ABSTRACT
Influenza virus is a prominent cause of respiratory illness in humans. Current influenza vaccines offer strain-specific immunity, while provide limited protection against drifted strains. Broad-spectrum influenza vaccines can induce broad and long-term immunity, and thus are regarded as a future direction for the development of next-generation influenza vaccines. In this study, we have conceptualized a novel mRNA-based multi-antigen influenza vaccine consisting of three conserved antigens of influenza A virus, including the ectodomain of the M2 ion channel (M2e), the long alpha helix of haemagglutinin stalk region (LAH), and nucleoprotein (NP). The vaccine design aims to enhance its potency and promote the development of a future broad-spectrum influenza vaccine. Our mRNA-based vaccine demonstrated potent humoral and cellular responses throughout the time points of the murine model, inducing viral neutralizing antibodies, antibody-dependent cell cytotoxicity effect mediating antibodies and cross-reactive CD8+ T cell immune responses. The vaccine conferred broad protection against H1N1, H3N2, and H9N2 viruses. Moreover, the single-cell transcriptional profiling of T cells in the spleens of vaccinated mice revealed that the mRNA-based vaccine significantly promoted CD8+ T cells and memory T cells by prime-boost immunization. Our results suggest that the mRNA-based influenza vaccine encoding conserved proteins is a promising approach for eliciting broadly protective humoral and cellular immunity against various influenza viruses.
Introduction
Influenza viruses infect a significant proportion of the global population annually, causing a large number of deaths due to respiratory diseases, according to the World Health Organization [Citation1,Citation2]. The development and use of influenza vaccines are critical measures for preventing and controlling influenza virus infections. Currently, available influenza vaccines induce antibody responses against influenza haemagglutinin (HA) that are limited in their ability to cross-protection against shifted and drifted viruses, as well as the emerging zoonotic viruses, owing to antigenic shift and/or drift of the influenza A virus. To overcome this problem, two approaches may be applied: improving the vaccine candidate virus selection to closely match circulating influenza viruses, and developing broad-spectrum influenza vaccines that can provide broadly protective efficacy against multiple strains of the influenza virus.
Several highly conserved antigens, such as the stalk domain of the HA protein, the ectodomain of the M2 ion channel (M2e), nucleoprotein (NP) and matrix protein (MP), have gained much interest as potential targets for broad-spectrum vaccines of influenza viruses [Citation3,Citation4]. Our team has also been working on the feasibility of conserved regions of influenza virus, such as HA stalk [Citation5], M2 [Citation6], and NP [Citation7,Citation8], leading to a comprehensive understanding of their significance in developing a candidate for broad-spectrum vaccine. The long alpha helix (LAH) domain (HA2 76-130aa), the most conserved influenza HA stalk regions, was also explored as a target for broad-spectrum influenza vaccines [Citation5]. Antibodies against the stalk are generally broadly cross-reactive and often neutralizing [Citation4,Citation9], and also can help to eliminate infected cells through Fcγ-mediated effector functions like antibody-dependent cell cytotoxicity (ADCC) [Citation10]. M2e, the extracellular N-terminal domain of M2, is highly conserved among human influenza A strains [Citation11,Citation12], and mediated protective immunity primarily relies on Fcγ-mediated effector mechanisms, such as ADCC or antibody-dependent cell phagocytosis [Citation13–15]. However, as the low immunogenicity of M2e, previous studies have approached M2e domain conjugate to carrier vehicles, novel adjuvants, and multiple immunizations with high vaccine doses [Citation16–18]. NP, a conserved internal antigen, is the major target recognized by cytotoxic T lymphocytes (CTL) during influenza virus infection [Citation19], having been confirmed to mediate cross-protection against heterosubtypic influenza viruses [Citation20,Citation21]. Here we combined these three conserved domains together to trigger more comprehensive immune responses against a spectrum of influenza viruses.
Nucleoside-modified mRNA-lipid nanoparticle (LNP) vaccines are increasingly promising as vaccine strategies in the prevention of cancer and infectious diseases [Citation22,Citation23]. The success of mRNA vaccines against COVID-19 has prompted the development of seasonal influenza vaccine candidates using this technology, which are currently in early-stage clinical development. mRNA-based vaccines exhibit the advantage of not only stimulating humoral responses that block viral expansion but also producing great T-cell immune responses. Several influenza vaccines based on mRNA-LNP are under development, and most vaccines contain diverse HA antigens primarily focusing on inducing humoral responses [Citation24–31]. Moreover, researchers are optimistic that mRNA vaccines will stimulate stronger or more diverse immune responses through a combination of conserved influenza proteins [Citation27,Citation32]. Nonetheless, most broad-spectrum vaccines are a combined mixture constituted by separated mRNA-LNPs for each of the vaccine's antigens, resulting in challenges of uniformed and consistent expression [Citation24,Citation27,Citation32]. In addition, the combination of variant antigens especially conserved antigens is of great significance to vaccine design. In this study, we have designed a broad-spectrum vaccine based on a single mRNA molecule with a tandem of multiple conserved antigens. The vaccine elicited broadly humoral and cellular responses, demonstrating its potential as a promising candidate for the development of a broad-spectrum vaccine.
Materials and methods
Ethics statement
All experiments involving animals were approved by Animal Care Committee of Shanghai Institute of Biological Products, in accordance guideline for ethical review of animal welfare.
Viruses, cells, peptides, proteins and animals
Mouse adapted influenza viruses H1N1 (A/Puerto Rico/8/1934, A/PR/8), H3N2 (A/Guizhou/54/1989, A/GZ), and H9N2 (A/Chicken/Jiangsu/11/2002, A/JS), were propagated in 8–10 day-old embryonated chicken eggs [Citation8]. All virus aliquots were frozen and stored at −80°C.
HEK293 T cells and Madin-Darby canine kidney (MDCK) cells were grown in complete Dulbecco’s modified Eagle medium (DMEM, Gibco, USA) supplemented with 10% fetal bovine serum (FBS, Gibco).
M2e peptide of human type and LAH peptide pool of A/PR/8 were custom-ordered from Nanjing GenScript Biotech Corporation, China. LAH (HA2 76-130aa) peptide pool contained 15-amino acid-long peptides with an overlap of 6 amino acids spanning the entire sequence. Before use, peptides were dissolved in sterile water, aliquoted, and stored at −80°C.
NP protein of influenza A/PR/8 virus feature a C-terminal hexahistidine purification tag was produced by BL21 expression system. Proteins were then purified by affinity chromatography using nickel nitrilotriacetic acid agarose (QIAGEN, Germany). NP protein was stored at −80°C.
Specific-pathogen-free (SPF) female BALB/c mice (3-4 weeks old) were purchased from Zhejiang Vital River Laboratory Animal Technology Co., Ltd., China.
mRNA-LNP production
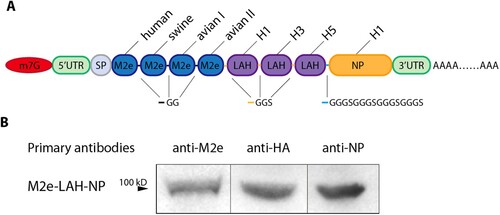
Encoding M2e-LAH-NP gene was synthesized (GenScript, China) and cloned into an mRNA production plasmid. The gene construct was genetically designed to contain a tPA signal peptide, four repeats of influenza virus M2e sequences from human influenza virus – SLLTEVETPIRNEWGSRSN, swine influenza virus – SLLTEVETPTRSEWESRSSDSS (A/California/4/2009, H1N1), avian influenza virus type I-SLLTEVETPTRNEWESRSSDSS (A/Vietnam/1203/04, H5N1) and avian influenza virus type II-SLLTEVETLTRNGWGCRCSDSS (A/Hong Kong/156/97, H5N1) [Citation33], three copies of LAH of HA2 sequences (HA2 76-130aa) from influenza A/PR/8, H3N2 (A/Hong Kong/1/1968) and H5N1 (A/Vietnam/1194/2004), a NP sequence from A/PR/8 virus with mutated the nuclear localization signals (NLSs) [Citation34], G5TKRSYXXM mutated to G5TAASYXXM, K213RTR mutated to A213ATA, R342V to A342A ((A)). T7-driven in vitro transcription reactions (Vazyme, China) using linearized plasmid templates were performed to generate mRNAs with N¹ -Me-Pseudo uridine. Capping of mRNAs was performed in concert with transcription through the addition of a trinucleotide cap1 analog, CleanCap (TriLink, USA). mRNAs was purified using mRNA Capture Beads (Vazyme). mRNAs were then tested on an agarose gel before storing at −80°C.
