ABSTRACT
Intratypic DNA polymorphism has been described for human papillomavirus (HPV) types infecting Inuit women in Nunavik, Quebec, a high-risk population for HPV infection and cervical cancer, but there is no previous research on the association between HPV polymorphism and infection persistence in Inuit women. Polymorphism of HPV types 16, 18 and 52 was described in a subset of 64 participants with multiple clinic visits within a cohort of 677 Nunavik Inuit women aged 15–69 recruited in 2002–2010 with testing results. Logistic regression and Cox proportional hazards models were used to assess the association between HPV variants and infection persistence and clearance. Infections with HPV16 lineage A3 variants cleared 3.13 times faster (95% CI: 1.10–8.97) than those with lineage A1 variants. HPV52 lineage C variants cleared slower than lineage A variants (HR = 0.28, 95% CI: 0.08–0.98). HPV polymorphism may be associated with viral persistence for certain HPV types in this population.
Introduction
Human papillomavirus (HPV), the most common sexually transmitted infection worldwide, is a necessary cause of cervical cancer [Citation1] and a key etiologic agent in other anogenital cancers and in head and neck cancers. There are over 160 types of HPV, of which over 40 infect the anogenital tract [Citation2–Citation5]. HPV types such as HPV16, 18 and 52 are classified as high-risk as they have oncogenic potential, particularly when the infection is persistent [Citation3, Citation6]. Although HPV infection persistence is not completely understood, both host characteristics such as older age, lifetime pregnancies, polymorphism in human-leukocyte antigen (HLA) genes, dietary factors and viral factors such as viral load, multiple type infection and viral polymorphism are associated with longer infection durations [Citation7–Citation12].
A viral isolate for any given HPV type that differs by less than 2% with the prototype isolate in the L1 gene is considered an intratypic variant [Citation2]. There is evidence that certain HPV types co-evolved with humans and that variants are geographically segregated. For HPV16 and 18, lineages of variants were originally described based on geographic clustering, indicative of human migration [Citation13]. HPV52 may show some signs of geographic clustering but this is unclear and research is limited [Citation14, Citation15]. HPV polymorphism may mediate HPV replication, transcriptional activity, protein function, and host immune system recognition that invoke differences both in the natural history of the virus and in its oncogenic potential [Citation16, Citation17, Citation18]. Overall, variants in non-European lineages of HPV16 and HPV18 persist longer and are more likely to be associated with cervical cancer than those in the European lineage [Citation19, Citation20,Citation21,Citation22] These associations with persistence may be a reflection of the adaptation of the virus to the host population but these mechanisms are not yet understood [Citation21].
The cervical cancer rate among Indigenous people in Canada is twice as large as the rate found in the general population [Citation22, Citation23]. The communities of Nunavik, the northernmost region of Quebec, Canada, are predominately inhabited with residents who self-identify as Inuit. Due to high rates of HPV infection and cervical cancer, Nunavik, is considered a high-risk area for cervical cancer and a potential target for public health initiatives concerning cervical cancer prevention [Citation24, Citation25].
As HPV persistence and associated cervical cancer risk have been shown to be dependent on variant prevalence and ethnic origin [Citation21], the previous literature on other high-risk communities may not be generalisable to Nunavik. Previous research in this area showed that there is a low diversity of variants present in this population and all HPV16 and 18 variants were of European lineage (lineage A) [Citation26]. Additionally, high-risk (HR) type HPV52 is relatively prevalent in this population [Citation24, Citation26]. The literature on HPV polymorphism and risk of persistence and cancer has primarily focused on HPV16 and 18 as they are implicated in the majority of cervical cancer cases. There is some evidence that other HR HPV types have polymorphisms that are associated with greater risk of persistence [Citation20,Citation27,Citation28], but it is not clear if this association is present for HPV52 in all populations. This study aims to assess the association between intratypic variants of HPV 16, 18 and 52 and infection persistence/clearance among Inuit women in Nunavik.
Methods
The prospective cohort of Inuit women living in Nunavik used for this study has been described elsewhere [Citation24]. Ethics approval from the Tulattavik Health Centre (which offers services to study participants) and support from the local women’s association and health board were obtained.
