ABSTRACT
Haemophilus influenzae serotype a (Hia) has recently emerged as an important cause of invasive disease in the North American Arctic and Sub-Arctic regions, mainly affecting young Indigenous children. In this study, we addressed the question of whether the prevalence of Hia and all H. influenzae in the nasopharynx differed between paediatric populations from regions with high versus low incidence of invasive Hia disease. Nasopharyngeal specimens from children with acute respiratory tract infections (ARTI) collected for routine diagnostic detection of respiratory viruses were analysed with molecular-genetic methods to identify and serotype H. influenzae. In Nunavut, a region with a high incidence of invasive Hia disease, all H. influenzae and particularly Hia were found in the nasopharynx of 60.6% and 3.0% children. In Southern Ontario (Hamilton region), where Hia invasive disease is rare, the frequencies of all H. influenzae and Hia detection were 38.5% and 0.6%, respectively. In both cohorts, non-typeable H. influenzae was prevalent (57.0% and 37.9%, respectively). Considering that Hia is an important cause of severe invasive disease in Nunavut children, 3% prevalence of Hia among children with ARTI can reflect continuing circulation of the pathogen in the Northern communities that may result in invasive disease outbreaks.
Introduction
Haemophilus influenzae is a gram-negative bacterial pathogen, which is classified based on the presence or absence of a polysaccharide capsule. Encapsulated strains are further categorised into six serotypes (a, b, c, d, e and f); non-encapsulated strains are denoted non-typeable (NTHi) [Citation1]. H. influenzae can cause local infections of the respiratory and genitourinary tracts, as well as invasive diseases such as meningitis and sepsis [Citation1]. Epidemiology of invasive H. influenzae disease has dramatically changed post introduction of paediatric immunisation against H. influenzae serotype b (Hib). In industrialised countries with universal immunisation programmes, a great reduction in the incidence of invasive Hib disease was followed by an increase in disease caused by NTHi and encapsulated non-Hib strains, predominantly of serotype f (Hif) [Citation2]. In contrast, in some North American regions with a high proportion of Indigenous Peoples, particularly in the Arctic, H. influenzae serotype a (Hia) became the significant cause of invasive disease, mainly affecting young children, with incidence rates approaching those of invasive Hib disease in the pre-vaccine era [Citation3,Citation4].
As H. influenzae is a human-restricted pathogen, it can only be acquired from other colonised individuals. Moreover, H. influenzae is a part of the respiratory microbiota, and asymptomatic colonisation with NTHi is very common, especially among children [Citation5–7]. Historical data demonstrated that nasopharyngeal Hib colonisation acted as a pre-requisite for invasive Hib disease and a reservoir for the pathogen transmission [Citation8]. The current knowledge of Hia carriage is limited by a few studies in Alaska and two Canadian regions, including central Canada [Citation9–11]. We have recently reported that in populations with higher rates of invasive Hia disease, Hia carriage rates among healthy children were close to those of Hib in Alaskan Indigenous children in the pre-Hib vaccine era [Citation11]. In Indigenous communities of Northern Ontario (Canada), with an invasive Hia disease incidence rate of 38.3/100,000 population <6 years of age (2013–2019), Hia was carried by 9% of healthy children as well as was isolated from cases of non-invasive disease, such as otitis media [Citation11,Citation12].
High incidence of invasive Hia has been documented in young children of the Northern Canada territory of Nunavut, where 85% of the population is Inuit [Citation13]. During 2000–2012, the incidence rates of invasive Hia disease in this region reached 274.8/100,000 population <1 year-old and 61.2/100,000 for 1–4-year-old children [Citation14]. The prevalence of Hia among children with non-invasive respiratory infections or H. influenzae carriage rates in Nunavut is unknown. To get insight into Hia prevalence in the paediatric population at high risk of invasive Hia disease, we analysed nasopharyngeal (NP) swabs collected for routine diagnostic virology testing in Nunavut children with acute respiratory tract infections (ARTI). The same analysis of NP swabs from age- and gender-matched children of diverse ethnic backgrounds residing in a region where invasive Hia disease is uncommon (Hamilton area, Southern Ontario) was conducted. A second control site included a group of children hospitalised with ARTI at the tertiary paediatric hospital serving the ethnically diverse population of Eastern Ontario. Our goal was to determine the presence of Hia and NTHi in the nasopharynx of children with symptoms of ARTI in a population with high rates of Hia disease and compare this to two other sites without high rates of Hia.