Figure 1. mRNA design and protein expression in transfected HEK293 T cells. (A) Schematic diagram of the design of MLN-mRNA vaccine. Codon-optimized M2e, LAH and NP sequences were inserted into the region between 5’UTR and 3’UTR. SP represents signal peptide. (B) Expression of MLN-mRNA-encoded proteins measured by immunoblotting. Samples were cell lysates from MLN-mRNA-transfected HEK293 T cells.
Purified mRNAs were formulated into LNP using a process wherein an ethanolic lipid mixture of an ionizable cationic lipid, phosphatidylcholine, cholesterol, and polyethylene glycol lipid (AVT Pharmaceutical Technology, China), at molar ratios of 46:10:42.5:1.5 with a microfluidic mixer (Micronano biologics, China). The average hydrodynamic diameter was ∼90 nm with a polydispersity index of <0.1 as measured by a Zetasizer Nano ZS (Malvern Instruments, UK) and an encapsulation efficiency of ∼90–95% as determined using a RiboGreen assay (Invitrogen, USA).
Cell transfections, western blot analysis and immunofluorescence assay of mRNA-LNPs
HEK293 T cells growing in 24 well plates were transfected with mRNAs combined with lipofectamine 3000 (Invitrogen). Cells were incubated at 37°C in 5% CO2 for 48 h. For Western blotting, mRNA-transfected and nontransfected HEK293 T cells were collected and lysed. The membrane was incubated with the anti-M2 (rabbit polyclonal, 1:1000, Abcam, UK), anti-H1N1 HA (rabbit polyclonal, 1:1000, Sino Biological, China), anti-NP (rabbit polyclonal, 1:1000, Invitrogen) or anti-β-actin (1:5000, rabbit polyclonal, Abcam) primary antibodies overnight at 4°C. Then the membrane was incubated with the horseradish peroxidase (HRP)-conjugated anti-rabbit (donkey polyclonal, 1:20000, Jackson ImmunoResearch, USA) secondary antibodies for 1 h at room temperature. The signal was developed with the tetramethylbenzidine (TMB) substrate solution (Beyotime, China) and imaged using Azure Imaging Systems (Azure Biosystem, USA). For Immunofluorescence assay, cells were fixed with 3.7% paraformaldehyde, permeabilized with 0.1% Triton X-100, blocked with 2% Bovine Serum Albumin (BSA)-Phosphate buffered saline (PBS) and then incubated with anti-NP antibodies (rabbit polyclonal, 1:1000, Invitrogen), followed by incubated with Alexa Fluor®488-conjugated anti-rabbit IgG (1:500, Jackson ImmunoResearch). DAPI (Sangon, China) was used as nuclear stain. Cells were then observed under immunofluorescence microscope (Olympus, Tokyo, Japan).
mRNA vaccination and virus challenge
Specific-pathogen-free female BALB/c mice were immunized via intramuscular injection of mRNA vaccine with a prime-boost strategy with a 3-week interval. PBS was used for the control group.
For challenge experiments, firstly, mice (n = 10) were infected from each dose ranging from 10 to 105 PFU in log interval to determine the 50% lethal dose (LD50) of each influenza virus. Mice were anesthetized with a ketamine/xylazine mixture, and 20 μL of virus at each dose was introduced through the intranasal route. Weight loss was monitored for 14 days, and mice losing 25% of their initial body weight were humanly sacrificed. The dose at which 50% of mice succumbed to infection was determined as the LD50 for future challenge studies. Then the immunized mice were anesthetized and infected with 20 μL 5×LD50 of H1N1 (A/PR/8), H3N2 (A/GZ), or H9N2 (A/JS) virus intranasally. The body weight and survival rate of infected mice were monitored for 14 days, and mice losing 25% of their initial body weight were humanly sacrificed.
Enzyme linked immunosorbent assay (ELISA)
Flat-bottom, 96-well plates were coated overnight with M2e peptide, LAH peptides or NP protein at 2 μg/ml (100 μl/well) at 4°C. Mice sera were diluted in a series of 2-fold dilutions and incubated at 37°C for 2 h. Then, the plates were washed and incubated with HRP-conjugated anti-mouse IgG, IgG1, IgG2a, IgG2b, IgG2c, IgA or IgM (1:1000, Southernbiotech, USA) at 37°C for 1 h. The signal was developed using TMB as the substrate. The reaction was stopped with sulfuric acid, and the optical density at 450 nm (OD450) was read on a microtitre plate reader SpectraMax i3x (Molecular Devices, USA). The area under the curve (AUC) was calculated using a baseline of the average of the control group plus 2 times the standard error of the mean (SEM), or 0.1 if the baseline was lower than 0.1.
Antibody-Dependent cell-mediated cytotoxicity (ADCC) assay
MDCK cells growing in 96 well plates overnight were added influenza virus A/PR/8 (MOI = 5) and incubated at 37°C in 5% CO2 for 16 h. Serum samples were serial diluted 2-fold in RPMI 1640 (Gibco) containing 0.1% FBS. Medium of infected MDCK cells was removed, followed by adding 50 μL diluted serum samples. After incubated at 37°C in 5% CO2 for 30 min, the plates were added 25 μL Jurkat cells expressing the mouse FcγRIV cell-surface receptor at a concentration of 6 × 106 cells/mL, and allowed to incubate at 37°C in 5% CO2 for 6 h. 75 μL of Bio-Glo™ Luciferase substrate (Promega, USA) were added to each well and luminescence was immediately read on SpectraMax i3x (Molecular Devices).
Microneutralization (MN) assay
Heat-inactivated serum was serially diluted on 96-well plates and mixed with 100-fold 50% tissue culture infection dose (TCID50) of virus at 37°C in 5% CO2 for 1 h. The serum/virus mixtures were transferred to plates containing MDCK cells and incubated at 37°C in 5% CO2 for 3 days. 50 μL of the culture was mixed with 50 μL 1% fresh chicken red blood cells. The mixture was incubated for 45 min at room temperature, after which agglutination was observed. Each sample was tested three times, and the results are reported as the geometric mean titre (GMT) of the three replicates.
Single cell suspension preparation
Spleens, lungs and lymph nodes were collected from mice and mashed through 40-micron Nylon cell strainers (Corning, USA). Flow-through cells were washed with RPMI 1640 (Gibco) containing 2% FBS and 100 U/mL penicillin–streptomycin (Gibco). Red blood cells were lysed with Mouse Erythrocyte Lysing Kit (R&D Systems, USA). Cells were counted by the Cellometre (Nexcelom Bioscience, USA).
Enzyme-Linked immunospot (ELISpot) assay
The ELISpot assay is a sensitive tool to measure antigen-specific T cell responses and antigen-specific antibody-secreting cells. For examining T cell responses, Mouse IFN-γ ELISpotPLUS kit (MabTech, Sweden) was used according to the manufacturer’s protocol. The plates were read by the ImmunoSpot reader (Cellular Technology, USA).
For the identification of antigen-specific antibody-secreting cells, M2e peptide, LAH peptide pool or NP protein at a final concentration of 5 μg/mL was pre-coated to MultiScreen-IP filter plates (Millipore, USA) overnight. 5 × 105 cells isolated from spleen, lung or lymph node were added in triplicates to plates and then incubated at 37°C in 5% CO2 for 48 h. After washing with PBS, the plates were added HRP-conjugated anti-mouse IgG (1:20000, Jackson ImmunoResearch) at room temperature for 1 h. Spots were exposed by TMB solution and were read by the ImmunoSpot reader (Cellular Technology).