The sampling frame included women (aged 15–69 years) who presented for a Pap test between January 2002 and November 2006. HPV DNA testing and cytology was performed opportunistically until the end of study follow-up in July 2010. To be eligible for this study, participants had to have at least two HPV-DNA test results of acceptable quality and had to have a variant test for HPV type 16, 18 and/or 52.
HPV DNA typing and PCR-sequencing of the long control region (LCR) for variant testing was previously described [Citation26]. The variant was assessed on the first positive specimen for HPV16, 18 or 52.
HPV16, HPV18 and HPV52 lineages were identified based on sequences obtained from high-quality electropherograms from the 3’ end of LCR. Variants of each type were classified into lineages and sublineages by phylogenetic analysis based on the neighbour-joining algorithm with the Mega version 5.0 including variant genomes of each sublineage or lineage [Citation29].
Four different lineages (A, B, C and D) of HPV16 have been described and are also subdivided in sublineages [Citation30], while HPV18 is divided in three lineages (A, B and C) [Citation31]. Four different lineages (A, B, C and D) of HPV52 have been defined in a study on isolates collected from multiple geographic regions [Citation32]. When comparisons were not feasible across lineages due to homogeneity in samples, comparisons were made by sublineages (A1 vs. A2, for example). Lack of genetic diversity means that only two categories were present for each HPV. The regression models used the first category (alpha-numerically) as the reference category.
A persistent infection was defined as a positive HPV test for the same HPV type on at least two consecutive visits [Citation6,Citation9,Citation19]. A negative test result that occurred at a visit in between two positive visits was treated as a false negative. The date of acquisition of an incident infection was calculated as the mid-point between the previous negative visit and the next positive visit. The time to clearance of an infection was calculated similarly, based on the midpoint between the last positive visit and the next negative one. For prevalent infections at study entry, infection duration was calculated from the first type-specific HPV-positive laboratory date. Subjects that remained positive at their last clinic visit were censored at the last available type-specific HPV-positive lab date.
We used actuarial analysis to infer the mean and median duration of combined prevalent and incident infections (in months) and 95% confidence intervals (CI) by variant category. A Shapiro-Wilk test for normality was also used.
Logistic regression was used to assess the odds of infection persistence by variant category. The following covariates measured at study entry were assessed: age, community, marital status, employment status, education, current smoker, alcohol use, birth control use, hormonal birth control use, age at first sexual intercourse, lifetime number of sexual partners, lifetime number of deliveries, pregnant, self-reported sexually transmitted infection (STI) history. The following HPV testing variables defined at the start of an infection were also assessed: coinfection with multiple HPV types or multiple HR-HPV types, and overall number of clinic visits. Univariate logistic regression models were performed and possible collinearity between variables was first assessed (data not shown). As the association between variant category and persistent infection was of interest, Bayesian (BIC) model selection was used to select final models so that variant category probability of inclusion could be set to 1.0. Analysis including, and not including imputed missing covariate data, was performed. For the multiple imputation, a prediction matrix was specified which indicated that all covariates predicted each other. Variant category was also used to predict missing covariates. Twenty imputed datasets were created, logistic regression models were run on each, and results were pooled.
For the Cox proportional hazard models, univariate models, age-adjusted models and as well as models adjusting for both age and number of clinic visits were run [Citation7, Citation8, Citation28]. Schönfeld residuals from all models were used to tests assumption of proportionality. The models were also tested for possible informative censoring. This was done (1) assuming that those censored cleared the infection and (2) by assuming that the time of clearance for those censored was the maximum found in the dataset. The results from these two scenarios were compared to the results obtained originally.
All data analyses were performed in R version 3.1.2 [Citation33] with the following packages: survival (for Cox PH), mice (for multiple imputations) and BMA (for model selection) [Citation34, Citation35, Citation36)]. Non-overlapping 95% CI were used to infer significant statistical differences between variant categories.