Methods
We analysed NP swabs collected from 200 young children residing in Qikiqtani, in the eastern region of Nunavut (NU), who presented with symptoms of ARTI (February 2018-May 2019); no clinical data were available on the patients. These specimens were collected for respiratory viral testing as part of routine clinical care at the hospital and health centres in the region. Immediately following collection, the specimens were placed in 3 mL of Universal Transport Medium (UTM, Copan, Italia) and shipped by courier mail to Virology, Hamilton Regional Laboratory Medicine Association, St. Joseph’s Healthcare (Hamilton, Ontario). For comparison, we selected 161 NP specimens collected from a cohort of children tested for respiratory viruses from the Hamilton area (HA) of Southern Ontario during the same time, using the same methodology. The specimens from HA were matched to 161 NU specimens by age, gender and season. In addition, 75 NP swabs were collected from children hospitalised at the Children’s Hospital of Eastern Ontario (CHEO, Ottawa, Ontario) with ARTI (October 2018-March 2020) (EO).
The specimens were analysed using a multiplex detection assay, with simultaneous identification of 10 respiratory viruses based on the detection of viral nucleic acids. The laboratory extracted total nucleic acids (RNA and DNA) using 200-μL aliquots into lysis buffer using a simple MAG extractor (bioMérieux). For the virology testing, only 20% of specimen is normally used, and the remaining material is routinely retained in the lab at −80°C for 1 year considering that the specimens may be called back for additional testing. The residual specimens were retrieved and anonymised (labels containing the child’s name, date of birth and community of residence were removed). The date of specimen collection, age and gender of children were recorded. The specimens, including unused UTM and extracted nucleic acids, were shipped by courier mail on dry ice to the National Microbiology Laboratory Branch (Winnipeg, Manitoba) for detection of H. influenzae. After the analyses had been completed, the remaining material was disposed according to the rule of handling the biohazardous materials.
Detection of H. influenzae was done by polymerase chain reaction (PCR) for the target genes hpd [Citation15,Citation16] and bexA [Citation17]. Those specimens tested positive for both hpd and bexA genes were regarded as positive for encapsulated H. influenzae and subjected to further PCR reactions to amplify the serotype-specific genes and if positive, confirmed by DNA sequencing of the amplicons [Citation17]. Those who tested positive for hpd but negative for bexA were regarded as positive for NTHi or H. haemolyticus. Gene sequencing of the hpd PCR amplicons was used to differentiate between H. influenzae and H. haemolyticus.
Statistical analysis was performed using Graph-Pad Prism 9 (GraphPad Prism Software Inc., San Diego, CA). The difference was determined with a one-way analysis of variance (ANOVA) with a Tukey post-hoc test. Nominal variables were compared using the Chi-Square Test. A p value of <0.05 was reported to be statistically significant.
Ethics approval
The study was performed in line with the principles of the Declaration of Helsinki. Approval was granted by the Hamilton Integrated Research Ethics Board (project No: 5367), Nunavut Research Institute (NRI 5 April 2019), CHEO Research Institute Research Ethics Board (protocol No: 18/128X; Romeo File No: 20180496) and Lakehead University Research Ethics Board (Romeo File No: 1465719).