Surface and intracellular cytokine staining
For surface staining, spleen cells were first incubated with an anti-CD16/32 antibody (1:200, BD Biosciences, USA) to block the Fc receptor. For T follicular helper (TFH) cells, cells were incubated with PE-conjugated anti-CD3 (1:100, Biolegend, USA), PE/Cyanine5-conjugated anti-CD4 (1:500, Biolegend), FITC-conjugated anti-CXCR5 (1:100, Biolegend), APC-conjugated anti-PD-1 (Biolegend), and the yellow LIVE/DEAD® dye (Invitrogen). For germinal centre (GC) B cells, cells were labeled with PE-conjugated anti-CD3 (1:100, Biolegend), PE/Cyanine7-conjugated anti-B220 (1:100, Biolegend), APC-conjugated anti-CD95 (1:200, Biolegend), Alexa Fluor® 488-conjugated anti-GL7 (1:200, Biolegend), and the yellow LIVE/DEAD® dye (1:500, Invitrogen). For central memory T cells (TCM) and effector memory T cells (TEM), cells were marked with PerCP/Cyanine5.5-conjugated anti-CD3 (1:100, Biolegend), FITC-conjugated anti-CD4 (1:100, Biolegend), BV650-conjugated anti-CD8α (1:100, Biolegend), APC/Cyanine7-conjugated anti-CD44 (1:100, Biolegend), PE-conjugated anti-CD62L (1:100, Biolegend), and the yellow LIVE/DEAD® dye (1:500, Invitrogen). For TRM, cells were stained with PE-conjugated anti-CD3 (1:100, Biolegend), PE/Cyanine5-conjugated anti-CD4 (1:500, Biolegend), BV650-conjugated anti-CD8α (1:100, Biolegend), APC-conjugated anti-CD69 (1:100, Biolegend), FITC-conjugated anti-CD103 (1:100, Biolegend), and the yellow LIVE/DEAD® dye (1:500, Invitrogen).
For intracellular cytokine staining (ICS), the cells isolated from spleens were stimulated with a mixture of M2e, LAH and NP (the molar ratio is 4:3:1) at 37°C in 5% CO2 for 6 h in the presence of Protein Transport Inhibitor (BD Biosciences). After fixation with Fixation/Permeabilization solution (BD Biosciences), cells were first incubated with anti-CD16/32 antibody (1:200, BD Biosciences). Half of cells were stained with PE-conjugated anti-CD3 (1:100, Biolegend), PE/Cyanine5-conjugated anti-CD4 (1:500, Biolegend), BV650-conjugated anti-CD8α (1:100, Biolegend), FITC-conjugated anti-IFN-γ (1:100, Biolegend), Alexa Fluor® 700-conjugated anti- interleukin (IL) −2 (1:100, BD Biosciences), BV421-conjugated anti-TNF-α (1:100, BD Biosciences), and APC-conjugated anti-IL-4 (1:100, Biolegend). The other half of cells were labeled with PE/Cyanine5-conjugated anti-CD3ϵ (1:100, Biolegend), FITC-conjugated anti-CD4 (1:500, Biolegend), BV650-conjugated anti-CD8α (1:100, Biolegend), PE-conjugated anti-IL-17A (1:100, Biolegend), APC-conjugated anti-IL-21 (1:100, Biolegend).
Flow cytometric analysis and cell sorting were performed on Attune NxT Acoustic Focusing Cytometer (Invitrogen). The raw data collected were gated in FlowJo™ 10.8.1 (TreeStar, USA).
Cytokine ELISA
The cells isolated from spleens were cultured and were stimulated with a mixture of M2e, LAH and NP (the molar ratio is 4:3:1). The culture supernatants were harvested after 72 h and secreted IFN-γ and IL-4 were measured by Mouse IFN-γ Precoated ELISA Kit and Mouse IL-4 Precoated ELISA Kit (Dakewe, China) based on the manufacturer’s instructions. All the tests were performed in duplicates. OD450 was read and cytokine level was measured for each sample based on standard curves.
Histopathology
Mice were euthanized at 3, 5 and 7 days following influenza virus challenge. The left lung lobe of mice were fixed with 4% paraformaldehyde, and embedded in paraffin according to standard histological assays. Then, lung tissues were sectioned and stained with hematoxylin and eosin (H&E). Images were captured using Slide Scanner Pannoramic Desk/Midi/250/1000 (3DHISTECH, Hungary) with slide viewing software CaseViewer2.4 (3DHISTECH, Hungary)
Viral load determination
The right lung lobe of mice were frozen and homogenized for determining level of viral load, granzyme B and perforin. All homogenate samples were stored at −80°C. For viral load determination, MDCK cells were grown in 96 well plates overnight. The lung homogenates were 10-fold serially diluted from 1:10 in DMEM (Gibco) supplemented with 2 µg/mL N-tosyl-L-phenylalanine chloromethyl ketone (TPCK)-trypsin (Sigma-Aldrich, USA) and 100 U/mL penicillin–streptomycin, and inoculated in MDCK cells for 3 days at 37°C in 5% CO2. 50 μL of the culture was mixed with 50 μL 1% fresh chicken red blood cells. The mixture was incubated for 45 min at room temperature, after which agglutination was observed. The virus titre was calculated by the Reed-Muench method, expressed as TCID50. The virus titre was represented as the mean ± SEM of the virus titre per ml of specimens from five mice in each group.
Granzyme B and perforin ELISA
Granzyme B and Perforin of homogenate samples above were measured by Mouse Granzyme B ELISA Kit (MultiSciences Biotech, China) and Mouse Perforin 1 (PF1) ELISA Kit (Jianglai Biological, China) according to the manufacturers’ instructions. All the tests were performed in duplicates. OD450 was read and cytokine level was measured for each sample based on standard curves.
T cell depletion experiments
mRNA vaccinated mice were intraperitoneally injected with 1 mg anti-CD4 (clone GK1.5, BD Biosciences) and 500 μg anti-CD8 (clone 2.43, BD Biosciences) depletion antibodies, respectively, at 2 days before infection and 1 d after A/PR/8 virus infection. Control mice received 500 μg mouse IgG2a produced in our lab.
Single-Cell RNA-Seq (scRNA-seq)
The fresh spleen tissues were stored in the sCelLive™ Tissue Preservation Solution (Singleron, China) on ice after the surgery within 30 min, and then digested with 3 mL sCelLive™ Tissue Dissociation Solution (Singleron) by PythoN™ Tissue Dissociation System (Singleron) at 37 °C for 15 min. The cell suspension was collected and filtered through a 40-micron sterile strainer. Afterwards, the red blood cell lysis buffer (RCLB, Singleron) was added and incubated at room temperature for 5-8 min. The mixture was then centrifuged to remove supernatant and suspended softly with PBS.
Single-cell suspensions (2 × 105 cells/mL) with PBS were loaded onto a microwell chip using the Matrix® Single Cell Processing System (Singleron). Barcoding beads are subsequently collected from the microwell chip, followed by reverse transcription of the mRNA captured by the beads to obtain cDNA for PCR amplification. The amplified cDNA is then fragmented and ligated with sequencing adapters. The scRNA-seq libraries were constructed according to the protocol of the Single Cell RNA Library Kits (Singleron). Individual libraries were diluted to 4 nM, pooled, and sequenced on an Illumina Novaseq 6000 (Illumina, USA) instrument with a 150 bp paired-end format. Full identified sequencing data are available in the gene expression omnibus (GEO) under accession number GSE234756.
Single-cell data processing
Cells were filtered by gene counts below 200 and the top 2% gene counts and the top 2% unique molecular identifier (UMI) counts. Cells with over 20% mitochondrial content were removed. We used functions from Seurat v3.1.2 [Citation35] for dimension-reduction and clustering. Then we normalized and scaled all gene expression, and selected the top 2000 variable genes for principal component analysis (PCA). Using the top 20 principle components, we separated cells into multiple clusters with FindClusters. Batch effect between samples was removed by Harmony [Citation36]. Finally, uniform manifold approximation and projection (UMAP) algorithm was applied to visualize cells in a two-dimensional space. FindAllMarkers was used to identify marker genes for each cluster, and cell types were manually annotated based on these marker genes. For gene set variation analysis (GSVA) pathway enrichment analysis, the average gene expression of each cell type was used as input data using the GSVA package.
