probe 3. RsmG mutation-associated loss of m7G modification mediates low-level streptomycin resistance [Citation42,Citation43]. In this study, rsmG mutation reduced susceptibility to several β-lactams, however, the resistance mechanism is unknown. RNA-seq revealed no resistance mechanism. The universally conserved 530 loops of 16S rRNA is crucial for translation [Citation44,Citation45]. Loss of the m7G modification may affect ribosomal function [Citation46], and the translation efficiency and fitness of bacterial pathogens [Citation47]. Therefore, we hypothesized that the RsmG mutation enhances β-lactam resistance by affecting ribosome function and protein translation. Indeed, rsmG mutation increased ribosomal translation efficiency, a rapid-response mechanism to environmental stress [Citation48,Citation49]. Stress can reduce the overall protein translation capacity of bacteria [Citation50], hampering their survival. Maintenance of translation efficiency guarantees timely synthesis of stress-responsive proteins, enabling long-term survival during colonization and infection [Citation51], Therefore, rsmG mutation-mediated upregulation of protein translation efficiency may promote bacterial survival under β-lactam stress.
β-lactam tolerance can also be mediated by overexpression of genes encoding efflux pumps and outer-membrane changes. Indeed, our data suggest that these were altered by rnaseI mutation. RNase I regulate bacterial biofilm formation, motility, acid resistance, β-lactam tolerance, and nucleotide metabolism [Citation52]. Under stress conditions, periplasmic RNase I enter the cell, leading to extensive RNA degradation [Citation53]. In this study, the mRNA stability of RND efflux pumps and outer membrane genes was upregulated by rnaseI mutation, enhancing the expression of RND efflux pumps, changing the morphology of the outer membrane, and decreasing its permeability, leading to β-lactam resistance. In addition, impaired iron utilization caused by the rsmG and rnaseI mutations promoted resistance to cefiderocol. This is unlike previously reported cefiderocol resistance mechanisms, such as mutations in genes encoding iron transport-related proteins and the transfer of an extended-spectrum β-lactamase-encoding gene [Citation54,Citation55]. This study demonstrates a mutagenic role for host factors previously thought to be insusceptible to resistance by mutation.
Conclusion
A. baumannii adapted to host-associated selective pressures during 9 days of co-culture with epithelial cells. Mucoid clones with β-lactam resistance and the ability to evade adherence and phagocytosis rapidly emerged. Our data reveal the complex influence of pulmonary epithelial cells on A. baumannii pathoadaptation, implicating the host-microbe interaction in A. baumannii survival and persistence. This pathoadaptation could explain the emergence of antibiotic resistance and tissue translocation in patients infected with A. baumannii. Our findings provide insight into the effect of the host response on the evolution of A. baumannii.
Authors’ contributions
W. Z. and Y.S.Y. wrote the main manuscript text, X.T.H., H.Z., and Z.H.Z. designed this study, Y.Y., J.T.H, J.F.W., L.L., M.S.G., Y.F., and Y.S. participated in part of the experiments, H.C., J.Z.L., X.T.H., and J.Z.L. revised the manuscript, Y.Y. and X.C.L. analyzed the data.
Supplemental Material
Download MS Word (927.8 KB)Acknowledgements
The work was supported by the National Natural Science Foundation of China (82072313, 31770142, 81971897) and the National Science Foundation of China International Cooperation and Exchange Programme (81861138054). We are grateful to Dr. Fengqin Wang from College of Animal Sciences, Zhejiang University for her great support to this work.
Data availability
The complete genome sequences of the evolved strains (ACD9 and HCD9) were deposited in NCBI under BioProject No. PRJNA867505. The RNA-seq data of A. baumannii ATCC17978, rnase I, and rsmG mutants were deposited in NCBI under BioProject No. PRJNA867119. The direct RNA-seq (ONT) data of A. baumannii rsmG WT/mutants were deposited in NCBI under BioProject No. PRJNA882509.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Olive AJ, Sassetti CM. Metabolic crosstalk between host and pathogen: sensing, adapting and competing. Nat Rev Microbiol. 2016 Apr;14(4):221–234.