Results
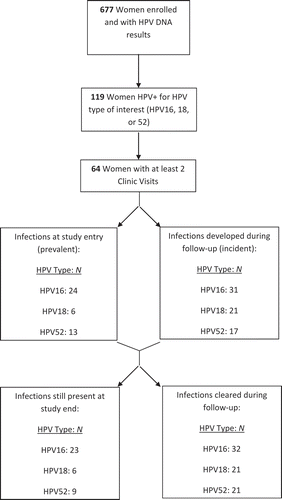
Of the 677 women in the Nunavik cohort with HPV DNA testing results, 119 tested positive for HPV16, 18, or 52 at study entry and/or at follow-up visits and 64 of these participants had at least two clinic visits and were included in our analysis. Among them, 112 HPV infections occurred, including 69 incident infections, which were included in the analysis ().
The average duration of infection with HPV16 isolates of the A1 sublineage was in general longer than that for infections with variants from the A3 sublineage (). Across all HPV types, as the 95% confidence intervals were overlapping, we were not able to detect statistically significant differences while comparing either median or mean duration.
Table 1. Comparison of length of infection (in months) for HPVs 16, 18, and 52.
The mean estimates were higher than the median estimates for most categories, suggesting a skewed distribution of infection duration times. For HPV18 and HPV52, infection duration within variant categories can be assumed normally distributed (p value <0.05 for Shapiro-Wilk test) but appears non-normal for HPV16.
The multivariable-adjusted model with the highest posterior probability (43%) included the following variables along with variant category: age at first sexual intercourse, lifetime number of partners at baseline, and number of clinic visits. HPV16 and 52 variant lineages were not significantly associated with persistent infection, with and without imputed covariates (). Due to small numbers and high variability we were unable to detect an association of HPV18 variant lineages with persistence.
Table 2. Logistic regression results for association between HPV16, 18 and 52 lineage category and persistent infection.
For each HPV type, no univariate model revealed an association between HPV polymorphism and clearing an infection (). From the age-adjusted model, the risk of clearance of an infection was 3.13 times higher (95% CI 1.10, 8.97) for individuals infected with HPV16 variants in lineage A3 than in those infected with variants in lineage A1. Between-lineage differences did not reach statistical significance in the age-adjusted models for HPV18 and 52. When all covariates were adjusted for, the risk of infection clearance in participants infected with lineage C HPV52 variants was 72% lower than those infected with lineage A variants.
Table 3. Cox proportional hazards regression results for association between variant category (HPV16: lineage A3 versus lineage A1; HPV18: lineage A4 versus lineage A2; HPV52: lineage C versus lineage A) and clearance.
Overall, there were no significant difference detected in infection duration for variants of HPV types 16, 18 and 52 when categorised in lineage groups.
Discussion
The duration of type-specific infections was longer than what has been typically reported for HR-HPV, less than 15 months, but reports of similarly long durations do exist [Citation7, Citation8, Citation37]. Previous estimates of infection duration from this cohort were similarly long despite being restricted to incident infections [Citation25]. This may be a reflection of the opportunistic follow-up of participants with an estimated median time between visits of 12.5 months [Citation25]. As time of onset/clearance of infection was calculated as the midpoint between visits, adoption of this long interval between visits may have led to an overestimation of infection duration. Alternatively, it is possible that the duration of infection was truly longer, as similar infection duration times have been shown in other high-risk cohorts. Indeed, in a study involving female university students in Montréal, the mean duration for HR-HPV (16.3 months) and for HPV16 (18.3 months) was similar to this study cohort despite the fact that the interval between visits in the former cohort was only 6 months [Citation38]. In fact, since our study included participants already infected by HPV at baseline, we may have underestimated the duration of HPV infection. Additionally, many infections were not cleared during follow-up and therefore may have persisted longer than what was accounted for.
We used Cox proportional hazards models to control for age and number of visits when assessing infection clearance differences. The infection clearance risk was significantly higher in individuals infected with HPV16 lineage A3 variants as compared to those infected with lineage A1 variants. For HPV52, infections, lineage C variants clearance was significantly less likely than lineage A variants.