Statistical analysis
Statistical analyses were performed using the Prism 9.4 program (GraphPad, San Diego, CA, USA). Single-cell data were performed using the R program. For comparing two groups, P-values were determined using Student’s t-tests (two-tailed). For comparing more than two groups, one-way ANOVAs followed by Tukey’s test were applied. All results are expressed as means ± SEM. All P values of < 0.05 were considered statistically significant. Asterisk markers for significance levels: * for P < 0.05, ** for P < 0.01, *** for P < 0.001, and **** for P < 0.0001.
Results
The design of MLN-mRNA vaccine
In this study, we sought to enhance the design of an mRNA vaccine for influenza by employing a transcript encoding tandem repeats of highly conserved protein domains present in various influenza A strains. Specifically, we utilized M2e (M2e4×) derived from human, swine, avian 1 and avian 2 influenza viruses respectively, LAH (LAH3×) derived from H1, H3, H5 strains, and the NP protein of A/PR/8 strain (H1N1) ((A)). Subsequently, we conducted codon optimization, selecting L04, which exhibited the highest in vitro expression level after preliminary screening, as the vaccine prototype (Supplementary Figure 1(A)). Immunoblotting analysis was employed to confirm the expression of each of the vaccine's antigens ((B)). Furthermore, to enable the conducive secretion of this fusion protein, we replaced the NLSs in NP [Citation34,Citation37–39], allowing for cytoplasmic localization, which was confirmed by immunofluorescence (Supplementary Figure 1(B)).
In addition, we compared the efficacy of our multi-antigen-based mRNA (MLN-mRNA) with single-antigen-based mRNAs (namely, M2e4×-mRNA, LAH3×-mRNA, and NP-mRNA, Supplementary Figure 2(A)). We intramuscularly immunized BALB/c mice with equimolar ratio of these vaccines and challenged them with A/PR/8 virus. Despite similar IgG response seen in mice immunized with MLN-mRNA and single-antigen vaccines (Supplementary Figure 2(B)–(D)), our MLN-mRNA vaccine provided the best protection with up to 80% survival rate and less body weight loss (Supplementary Figure 2(E)–(H)). These results suggest the potential of our MLN-mRNA vaccine prototype.
MLN-mRNA vaccine elicited humoral immune responses
We utilized a prime-boost strategy to immunize BALB/c mice at three-week intervals to assess the humoral immune response elicited by the MLN-mRNA vaccine against influenza virus ((A)). After determining the best sub-optimal dose for subsequent experiments (Supplementary Figure 3), we found that the vaccine induced IgGs reactive to M2e, LAH peptides, or NP proteins, with a significant dose-dependency ((B)). While the prime alone induced higher levels of IgM, it was not sufficient to increase IgA or IgG levels, unlike the boost procedure, which significantly and systemically increased IgG1, IgG2a, and IgG2b ((C), Supplementary Figure 4). We observed a positive correlation between the increase in M2e/LAH/NP-specific antibody levels and the number of antibody-secreting spleen B cells induced by the vaccine after both prime and boost procedures ((D)). Similar results were obtained for lungs and lymph nodes after the boost procedure ((E)).
Figure 2. MLN-mRNA vaccination elicited humoral immune responses. (A) Schematic of the immunization, virus challenge and sampling strategy. BALB/c mice were immunized intramuscularly with MLN-mRNA vaccine (5, 10, 20 μg) following a prime-boost program, followed by viral challenge and tissue sample collection. (B) Measurement of M2e-/LAH-/NP- specific IgG antibodies post-prime or boost. IgG levels against M2e/LAH peptides or NP protein in mice serum were analysed by ELISA. (C) Isotypes of NP-specific IgG antibodies were determined using anti-mouse immunoglobin isotype-specific antibodies. (D and E) Antigen-specific antibody-secreting cells were analysed by B-cell ELISpot assay. Spot-counting for M2e-/LAH-/NP-specific antibody-secreting cells in spleen seven days post-prime or boost (D). Spot-counting for M2e-/LAH-/NP-specific antibody-secreting cells in spleens, lungs, and lymph nodes seven days post boost (E). (F) ADCC activities of sera were measured using a reporter assay to determine the engagement with mouse FcγR. Three technical replicates were conducted for each sample, and curves were fit using a non-linear regression formula (four parameter logarithmic). (G) The proportion of splenic GC B cells (GL7 + CD95+) in B cells collected from the 20 μg vaccine-treated mice seven days post-prime or boost. (H) The proportion of splenic TFH cells (CD4 + PD1 + CXCR5) in the CD4+ T population collected from the 20 μg vaccine-treated mice seven days post-prime or boost. Statistical significance was calculated by one-way ANOVA; *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. Data represents mean value and error bars for SEM.
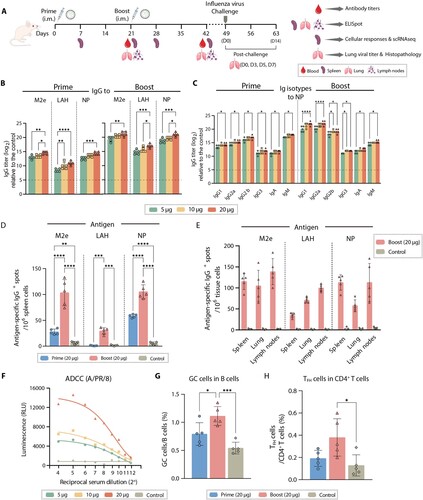
Given that antibodies against M2e and LAH can elicit Fc-mediated effector functions [Citation10,Citation13–15], we evaluated the ADCC effect of the serum derived from vaccine immunized mice and found that it demonstrated a strong antibody-mediated cytotoxicity against cells infected with A/PR/8 virus, particularly at the highest dose (20 μg) ((F)). Additionally, we used a MN assay to evaluate the neutralization activity of antibodies induced by the mRNA vaccines. Our results revealed that the mRNA vaccine containing LAH peptides displayed neutralizing activity against A/PR/8 virus, whereas vaccines containing only M2e peptides or NP exhibited negligible activity (Supplementary Figure 5). In addition to serological responses, we also examined GC formation in spleens by measuring the proportion of GC B cells (GL7 + CD95+) and CD4+ TFH cells (CD4 + PD1 + CXCR5+) which were critical to antibody production (Supplementary Figure 6). We observed that the MLN-mRNA vaccine elicited higher levels of induction for GC B cells and TFH cells after boost injections, while such an effect was less pronounced after prime injections, consistent with previous reports that nucleoside-modified mRNA vaccines generate stronger GC B cell reactions [Citation40] ((G) and (H)). In conclusion, our study demonstrated that the MLN-mRNA vaccine can elicit robust humoral immune responses against influenza viruses.
MLN-mRNA vaccine induced strong cellular immune responses
To assess the cellular-based immune response elicited by the MLN-mRNA vaccine, we measured the secretion profiles of IFN-γ and IL-4 in splenic cells treated with MLN-mRNA-encoding peptides as a stimulant. Our findings indicated that these peptides induced CD4+ T helper type 1 (TH1)-biased cellular immunity, as demonstrated by a high IFN-γ/IL-4 ratio ((A)–(C)). Furthermore, we observed a robust dose-dependent increase in IFN-γ-secreting cells in spleens, lungs, and lymph nodes when stimulated with M2e and NP, but not with LAH ((D)). This indicates a lack of T cell epitopes on the LAH peptide, which was similar to the data from the Immune Epitope Database (www.iedb.org).
Figure 3. MLN-mRNA vaccination induced cellular immunity. (A to C) Splenic lymphocytes were collected 21 days post boost, followed by re-stimulation using a mixture of M2e, LAH and NP (4:3:1, molar ratio). IFN-γ (A) and IL-4 (B) in the conditional media were detected using ELISA. (C) Ratio between IFN-γ and IL-4 level is used to represent TH1-biased immune response. (D) Quantification of re-stimulated IFN-γ-secreting T cells by ELISpot assay in spleens, lungs, lymph nodes 21 days post boost after a two-day-stimulation with M2e-LAH-NP mixture. (E and F) Multifunctional cytokine analysis of CD4+ and CD8+ T cells in spleens of immunized mice on day 7 post-prime or boost in the high dosage group (20 μg). After stimulated by M2e-LAH-NP mixture for 6 h, spleen cells were stained for intracellular cytokines and analysed by flow cytometry. (G to I) T cell memory induced by MLN-mRNA vaccine. The proportion of CD4+ and CD8+ T cells (G), CD8+ TCM (CD8 + CD44 + CD62L+) and CD8+ TEM (CD8 + CD44 + CD62L-) (H), CD4 + TRM (CD4 + CD69 + CD103+) and CD8 + TRM (CD8 + CD69 + CD103+) cells (I) at days 7 post-prime or boost in splenic lymphocytes. Statistical significance was calculated by one-way ANOVA; *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. Data represents mean value and error bars for SEM.