- Gatt YE, Margalit H. Common adaptive strategies underlie within-host evolution of bacterial pathogens. Mol Biol Evol. 2021 Mar 9;38(3):1101–1121.
- Proenca JT, Barral DC, Gordo I. Commensal-to-pathogen transition: one-single transposon insertion results in two pathoadaptive traits in Escherichia coli -macrophage interaction. Sci Rep. 2017 Jul 3;7(1):4504.
- Diaz Caballero J, Clark ST, Coburn B, et al. Selective sweeps and parallel pathoadaptation drive Pseudomonas aeruginosa evolution in the cystic fibrosis lung. mBio. 2015 Sep 1;6(5):e00981-15.
- Mwangi MM, Wu SW, Zhou Y, et al. Tracking the in vivo evolution of multidrug resistance in Staphylococcus aureus by whole-genome sequencing. Proc Natl Acad Sci U S A. 2007 May 29;104(22):9451–9456.
- Magret M, Lisboa T, Martin-Loeches I, et al. Bacteremia is an independent risk factor for mortality in nosocomial pneumonia: a prospective and observational multicenter study. Critical Care (London, England). 2011;15(1):R62.
- Brotfain E, Borer A, Koyfman L, et al. Multidrug resistance Acinetobacter bacteremia secondary to ventilator-associated pneumonia: risk factors and outcome. J Intensive Care Med. 2017 Oct;32(9):528–534.
- Eijkelkamp BA, Stroeher UH, Hassan KA, et al. H-NS plays a role in expression of Acinetobacter baumannii virulence features. Infect Immun. 2013 Jul;81(7):2574–2583.
- Zhang W, Zhou H, Jiang Y, et al. Acinetobacter baumannii outer membrane protein A induces pulmonary epithelial barrier dysfunction and bacterial translocation through the TLR2/IQGAP1 axis. Front Immunol. 2022;13:927955.
- Geisinger E, Huo W, Hernandez-Bird J, et al. Acinetobacter baumannii: envelope determinants that control drug resistance, virulence, and surface variability. Annu Rev Microbiol. 2019 Sep 8;73:481–506.
- Campos MA, Vargas MA, Regueiro V, et al. Capsule polysaccharide mediates bacterial resistance to antimicrobial peptides. Infect Immun. 2004 Dec;72(12):7107–7114.
- Llobet E, Tomas JM, Bengoechea JA. Capsule polysaccharide is a bacterial decoy for antimicrobial peptides. Microbiology (Reading). 2008 Dec;154(Pt 12):3877–3886.
- Jones A, Georg M, Maudsdotter L, et al. Endotoxin, capsule, and bacterial attachment contribute to Neisseria meningitidis resistance to the human antimicrobial peptide LL-37. J Bacteriol. 2009 Jun;191(12):3861–3868.
- Smith MG, Gianoulis TA, Pukatzki S, et al. New insights into Acinetobacter baumannii pathogenesis revealed by high-density pyrosequencing and transposon mutagenesis. Genes Dev. 2007 Mar 1;21(5):601–614.
- Wong D, Nielsen TB, Bonomo RA, et al. Clinical and pathophysiological overview of Acinetobacter infections: a century of challenges. Clin Microbiol Rev. 2017 Jan;30(1):409–447.
- McConnell MJ, Actis L, Pachon J. Acinetobacter baumannii: human infections, factors contributing to pathogenesis and animal models. FEMS Microbiol Rev. 2013 Mar;37(2):130–155.
- Naumann M. Nuclear factor-kappa B activation and innate immune response in microbial pathogen infection. Biochem Pharmacol. 2000 Oct 15;60(8):1109–1114.