Although our study was small, the cohort had a high participation rate by enrolling a large proportion of women in Nunavik, which makes it nearly population-based which means it is highly representative of this population [Citation39]. Nevertheless, constrained by the small size, we had low power to detect an association between individual intratypic variants and persistence. Moreover, there were a very limited number of HPV variants detected in this population which is expected as Nunavik is a relatively geographically isolated region with limited in-migration. This has also been a common finding in other cohort studies that have assessed intratypic variants, particularly for types other than 16 and 18 [Citation20,Citation27]. Due to sample size constraints, prevalent infections at study entry were included in the analysis. Analysis restricted to incident infections was also performed and compared to ensure that including prevalent infections which leads to left censoring and underestimating infection duration did not significantly change the trends. Although right censoring was also present, it is important to include those infections that did not clear by study end to minimise bias[Citation3]. Detection opportunity bias is possible for those who had fewer visits and this was adjusted for in the analysis. The time interval between visits varied across the cohort and this may have influenced the infection onset and clearance times but more consistent and frequent follow-up was not feasible and number of clinic visits was adjusted for. Additionally, as in every study, low viral load infections may not be detected which results in misclassification of infection time but this is similar to other HPV persistence studies.
Only the first type-specific infection per participant was included for molecular variant testing. Other studies have shown that the same variant is detected in consecutive HPV-positive visits and have also found that co-infection with multiple variants is rare so this seemed to be a reasonable assumption [Citation19, Citation20, Citation40]. Additionally, the same variant was detected for the subset of participants for whom multiple samples were tested for variant analysis (N = 30 total) [Citation26]. Most reports categorise variants into phylogenetic lineages which was not feasible here for HPV16 and 18 due to high homogeneity of sequences in this cohort. Variants were all in the same lineage and therefore sublineage comparisons were made instead [Citation26]. The impact on cervical cancer and pre-cancer outcomes could not be assessed because of the small number of these outcomes (N = 16).
Conclusion
Our findings add to the understanding of HPV intratypic variants by demonstrating the association between viral polymorphism with clearance of infection in a population that had not been studied yet. Additionally, these findings add support to the universal role of HPV polymorphism in pathogenesis. More research into the application of information on variants is necessary, especially with larger cohorts with higher statistical power. This work contributes to the understanding of the natural history of HPV infection within Nunavik and supports the potential transition to HPV screening as the primary means of cervical cancer detection [Citation41]. Indeed, improved screening for early detection and treatment remains the best option for reducing cervical cancer burden. With HPV screening as a primary screening method in Nunavik, health care providers will be able to identify and encourage HR-HPV positive women to undergo subsequent evaluation. By directing limited resources in such remote communities towards women who are at higher risk of cervical cancer should improve the public health response for cervical cancer prevention.
Ethical statement
Informed consent was obtained from all study participants. Ethical approval was obtained from the Institutional Review Boards at McGill University and the Tulattavik Health Centre.
Acknowledgments
We gratefully acknowledge the Tulattavik Health Centre, collaborating nurse practitioners, participating communities, and the Nunavik Regional Board of Health and Social Services.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Walboomers JM, Jacobs MV, Manos MM, et al. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J Pathol. 1999;189(1):12–7.
- De Villiers E-M, Fauquet C, Broker TR, et al. Classification of papillomaviruses. Virol. 2004;324(1):17–27.
- Trottier H, Franco EL. The epidemiology of genital human papillomavirus infection. Vaccine. 2006;24:S4–S15.
- Smith B, Chen Z, Reimers L, et al. Sequence imputation of HPV16 genomes for genetic association studies. PLoS ONE. 2011;6(6):e21375. DOI:10.1371/journal.pone.0021375
- Zur Hausen H. Papillomaviruses and cancer: from basic studies to clinical application. Nat Rev Cancer. 2002;2(5):342–350DOI:10.1038/nrc798
- Koshiol J, Lindsay L, Pimenta JM, et al. Persistent human papillomavirus infection and cervical neoplasia: a systematic review and meta-analysis. Am J Epidemiol. 2008;168(2):123–137.
- Goodman MT, Shvetsov YB, McDuffie K, et al. Prevalence, acquisition, and clearance of cervical human papillomavirus infection among women with normal cytology: hawaii human papillomavirus cohort study. Cancer Res. 2008;68(21):8813–8824.