Consent to participate was not required because in this study we used de-identified specimens collected for diagnostic purposes (according to Article 12.3A, TCPS 2, Government of Canada, 2022, https://ethics.gc.ca/eng/tcps2-eptc2_2022_chapter12-chapitre12.html#c)
Results
The age of NU and HA children was similar (median 12.0 months); no significant difference in gender was observed (46.8% and 41.0% of girls, p > 0.05) (). H. influenzae was more common in NU than in HA children, 121/200, or 60.5%, versus 62/161, or 38.5%, respectively, p < 0.0001 (; Supplementary Table S1). In both cohorts, NTHi was dominant among all detected H. influenzae. In NU children, NTHi represented 94.2% (114/121) of all H. influenzae found; the prevalence of NTHi was 57% (114/200) (Supplementary Table S1). In HA children, out of 62 H. influenzae, 61 were NTHi (98.4%); the prevalence of NTHi was 37.9%; the difference in prevalence of NTHi between two cohorts was significant (p < 0.001). Hia was identified in six children in NU (3%) and one child in HA (0.6%), p > 0.05. In one NU child, Hie was identified (0.5% prevalence) (Supplementary Table S1). In comparison, EO children were older than NU and HA children (median 17.7 months), p < 0.01; 45.3% were female (p > 0.05); 59/75 (78.7%) had H. influenzae (NTHi) (Supplementary Table S2). No serotypes b, c, d, or f were identified.
Figure 1. Prevalence of H. influenzae in nasopharyngeal swabs collected from children with acute respiratory tract infections in Eastern Nunavut and Southern Ontario/Hamilton area (Canada).
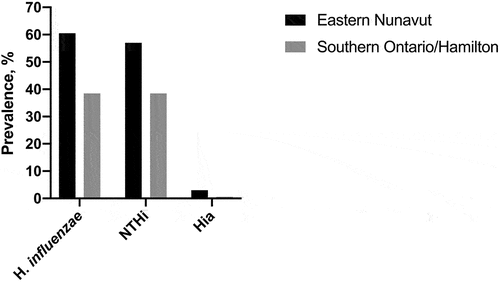
Table 1. Demographics of study participants.
The collection of specimens was not uniform over the seasons; 121/200 (60.5%) of NU swabs were obtained during the spring (March to May) that corresponded to the RSV infection season. Outside the spring time, fewer specimens were collected, i.e. 43/200 (21.5%) in December–February; 23/200 (11.5%) in June–August and 13/200 (6.5%) in September–November. The frequency of H. influenzae detection did not significantly depend on the season, i.e. varied between 56.2% (spring), 60.5% (winter), 69.2% (autumn) to 78.3% (summer). In NU children, H. influenzae prevalence decreased with age: 72/105 (68.6%) in <2 years old, 33/58 (56.9%) in 2–3-year-old and 16/37 (43.2%) in ≥4 years old (p = 0.02) (Supplementary Table S3). No dependence of H. influenzae on age was found in HA or EO children (Supplementary Tables 2, 4). In NU, Hia was identified in two <2 years old, two 2–3-year-old and two ≥4 years old; in HA, the only child with Hia was 3 years old (Supplementary Tables 3–4). Although the percentage of children found to carry Hia appeared to be increasing with age, the number of older children sampled was too small to judge if this was a random finding.
Discussion
Although invasive H. influenzae disease is reportable in Canada [Citation18], the overall prevalence of H. influenzae, including asymptomatic colonisation of mucosal surfaces and non-invasive infections, is unknown. In populations with high incidence of invasive disease, the latter usually represents just the tip of the iceberg as only some individuals exposed to the virulent microorganism would develop a serious disease. In this study, we addressed the question of whether the prevalence of H. influenzae in the nasopharynx differed between paediatric populations from regions with high versus low incidence of invasive Hia disease. We used NP specimens collected for routine diagnostic detection of respiratory viruses; those were saved and further analysed with PCR to identify and serotype H. influenzae. The presence of H. influenzae DNA in NP specimens of children with symptoms of ARTI could be associated with various roles of H. influenzae in the pathogenesis. H. influenzae could have been a coloniser that was present before the child developed clinical infection caused by another organism and remained a coloniser during the illness, rather than being a part of the pathogenesis. Alternatively, H. influenzae could be pathogenic, acting as an initial cause or “co-cause” of the current illness, or have not been initially present but rather acquired following viral infection. Because we were unable to collect clinical data on the study participants, we could not determine the significance of H. influenzae in individual cases. Nevertheless, finding these bacteria in the paediatric population with a high incidence of invasive Hia disease can potentially shed light on the epidemiology of this infection.