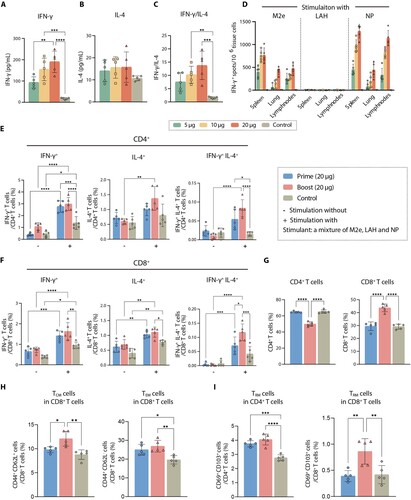
To examine cytokine-secreting CD4+ and CD8+ T cell responses independently, we utilized an ICS assay to measure the proportion of IFN-γ, TNF-α, IL-4, IL-2, IL-17A, and IL-21 on cells (Supplementary Figure 7). Mice were immunized with 20 μg of MLN-mRNA vaccine via prime or boost immunization, and their splenocytes were collected and cytokine expression was measured after stimulation with M2e peptides and NP for 6 h. While prime immunization elicited limited but detectable antigen-specific CD4+ and CD8+ T cell responses, boost immunization had a pronounced effect. Consistent with our previous results, CD4+ T cell responses were primarily TH1-type, with less induction of IL-4 response. There were increases in IFN-γ+, TNF-α+, and IL-2+ cytokine-secreting CD4+ T cells, as well as IFN-γ+, IL-4+, IL7A+, and IL-21+ CD8+ T cells in spleens after boost vaccinations ((E) and 3(F), Supplementary Figure 8 and 9), and multifunctional CD4+ and CD8+ T cells producing both IFN-γ and IL-4 were also stimulated. These results demonstrate that the MLN-mRNA vaccine produces a robust cellular response, which promotes combating influenza infection.
Moreover, we characterized the long-living memory T cells induced by the MLN-mRNA vaccine (Supplementary Figure 10). CD8+ T cells were extensively mobilized and recruited into spleens upon antigen re-exposure by booster injection ((G)). Long-living memory cells can be classified into two classes: TCM and TEM [Citation41]. TCM exhibits significant proliferative potential upon re-infection, develops into protective TEM, and confers long-lasting protective immunity against infection [Citation42], while the prevention of local infection is governed by tissue-resident memory T cells (TRM) [Citation43,Citation44]. Our results showed that re-immunization with MLN-mRNA vaccine significantly induced both TCM and TEM in CD8+ T cells ((H)) and caused a significant expansion of CD8+ TRM cells marked by CD69+ and CD103+ within spleen tissue ((I)). Thus, the MLN-mRNA vaccine promotes various long-living memory T cells in the spleen.
Protective efficacy of MLN-mRNA vaccine against influenza virus challenge in mice
We aimed to evaluate the protective efficacy of the MLN-mRNA vaccine against influenza virus challenge in vivo. To this end, we immunized mice intramuscularly with varying doses of the MLN-mRNA vaccine (5, 10, or 20 μg) and challenged them with three different strains of influenza A virus (A/PR/8 (H1N1), A/GZ (H3N2) and A/JS (H9N2)). Weight loss was monitored for 14 days post-infection. The control group exhibited severe sickness after the H1N1 (A/PR/8) virus challenge, resulting in significant weight loss (approximately 25%) and euthanasia at day 7–9 ((A)). In contrast, all mice vaccinated with MLN-mRNA were protected from infection, with the 20 μg dose showing the best efficacy, and no mortality observed. The same trend was observed in mice challenged with H3N2 (A/GZ) and H9N2 (A/JS) viruses, except for the development of visible morbidity after the challenge ((B) and 4(C)). We also measured residual viral load in the lungs of mice post-challenge with A/PR/8 virus by TCID50 and found that MLN-mRNA-immunized mice had significantly lower viral titres on day 5 or 7 post-challenge compared to the control cohort, where the viral load peaked on day 5 and then decreased ((D)).
Figure 4. MLN-mRNA vaccination protected mice from influenza virus challenge. (A to C) Body weight measurement and survival curves of mice. Adult mice (n = 10) were immunized twice with 5, 10, 20 μg of the MLN-mRNA vaccine and were challenged after 4 weeks post boost injection with a lethal doses (5×LD50) of H1N1 (A/PR/8), H3N2 (A/GZ), or H9N2 (A/JS) virus. Body weight was monitored daily for 14 days post-infection. Statistical significance was calculated by Student’s t-tests (two-tailed). (D) Mice (n = 5) were euthanized at days 3, 5 and 7 post-challenge with H1N1 (A/PR/8), and their lungs were harvested and homogenized for viral titre determination by TCID50. The homogenates were also used for determination of granzyme B (E) and perforin 1 (F) by ELISA. (G) Hematoxylin and eosin (H&E) staining of the lung sections. Lungs were collected 3, 5 and 7 days post-infection. (H and I) Mice (n = 10) were immunized twice with 20 μg of the MLN-mRNA vaccine and were intraperitoneally received with control, CD4- or CD8-depleting antibodies at −2 days and +1 d of challenge. All mice were challenged with a lethal dose (5×LD50) of H1N1 (A/PR/8). Body weight, survival curves for mice (H), and lung viral titres (I) at day 5 post-infection. Statistical significance was calculated by one-way ANOVA; *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. Data represents mean value and error bars for SEM.
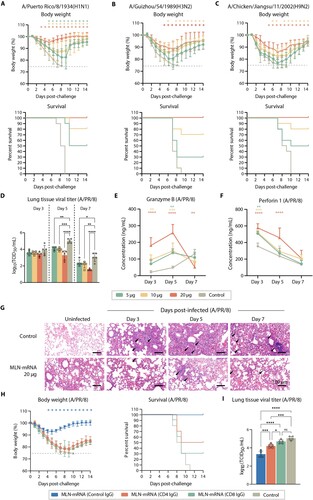
Since it is well-established that M2e/LAH-specific antibodies induced by MLN-mRNA vaccine are involved in cross-protection via ADCC or antigen neutralization [Citation27], to delve deeper into the mechanism of immune response underlying the protective effect of MLN-mRNA vaccine we investigated the functions of T cells during this process. We examined the levels of granzyme B and perforin 1 which were the main factors in T cells anti-virus cytotoxicity in the lungs of mice post-challenge, to investigate the mechanism underlying the protective effect of MLN-mRNA vaccine ((E) and (F)). We found a significant increase in the levels of granzyme B and sustained perforin 1 levels in the lungs only in mice within the 20 μg dose group after 5 days of immunization, indicating the role of CD8+ T cell homing to the lungs post virus infection and the clearance of viruses by perforin/granzyme pathway in CD8+ T cells. Additionally, we performed lung histopathology analysis on days 3, 5, and 7 post-challenge, and observed thickening of the alveolar septa surrounded by a mixed inflammatory cell population, diffused perivascular necrosis with neutrophils/macrophages, necrotic cell fragments, and bronchiolar, peribronchiolar, and perivascular inflammations in samples from the control group (Figure G). Meanwhile, a visible reduction in inflammation surrounding the bronchi and arterioles was seen in the samples from the 20 μg MLN-mRNA-treated group. These results indicated a potent protective effect against influenza viruses by the MLN-mRNA vaccine in murine model.