- Mortensen BL, Skaar EP. Host-microbe interactions that shape the pathogenesis of Acinetobacter baumannii infection. Cell Microbiol. 2012 Sep;14(9):1336–1344.
- Wright MS, Jacobs MR, Bonomo RA, et al. Transcriptome remodeling of Acinetobacter baumannii during infection and treatment. mBio. 2017 Mar 7;8(2):e02193-16.
- Quinn B, Rodman N, Jara E, et al. Human serum albumin alters specific genes that can play a role in survival and persistence in Acinetobacter baumannii. Sci Rep. 2018 Oct 3;8(1):14741.
- Martinez J, Fernandez JS, Liu C, et al. Human pleural fluid triggers global changes in the transcriptional landscape of Acinetobacter baumannii as an adaptive response to stress. Sci Rep. 2019 Nov 21;9(1):17251.
- Hua X, He J, Wang J, et al. Novel tigecycline resistance mechanisms in Acinetobacter baumannii mediated by mutations in adeS, rpoB and rrf. Emerg Microbes Infect. 2021 Dec;10(1):1404–1417.
- Deatherage DE, Barrick JE. Identification of mutations in laboratory-evolved microbes from next-generation sequencing data using breseq. Methods Mol Biol. 2014;1151:165–188.
- Li Y, Zhu Y, Zhou W, et al. Alcaligenes faecalis metallo-β-lactamase in extensively drug-resistant Pseudomonas aeruginosa isolates. Clin Microbiol Infect. 2022 Jun;28(6):880.e1–880.e8.
- CLSI. Performance standards for antimicrobial susceptibility testing. 29th ed. Wayne (PA): Clinical and Laboratory Standards Institute; 2019; p. 1–282.
- Hu L, Shi Y, Xu Q, et al. Capsule thickness, not biofilm formation, gives rise to Mucoid Acinetobacter baumannii phenotypes that are more prevalent in long-term infections: a study of clinical isolates from a hospital in China. Infect Drug Resist. 2020;13:99–109.
- Xu Q, Chen T, Yan B, et al. Dual role of gnaA in antibiotic resistance and virulence in Acinetobacter baumannii. Antimicrob Agents Chemother. 2019 Oct;63(10):e00694-19.
- Liu H, Begik O, Lucas MC, et al. Accurate detection of m(6)A RNA modifications in native RNA sequences. Nat Commun. 2019 Sep 9;10(1):4079.
- Liu J, Yue Y, Han D, et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol. 2014 Feb;10(2):93–95.
- Jia G, Fu Y, Zhao X, et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol. 2011 Oct 16;7(12):885–887.
- Li W, Li L, Zhang C, et al. Investigations into the antibacterial mechanism of action of Viridicatumtoxins. ACS Infect Dis. 2020 Jul 10;6(7):1759–1769.
- San Millan A. Evolution of plasmid-mediated antibiotic resistance in the clinical context. Trends Microbiol. 2018 Dec;26(12):978–985.
- Hua X, Liu L, Fang Y, et al. Colistin resistance in Acinetobacter baumannii MDR-ZJ06 revealed by a multiomics approach. Front Cell Infect Microbiol. 2017;7(45). doi:10.3389/fcimb.2017.00045
- Sokurenko EV, Hasty DL, Dykhuizen DE. Pathoadaptive mutations: gene loss and variation in bacterial pathogens. Trends Microbiol. 1999 May;7(5):191–195.
- Miskinyte M, Sousa A, Ramiro RS, et al. The genetic basis of Escherichia coli pathoadaptation to macrophages. PLoS Pathog. 2013;9(12):e1003802.
- Yu H, Hanes M, Chrisp CE, et al. Microbial pathogenesis in cystic fibrosis: pulmonary clearance of mucoid Pseudomonas aeruginosa and inflammation in a mouse model of repeated respiratory challenge. Infect Immun. 1998 Jan;66(1):280–288.