- Ho GY, Bierman R, Beardsley L, et al. Natural history of cervicovaginal papillomavirus infection in young women. N Engl J Med. 1998;338(7):423–428.
- Baseman JG, Koutsky LA. The epidemiology of human papillomavirus infections. J Clin Virol. 2005;32:16–24.
- Broccolo F. A multiplex real-time PCR-platform integrated into automated extraction method for the rapid detection and measurement of oncogenic HPV type-specific viral DNA load from cervical samples. Methods Mol Biol Epub 2014/ 04/18 PubMed PMID: 24740223. 2014;1160:87–97. DOI:10.1007/978-1-4939-0733-5_8
- Miranda PM, Silva NN, Pitol BC, et al. Persistence or clearance of human papillomavirus infections in women in Ouro Preto, Brazil. Biomed Res Int. 2013;2013:578276. Epub 2013/12/04. PubMed PMID: 24298551; PubMed Central PMCID: PMCPmc3835752. DOI:10.1155/2013/578276
- Metcalfe S, Roger M, Faucher MC, et al. The association between human leukocyte antigen (HLA)-G polymorphisms and human papillomavirus (HPV) infection in Inuit women of northern Quebec. PubMed PMID: 23994586 Hum Immunol. 2013;74(12):1610–1615. DOI:10.1016/j.humimm.2013.08.279
- Ho L, Chan S, Burk R, et al. The genetic drift of human papillomavirus type 16 is a means of reconstructing prehistoric viral spread and the movement of ancient human populations. J Virol. 1993;67(11):6413–6423
- Raiol T, Wyant PS, de Amorim RMS, et al. Genetic variability and phylogeny of the high-risk HPV-31, −33, −35, −52, and −58 in central Brazil. J Med Virol. 2009;81(4):685–692. DOI:10.1002/jmv.21432
- Zhang C, Park J-S, Grce M, et al. Geographical distribution and risk association of human papillomavirus genotype 52–variant lineages. J Infect Dis. 2014;210(10):1600–1604. DOI:10.1093/infdis/jiu310
- Hildesheim A, Schiffman M, Bromley C, et al. Human papillomavirus type 16 variants and risk of cervical cancer. J Natl Cancer Inst. 2001;93(4):315–318.
- Cuzick J, Arbyn M, Sankaranarayanan R, et al. Overview of human papillomavirus-based and other novel options for cervical cancer screening in developed and developing countries. Vaccine. 2008;26:K29–K41.
- Mirabello L, Yeager M, Cullen M, et al. HPV16 sublineage associations with histology-specific cancer risk using HPV whole-genome sequences in 3200 women. Jnci. 2016;108:9.
- Villa LL, Sichero L, Rahal P, et al. Molecular variants of human papillomavirus types 16 and 18 preferentially associated with cervical neoplasia. J Gen Virol. 2000;81(Pt 12):2959–2968. Epub 2000/11/22.PubMed PMID: 11086127.
- Schiffman M, Rodriguez AC, Chen Z, et al. A population-based prospective study of carcinogenic human papillomavirus variant lineages, viral persistence, and cervical neoplasia. Cancer Res. 2010;70(8):3159–3169.
- Xi LF, Kiviat NB, Hildesheim A, et al. Human papillomavirus type 16 and 18 variants: race-related distribution and persistence. J Natl Cancer Inst. 2006;98(15):1045–1052.
- Louchini R, Beaupre M. Cancer incidence and mortality among Aboriginal people living on reserves and northern villages in Quebec, 1988–2004. Int J Circumpolar Health. 2008;67(5): 445–451. Epub 2009/ 02/04.PubMed PMID: 19186765.
- Decker KM, Demers AA, Kliewer EV, et al. Pap test use and cervical cancer incidence in first nations women living in Manitoba. Cancer Prev Res (Phila). 2014. Epub 2014/ 11/19. PubMed PMID: 25403849. DOI:10.1158/1940-6207.capr-14-0277.