To further validate the role of T cell immune response in MLN-mRNA vaccine induced protection to influenza infection. We administered CD4- and CD8-depleting antibodies or control IgG to MLN-mRNA vaccinated mice before and during the challenge with A/PR/8 virus. Following the confirmation of CD4+ and CD8+ T cells depletion (Supplementary Figure 11), we observed a significant rescue effect for mice administered with MLN-mRNA vaccine, which was dependent on the presence of CD4+ and CD8+ T cells ((H)). CD4+ T cells are important for class switching of B cells and therefore the development of anti-influenza antibody responses, while CD8+ T cells was the major contributor to clearance of influenza demonstrated by the measurement of viral titre in the lung ((I)). These results suggest that the MLN-mRNA vaccine induces not only ADCC or neutralizing antibodies, but also robust T cell responses to achieve protective efficacy against influenza virus.
The impact of the MLN-mRNA vaccine on T cell transcriptome
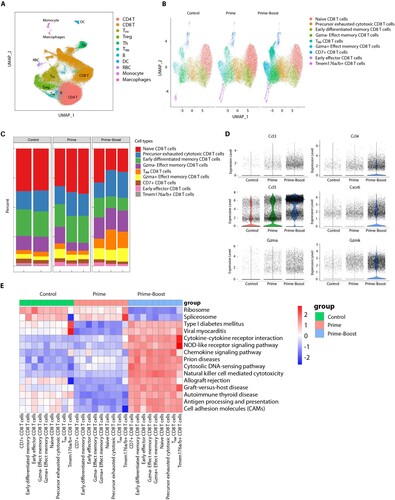
To delve deeper into the intricate interplay between the vaccine and T cell subpopulations, we conducted single-cell sequencing analysis of CD3+ T cells obtained from the spleen. Our analysis comprised of 140,000 cells from each of the three experimental groups, the results of which revealed that CD8+ (37%) and CD4+ (33%) T cells constituted the majority of the analysed population ((A)). Our findings are consistent with prior observations, demonstrating that the proportion of CD8+ T cells was substantially higher in the prime-boost group compared to the prime-alone group (46% vs. 32%, P = 0.007), whereas the converse held for CD4+ T cells (35%, prime-boost vs. 28% prime-alone, P = 0.01). Further, we categorized CD8+ T cells into nine subtypes based on CD8+ T cell marker expression levels ((B)). We noted that three CD8+ T cell subtypes, namely TRM CD8+ T cells (13% vs. 4%, P = 0.03), Gzma- effect memory CD8+ T cells (18% vs. 9%, P = 0.02), and Gzma+ effect memory CD8+ T cells (10% vs. 6%, P = 0.04, (C)) were significantly increased in the prime-boost group compared to the prime-alone group. Our analysis of effector gene expression patterns for these cells demonstrated significantly increased expression of Ccl3, Ccl4, Ccl5, Gzmk, and Gzma for TEM cells and Cxcr6 for TRM cells in the prime-boost group ((D)). Moreover, GSVA demonstrated that immune activity-regulating gene sets, such as cytokine-cytokine receptor interaction, NOD-like receptor signaling pathway, chemokine signaling pathway, cytosolic DNA-sensing pathway, and antigen processing/presentation pathway, were enriched in CD8+ T cells from the prime-boost group, while ribosome and spliceosome-related pathways were less enriched ((E)). These results support our hypothesis that the MLN-mRNA vaccine enhances immune activity during reinfection by promoting the maturation of TEM and TRM CD8+ cells.
Figure 5. Single-cell sequencing analysis of spleen CD3+ T cells. (A) UMAP clustering of sequenced splenic cells. (B) Clustering of CD8+ T cell subtypes within each group. CD8+ T cells are classified into nine subtypes as indicated. (C) Histogram showing the proportion of different CD8+ T cell subtypes in each sample. (D) Violin plots showing the expression of effect and tissue resident memory CD8+ T cell markers being significantly upregulated in the prime-boost group. (E) Heatmap showing the pathways that are significantly different between the prime-boost group and the other two groups in the CD8+ cells, which are the results of variation analysis using the GSVA method.
MLN-mRNA vaccine induced sustained antigen-specific antibody responses
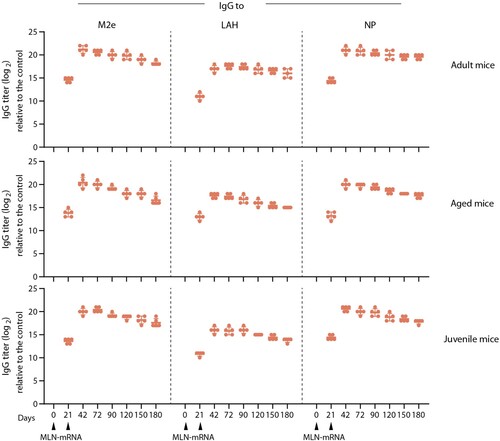
To ascertain the duration of antigen-specific antibody responses induced by the MLN-mRNA vaccine, we conducted a study to measure IgG titres against M2e, LAH and NP in vaccinated mice from varying age cohorts. As presented in , consistent anti-M2e, -LAH, and -NP IgG levels were observed in all age groups at six months after the prime immunization. Correspondingly, similar kinetics profiles for serum IgG titres were noticed, whereby the IgG level peaked at 42 days and maintained a relatively high level thereafter. Besides adult mice, we also evaluated the efficiency of the MLN-mRNA vaccine immunization in aged (8-month-old) and juvenile (3 or 4-week-old) mice, as the elderly and the young individuals are usually considered vulnerable groups with sub-optimal immune responses to infection and vaccination [Citation45]. In order to simulate real-life conditions, where elderly people are likely to be exposed to multiple influenza infections over their lifetime, we pre-exposed the aged mice to a sub-lethal dose of influenza virus (A/PR/8) before vaccination. Our findings revealed that the MLN-mRNA vaccine was capable of effectively eliciting potent immune responses in both age groups, which were comparable to those of adult mice. This suggested that the MLN-mRNA vaccine has the potential to induce long-lasting antigen-specific antibody responses across all human age groups.
Figure 6. Immunogenicity of MLN-mRNA vaccine in adult, aged and juvenile mice. Adult (6-8 weeks old, upper panel, n = 5) or juvenile (3-4 weeks old, lower panel, n = 5) were immunized twice as indicated with 20 μg of MLN-mRNA. Aged mice (8 months old, middle panel, n = 5) were exposed to sublethal dosage of A/PR/8 infections one month before the primary immunization and then vaccinated with 20 μg MLN-mRNA twice as indicated. Anti-M2e/LAH/NP antibody titres were monitored by ELISA for 6 months.
Discussion
The successful of the mRNA vaccines for COVID-19 pandemic has prompted numerous clinical trials of seasonal influenza vaccines utilizing the same platform, most of which encoded influenza virus HA antigens [Citation24,Citation25,Citation29]. However, HA antigen often undergoes antigenic drift and/or shift, resulting in antigenic mismatches between vaccine strains and circulating strains, which poses a challenge to the current production and application of influenza vaccines, and thus an influenza vaccine with broad-spectrum protection against different influenza viruses is urgently needed [Citation46].
In the pursuit of a broad-spectrum influenza vaccine, several conserved influenza proteins such as the stalk of HA, NP, M2, M1, and polymerase basic protein 1 (PB1) have been subjected to various vaccine designs including adjuvanted recombinant protein-based vaccines, viral vector and nucleic acid vaccines, with promising results in animal models and early clinical trials [Citation4,Citation5,Citation27,Citation33,Citation47]. Aiming to enhance both the potency and breadth of protection, our approach, like that of several other research groups, employed multi-targeting vaccines to elicit comprehensive immune responses [Citation24,Citation25,Citation27,Citation31,Citation32]. We observed that combinations of antigens consistently yielded superior protection against different subtypes of influenza A viruses, while vaccines utilizing single antigens delivered suboptimal protection. This finding is in concordance with the previous studies, underscoring the essential role of a combinatorial approach in influenza vaccine design. For instance, Nachbagauer et al. developed a universal influenza vaccine candidate using four mRNA-LNPs targeting specific segments of HA, NA, M2, and NP, and demonstrated robust immune responses in mice and protection against pandemic H1N1 [Citation27]. Similarly, De Jonge et al. crafted a three-component mRNA-LNP universal vaccine from NP, M1, and PB1 of an H1N1 strain, which enhanced T cell immune responses and protection against H7N9 influenza in a ferret model [Citation32]. Furthermore, Hensley et al. engineered a multivalent mRNA-LNP vaccine encoding twenty different HA antigens, spanning nearly all influenza virus subtypes and eliciting high levels of cross-reactive and subtype-specific antibodies in both mice and ferrets [Citation24]. However, the use of mixtures of multiple mRNA-LNPs in these studies may pose a challenge in large-scale production, thus may limit their application. Our vaccine, distinct in its design, introduces a fusion protein consisting of tandem repeats that link the most promising influenza antigens, streamlining both production and quality control processes.