- Grana-Miraglia L, Lozano LF, Velazquez C, et al. Rapid gene turnover as a significant source of genetic variation in a recently seeded population of a healthcare-associated pathogen. Front Microbiol. 2017;8:1817.
- Sarantis H, Grinstein S. Subversion of phagocytosis for pathogen survival. Cell Host Microbe. 2012 Oct 18;12(4):419–431.
- Hua X, Zhou Z, Yang Q, et al. Evolution of Acinetobacter baumannii in vivo: international clone II, more resistance to ceftazidime, mutation in ptk. Front Microbiol. 2017;8:1256.
- Geisinger E, Isberg RR. Antibiotic modulation of capsular exopolysaccharide and virulence in Acinetobacter baumannii. PLoS Pathog. 2015 Feb;11(2):e1004691.
- Russo TA, Luke NR, Beanan JM, et al. The K1 capsular polysaccharide of Acinetobacter baumannii strain 307-0294 is a major virulence factor. Infect Immun. 2010 Sep;78(9):3993–4000.
- Okamoto S, Tamaru A, Nakajima C, et al. Loss of a conserved 7-methylguanosine modification in 16S rRNA confers low-level streptomycin resistance in bacteria. Mol Microbiol. 2007 Feb;63(4):1096–1106.
- Nishimura K, Hosaka T, Tokuyama S, et al. Mutations in rsmG, encoding a 16S rRNA methyltransferase, result in low-level streptomycin resistance and antibiotic overproduction in Streptomyces coelicolor A3(2). J Bacteriol. 2007 May;189(10):3876–3883.
- Powers T, Noller HF. A functional pseudoknot in 16S ribosomal RNA. EMBO J. 1991 Aug;10(8):2203–2214.
- Wong SY, Javid B, Addepalli B, et al. Functional role of methylation of G518 of the 16S rRNA 530 loop by GidB in Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2013 Dec;57(12):6311–6318.
- Benitez-Paez A, Villarroya M, Armengod ME. Regulation of expression and catalytic activity of Escherichia coli RsmG methyltransferase. RNA. 2012 Apr;18(4):795–806.
- Lioy VS, Goussard S, Guerineau V, et al. Aminoglycoside resistance 16S rRNA methyltransferases block endogenous methylation, affect translation efficiency and fitness of the host. RNA. 2014 Mar;20(3):382–391.
- Grenga L, Little RH, Chandra G, et al. Control of mRNA translation by dynamic ribosome modification. PLoS Genet. 2020 Jun;16(6):e1008837.
- Zhu M, Dai X. Bacterial stress defense: the crucial role of ribosome speed. Cellular and Molecular Life Sciences: CMLS. 2020 Mar;77(5):853–858.
- Dai X, Zhu M, Warren M, et al. Slowdown of translational elongation in Escherichia coli under hyperosmotic stress. mBio. 2018 Feb 13;9(1):e02375-17.
- Imlay JA. The molecular mechanisms and physiological consequences of oxidative stress: lessons from a model bacterium. Nat Rev Microbiol. 2013 Jul;11(7):443–454.
- Duggal Y, Fontaine BM, Dailey DM, et al. RNase I modulates Escherichia coli motility, metabolism, and resistance. ACS Chem Biol. 2020 Jul 17;15(7):1996–2004.
- Bechhofer DH, Deutscher MP. Bacterial ribonucleases and their roles in RNA metabolism. Crit Rev Biochem Mol Biol. 2019 Jun;54(3):242–300.
- Simner PJ, Patel R. Cefiderocol antimicrobial susceptibility testing considerations: the Achilles’ heel of the Trojan horse? J Clin Microbiol. 2020 Dec 17;59(1):e00951-20.
- McCreary EK, Heil EL, Tamma PD. New perspectives on antimicrobial agents: cefiderocol. Antimicrob Agents Chemother. 2021 Jul 16;65(8):e0217120.