- Hamlin-Douglas LK, Coutlee F, Roger M, et al. Prevalence and age distribution of human papillomavirus infection in a population of Inuit women in Nunavik, Quebec. Cancer Epidemiol Biomarkers Prev. 2008;17(11):3141–3149. Epub 2008/ 11/08 PubMed PMID: 18990756. DOI:10.1158/1055-9965.epi-08-0625
- Bennett R, Cerigo H, Coutlée F, et al. Incidence, persistence, and determinants of human papillomavirus infection in a population of Inuit women in Northern Quebec. Sex Transm Dis. 2015;42(5):272–278.
- Gauthier B, Coutlée F, Franco E, et al. Human papillomavirus (HPV) variants among Inuit women in Northern Quebec, Canada. Int J Circumpolar Health. 2015;74. DOI:10.3402/ijch.v74.29482
- Xi LF, Schiffman M, Koutsky LA, et al. Lineages of oncogenic human papillomavirus types other than type 16 and 18 and risk for cervical intraepithelial neoplasia. J Natl Cancer Inst. 2014;106(10):dju270.
- Xi LF, Schiffman M, Koutsky LA, et al. Persistence of newly detected human papillomavirus type 31 infection, stratified by variant lineage. Int J Cancer. 2013;132(3):549–555.
- Tamura K, Peterson D, Peterson N, et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28(10):2731–2739. DOI:10.1093/molbev/msr121
- Burk RD, Harari A, Chen Z. Human papillomavirus genome variants. Virology. 2013;445(1–2):232–243. Epub 2013/09/04. PubMed PMID: 23998342; PubMed Central PMCID: PMCPmc3979972. DOI:10.1016/j.virol.2013.07.018
- Arias-Pulido H, Peyton CL, Torrez-Martinez N, et al. Human papillomavirus type 18 variant lineages in USA populations characterized by sequence analysis of LCR-E6, E2, and L1 regions. Virology. 2005;338(1):22–34. Epub 2005/06/07. PubMed PMID: 15936050. DOI:10.1016/j.virol.2013.07.018
- Chen Z, Schiffman M, Herrero R, et al. Evolution and taxonomic classification of human papillomavirus 16 (HPV16)-related variant genomes: HPV31, HPV33, HPV35, HPV52, HPV58 and HPV67. PloS one. 2011;6(5):e20183. DOI:10.1016/j.virol.2005.04.022
- R; A language and environment for statistical computing. R. Founcdation for Statistical Computing [Internet]. 2014.
- BMA: Bayesian Model Averaging. R. package version 3.18.1 [Internet]. 2014.
- A package for survival analysis in S_. R package version 2.37-7. [Internet]. 2014.
- van Burren S, Groothuis-Oudshoorn K. Mice: multivariate imputation by chained equations in R. J Stat Softw. 2011;45(3):1–67.
- Muñoz N, Méndez F, Posso H, et al. Incidence, duration, and determinants of cervical human papillomavirus infection in a cohort of Colombian women with normal cytological results. J Infect Dis. 2004;190(12):2077–2087.
- Richardson H, Kelsall G, Tellier P, et al. The natural history of type-specific human papillomavirus infections in female university students. Cancer Epidemiol Biomarkers Prev. 2003;12(6):485–490.
- Hamlin-Douglas LK, Coutlee F, Roger M, et al. Determinants of human papillomavirus infection among Inuit women of northern Quebec, Canada. Sex Transm Dis. 2010;37(6):377–381. Epub 2010/05/18. PubMed PMID: 20473246. DOI:10.1097/OLQ.0b013e3181cc4d22
- Sichero L, Ferreira S, Trottier H, et al. High grade cervical lesions are caused preferentially by non-European variants of HPVs 16 and 18. Int J Cancer. 2007;120(8):1763–1768. Epub 2007/01/19. PubMed PMID: 17230525. DOI:10.1002/ijc.22481
- Ogilvie GS, van Niekerk D, Krajden M, et al. Effect of screening with primary cervical HPV testing vs cytology testing on high-grade cervical intraepithelial neoplasia at 48 months. JAMA. 2018;320(1):43. DOI:10.1001/jama.2018.7464