Our evaluation efforts showed that this vaccine design preserved the immunogenicity of the antigens and elicited strong protective efficacy primarily via ADCC, neutralization and CD8+ T cell immune responses [Citation19], in line with the function of the M2e, LAH, and NP domains (Supplementary Figure 12) [Citation10,Citation13–15]. Additionally, the MLN-mRNA vaccine induced a strong TH1-driven T cell response leading to the secretion of type-1 cytokines, such as IFN-γ and IL-2, that activated CD8+ T cell-mediated immune responses [Citation48]. Moreover, we found that the proportion of TRM in CD4+/8+ T cells and TCM/TEM cells in CD8+ T cells conferred long-lasting protective immunity against viral infections.
Specific cellular immune responses targeting conservative internal antigens of the influenza virus are widely recognized as the primary mechanisms mediating cross-protection against influenza infection [Citation49]. NP-specific CTLs, for instance, can rapidly proliferate, differentiate, and be recruited to sites of infection following viral exposure. Single-cell RNA-seq analyses provided more insights into the underlying mechanism involved in cellular immune responses. Some immune processes, such as cytokine-cytokine receptor interaction, NOD-like receptor signaling pathway, chemokine signaling pathway, cytosolic DNA-sensing pathway and antigen processing/presentation pathway, were disturbed in mice immunized with MLN-mRNA vaccine. Moreover, the expression levels of genes related to chemokine ligand (e.g. Ccl3, Ccl4 and Ccl5), chemokine receptor (e.g. Cxcr6) and granzyme (e.g. Gzmk and Gzma) in CD8+ T cells were significantly increased, which indicated that the enhancement of protection activity was promoted by the activation and cytotoxic activity of T cells.
However, relying solely on cellular immune responses for protection against viral diseases can be limiting since their effect is restricted to infected cells. Therefore, vaccines that are capable of producing virus-neutralizing antibodies are preferred to prevent viral infections at earlier stages of the disease. It is noteworthy that our LAH-containing vaccine, which promotes virus-neutralizing antibodies, including IgG1 and IgG2a, has been able to reduce viral load efficiently [Citation15,Citation50]. A robust TFH response on GC B cells driven by the MLN-mRNA vaccine led to this outcome. The synergy between vaccine-induced cellular and humoral immune responses by MLN-mRNA has been seen in our study, which showed a strong vaccine potency. These results indicate that our design could serve as a starting point for the development of a future broad-spectrum influenza vaccine that can be tested with more viral strains for cross-reactivity.
In conclusion, our proof-of-concept model has shown that the MLN-mRNA vaccine, a nucleoside-modified mRNA-LNP vaccine encoding a fusion protein of multiple influenza virus antigens, has great potential to deliver comprehensive immune responses and provide cross-protection against distal influenza virus strains, presenting a promising strategy for developing a broad-spectrum influenza vaccine candidate.
Acknowledgments
We thank Dr. Qinghua Zhang, Dr. Boquan Ji, Danfeng Li, Xin Kuang, Dingfang Liang from Wayen Biotechnologies (Shanghai), Inc. for expert technical assistance and bioinformatic analysis for scRNA-seq.
Author contributions
F. X., J. L., and X. L. conceived project and designed experiments. F. X., B. S., M. Z., Q. W. and Y. D. performed experiments. F. X. analyzed data. F. X. and C. Z. wrote and edited the original manuscript. All coauthors reviewed the manuscript.
Supplemental Material
Download MS Word (1.6 MB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Iuliano AD, Roguski KM, Chang HH, et al. Estimates of global seasonal influenza-associated respiratory mortality: a modelling study. Lancet. 2018;391:1285–1300. doi:10.1016/S0140-6736(17)33293-2
- Paget J, Spreeuwenberg P, Charu V, et al. Global mortality associated with seasonal influenza epidemics: New burden estimates and predictors from the GLaMOR project. J Glob Health. 2019;9(2):020421. doi:10.7189/jogh.09.020421
- Rolfes MA, Flannery B, Chung JR, et al. Effects of influenza vaccination in the United States during the 2017-2018 influenza season. Clin Infect Dis. 2019;69(11):1845–1853. doi:10.1093/cid/ciz075
- Nachbagauer R, Krammer F. Universal influenza virus vaccines and therapeutic antibodies. Clin Microbiol Infect. 2017;23(4):222–228. doi:10.1016/j.cmi.2017.02.009
- Chen S, Zheng D, Li C, et al. Protection against multiple subtypes of influenza viruses by virus-like particle vaccines based on a hemagglutinin conserved epitope. Biomed Res Int. 2015; 901817. doi:10.1155/2015/901817
- Sui Z, Chen Q, Wu R, et al. Cross-protection against influenza virus infection by intranasal administration of M2-based vaccine with chitosan as an adjuvant. Arch Virol. 2010;155(4):535–544. doi:10.1007/s00705-010-0621-4
- Guo L, Zheng M, Ding Y, et al. Protection against multiple influenza A virus subtypes by intranasal administration of recombinant nucleoprotein. Arch Virol. 2010;155(11):1765–1775. doi:10.1007/s00705-010-0756-3
- Luo J, Zheng D, Zhang W, et al. Phylogenetic relationships of the glycoprotein gene of bovine ephemeral fever virus isolated from mainland China, Taiwan, Japan, Turkey, Israel and Australia. Virol J. 2012;9:286. doi:10.1186/1743-422X-9-268
- Bullard BL, Weaver EA. Strategies targeting hemagglutinin as a universal influenza vaccine. Vaccines (Basel). 2021;9(3):257. doi:10.3390/vaccines9030257
- DiLillo DJ, Tan GS, Palese P, et al. Broadly neutralizing hemagglutinin stalk–specific antibodies require FcγR interactions for protection against influenza virus in vivo. Nat Med. 2014;20(2):143–151. doi:10.1038/nm.3443
- Neirynck S, Deroo T, Saelens X, et al. A universal influenza A vaccine based on the extracellular domain of the M2 protein. Nat Med. 1999;5(10):1157–1163. doi:10.1038/13484
- Liu W, Zou P, Ding J, et al. Sequence comparison between the extracellular domain of M2 protein human and avian influenza A virus provides new information for bivalent influenza vaccine design. Microbes Infect. 2005;7(2):171–177. doi:10.1016/j.micinf.2004.10.006
- Lee YN, Kim MC, Lee YT, et al. Mechanisms of cross-protection by influenza virus M2-based vaccines. Immune Netw. 2015;15(5):213–221. doi:10.4110/in.2015.15.5.213
- El Bakkouri K, Descamps F, De Filette M, et al. Universal vaccine based on ectodomain of matrix protein 2 of influenza A: Fc receptors and alveolar macrophages mediate protection. J Immunol. 2011;186(2):1022–1031. doi:10.4049/jimmunol.0902147
- Van den Hoecke S, Ehrhardt K, Kolpe A, et al. Hierarchical and redundant roles of activating FcgammaRs in protection against influenza disease by M2e-specific IgG1 and IgG2a antibodies. J Virol. 2017;91(7):e02500-16. doi:10.1128/JVI.02500-16
- Tompkins SM, Zhao ZS, Lo CY, et al. Matrix protein 2 vaccination and protection against influenza viruses, including subtype H5N1. Emerg Infect Dis. 2007;13(3):426–435. doi:10.3201/eid1303.061125
- Fan J, Liang X, Horton MS, et al. Preclinical study of influenza virus A M2 peptide conjugate vaccines in mice, ferrets, and rhesus monkeys. Vaccine. 2004;22:2993–3003. doi:10.1016/j.vaccine.2004.02.021
- Denis J, Acosta-Ramirez E, Zhao Y, et al. Development of a universal influenza A vaccine based on the M2e peptide fused to the papaya mosaic virus (PapMV) vaccine platform. Vaccine. 2008;26:3395–3403. doi:10.1016/j.vaccine.2008.04.052
- Grant E, Wu C, Chan KF, et al. Nucleoprotein of influenza A virus is a major target of immunodominant CD8+T-cell responses. Immunol Cell Biol. 2013;91(2):184–194. doi:10.1038/icb.2012.78
- Eickhoff CS, Terry FE, Peng L, et al. Highly conserved influenza T cell epitopes induce broadly protective immunity. Vaccine. 2019;37:5371–5381. doi:10.1016/j.vaccine.2019.07.033
- Grebe KM, Yewdell JW, Bennink JR. Heterosubtypic immunity to influenza A virus: where do we stand? Microbes Infect. 2008;10:1024–1029. doi:10.1016/j.micinf.2008.07.002
- Chaudhary N, Weissman D, Whitehead KA. mRNA vaccines for infectious diseases: principles, delivery and clinical translation. Nat Rev Drug Discov. 2021;20:817–838. doi:10.1038/s41573-021-00283-5
- Qin S, Tang X, Chen Y, et al. mRNA-based therapeutics: powerful and versatile tools to combat diseases. Signal Transduct Target Ther. 2022;7(1):166), doi:10.1038/s41392-022-01007-w
- Arevalo CP, Bolton MJ, Le Sage V, et al. A multivalent nucleoside-modified mRNA vaccine against all known influenza virus subtypes. Science. 2022;378(6622):899–904. doi:10.1126/science.abm0271
- Chivukula S, Plitnik T, Tibbitts T, et al. Development of multivalent mRNA vaccine candidates for seasonal or pandemic influenza. NPJ Vaccines. 2021;6(1):153. doi:10.1038/s41541-021-00420-6
- Pardi N, Parkhouse K, Kirkpatrick E, et al. Nucleoside-modified mRNA immunization elicits influenza virus hemagglutinin stalk-specific antibodies. Nat Commun. 2018;9(1):3361. doi:10.1038/s41467-018-05482-0
- Freyn AW, da Silva R, Rosado J, et al. A multi-targeting, nucleoside-modified mRNA influenza virus vaccine provides broad protection in mice. Mol Ther. 2020;28(7):1569–1584. doi:10.1016/j.ymthe.2020.04.018
- Feldman RA, Fuhr R, Smolenov I, et al. mRNA vaccines against H10N8 and H7N9 influenza viruses of pandemic potential are immunogenic and well tolerated in healthy adults in phase 1 randomized clinical trials. Vaccine. 2019;37(25):3326–3334. doi:10.1016/j.vaccine.2019.04.074
- Bahl K, Senn JJ, Yuzhakov O, et al. Preclinical and clinical demonstration of immunogenicity by mRNA vaccines against H10N8 and H7N9 influenza viruses. Mol Ther. 2017;25(6):1316–1327. doi:10.1016/j.ymthe.2017.03.035
- Zhuang X, Qi Y, Wang M, et al. mRNA vaccines encoding the HA protein of influenza A H1N1 virus delivered by cationic lipid nanoparticles induce protective immune responses in mice. Vaccines (Basel). 2020;8(1):123. doi:10.3390/vaccines8010123
- Pardi N, Carreno JM, O'Dell G, et al. Development of a pentavalent broadly protective nucleoside-modified mRNA vaccine against influenza B viruses. Nat Commun. 2022;13(1):4677. doi:10.1038/s41467-022-32149-8
- van de Ven K, Lanfermeijer J, van Dijken H, et al. A universal influenza mRNA vaccine candidate boosts T cell responses and reduces zoonotic influenza virus disease in ferrets. Sci Adv. 2022;8(50):eadc9937. doi:10.1126/sciadv.adc9937
- Lee YT, Ko EJ, Lee Y, et al. Intranasal vaccination with M2e5x virus-like particles induces humoral and cellular immune responses conferring cross-protection against heterosubtypic influenza viruses. PLoS One. 2018;13(1):e0190868. doi:10.1371/journal.pone.0190868
- Cros JF, Garcia-Sastre A, Palese P. An unconventional NLS is critical for the nuclear import of the influenza A virus nucleoprotein and ribonucleoprotein. Traffic. 2005;6(3):205–213. doi:10.1111/j.1600-0854.2005.00263.x
- Korsunsky I, Millard N, Fan J, et al. Fast, sensitive and accurate integration of single-cell data with harmony. Nat Methods. 2019;16(12):1289–1296. doi:10.1038/s41592-019-0619-0
- Hao Y, Hao S, Andersen-Nissen E, et al. Integrated analysis of multimodal single-cell data. Cell. 2021;184(13):3573–3587.e29. doi:10.1016/j.cell.2021.04.048
- Wang P, Palese P, Neill O, et al. The NPI-1/NPI-3 (karyopherin alpha) binding site on the influenza a virus nucleoprotein NP is a nonconventional nuclear localization signal. J Virol. 1997;71(3):1850–1856. doi:10.1128/jvi.71.3.1850-1856.1997
- Weber F, Kochs G, Gruber S, et al. A classical bipartite nuclear localization signal on thogoto and influenza A virus nucleoproteins. Virology. 1998;250(1):9–18. doi:10.1006/viro.1998.9329
- Bullido R, Gomez-Puertas P, Albo C, et al. Several protein regions contribute to determine the nuclear and cytoplasmic localization of the influenza A virus nucleoprotein. J Gen Virol. 2000;81(Pt 1):135–142. doi:10.1099/0022-1317-81-1-135
- Pardi N, Hogan MJ, Naradikian MS, et al. Nucleoside-modified mRNA vaccines induce potent T follicular helper and germinal center B cell responses. J Exp Med. 2018;215(6):1571–1588. doi:10.1084/jem.20171450
- Stemberger C, Neuenhahn M, Buchholz VR, et al. Origin of CD8 + effector and memory T cell subsets. Cell Mol Immunol. 2007;4(6):399–405.
- Huster KM, Stemberger C, Busch DH. Protective immunity towards intracellular pathogens. Curr Opin Immunol. 2006;18(4):458–464. doi:10.1016/j.coi.2006.05.008
- Arunachalam PS, Charles TP, Joag V, et al. T cell-inducing vaccine durably prevents mucosal SHIV infection even with lower neutralizing antibody titers. Nat Med. 2020;26(6):932–940. doi:10.1038/s41591-020-0858-8
- Schenkel JM, Fraser KA, Beura LK, et al. Resident memory CD8 T cells trigger protective innate and adaptive immune responses. Science. 2014;346(6205):98–101. doi:10.1126/science.1254536
- Osterholm MT, Kelley NS, Sommer A, et al. Efficacy and effectiveness of influenza vaccines: a systematic review and meta-analysis. Lancet Infect Dis. 2012;12(1):36–44. doi:10.1016/S1473-3099(11)70295-X
- Zheng M, Luo J. Progress in research of broad-spectrum influenza vaccines based on conserved antigens of influenza virus. Int J Biologicals. 2019;42(4):198–202.
- Kavishna R, Kang TY, Vacca M, et al. A single-shot vaccine approach for the universal influenza A vaccine candidate M2e. Proc Natl Acad Sci U S A. 2022;119(13):e2025607119. doi:10.1073/pnas.2025607119
- Liu JX, Zhang Y, An M, et al. Diversity of Th1/Th2 immunity in mice with acute lung injury induced by the H1N1 influenza virus and lipopolysaccharides. J Infect Dev Ctries. 2019;13(6):536–544. doi:10.3855/jidc.10338
- Cerwenka A, Morgan TM, Dutton RW. Antigen concentration and precursor frequency determine the rate of CD8+ T cell tolerance to peripherally expressed antigens. J Immunol. 1999;163:723–727. doi:10.4049/jimmunol.163.2.723
- Huber VC, McKeon RM, Brackin MN, et al. Distinct contributions of vaccine-induced immunoglobulin G1 (IgG1) and IgG2a antibodies to protective immunity against influenza. Clin Vaccine Immunol. 2006;13(9):981–990. doi:10.1128/CVI.00156-06