The prevalence of all H. influenzae, particularly NTHi, was significantly higher in NP specimens of children from Nunavut compared to children from Southern Ontario. Invasive H. influenzae disease is also much more common in children from Northern Canada, including Nunavut, than in the whole province of Ontario, illustrated by recently reported incidence rates of 195.15/100,000 versus 10.17/100,000 for <1-year old, respectively [Citation19,Citation20]. Moreover, the serotype distribution of invasive H. influenzae isolates differs strikingly between the regions, with Hia greatly offsetting NTHi in the North (60.5% Hia and 17.8% NTHi), in contrast to Ontario where NTHi predominates (8.9% Hia and 74.2%, NTHi) [Citation19,Citation20]. In Ontario children <1 year of age, in 2014–2018, the incidence rates of invasive disease caused by Hia and NTHi were 2.79/100,000 and 5.85/100,000, respectively [Citation20]. Disproportionally high incidence of severe infections in the North is not specific to H. influenzae but has been reported for other invasive bacterial diseases (Streptococcus pneumoniae, Neisseria meningitidis, group A and B streptococci) that are commonly explained by unfavourable environmental conditions and specific demographics in the Arctic [Citation19]. However, the reasons for the prevalence of Hia among invasive H. influenzae isolates in Northern Canada are uncertain. Intriguingly, Hia has not been reported from Greenland, where the climate and demographics are similar to North American Arctic [Citation21].
Historical studies conducted in the pre-Hib vaccine era found that carriage of Hib in children was higher in North American Indigenous communities, such as Alaskan Native, Navajo and White Mountain Apache, than in non-Indigenous populations [Citation8, Citation22–24]. Among various ethnic groups living in Alaska, Inuit children had the highest incidence of invasive Hib disease [Citation23]. Populations with a high incidence of invasive Hib disease were also characterised by high carriage rates [Citation8]. Since the 1980s, the use of Hib conjugate vaccines greatly decreased the rates of Hib invasive disease, as well as reduced oropharyngeal (OP) colonisation among populations at high risk of invasive disease, such as Navajo and Apache children [Citation8,Citation25]. Previous studies have identified the importance of unfavourable environmental and socio-economic conditions as risk factors for Hib carriage in the pre-Hib vaccine era. Disadvantaged living conditions, such as overcrowded housing, lack of access to clean water and poor indoor ventilation have been implicated as factors favouring increased transmission of respiratory pathogens, including H. influenzae [Citation26]. Although Hia carriage in Indigenous children has hardly been studied, Nolen et al. (2021) identified crowding and presence of several tobacco users in households as risk factors for Hia carriage in a rural Alaska community [Citation10]. However, although Inuit are substantially different from other Indigenous populations in the North, these studies did not distinguish Inuit from other groups of Indigenous Peoples, collectively referred to as “Alaska Native”. Recently published Canadian data on Hia carriage in healthy Indigenous children only included First Nations [Citation11]. More studies on Hia epidemiology with analysis involving specific ethnic groups are needed as they may help to understand the reasons for the uneven distribution of Hia disease among geographic areas and populations.
We found that NTHi dominated in NP specimens from both northern and southern regions accounting for 94.2% and 98.4% of all H. influenzae. High prevalence of NTHi in NP swabs from children with ARTI has been reported by others suggesting that inflammatory responses triggered by viral infections may favour colonisation of mucosal surfaces by NTHi [Citation27–29]. Experimental studies demonstrated that disruption of epithelial barrier function by rhinovirus facilitated NTHi colonisation [Citation30]. Despite considerably higher presence of Hia among invasive clinical isolates in Nunavut than in Ontario [Citation19,Citation20], there was no significant difference in Hia prevalence in non-invasive specimens between NU and HA cohorts (3.0% and 0.6%, respectively). These findings substantiate our recent data collected in Northern Ontario Indigenous communities where Hia was overrepresented among invasive H. influenzae isolates (60%) but present only in 3% of non-invasive isolates, in clear contrast to NTHi associated with 25% of invasive and 91% non-invasive isolates [Citation12]. Superior abilities of NTHi to colonise mucosal surfaces as well as potential competition among encapsulated and non-encapsulated strains in the same ecological niche may explain these observations [Citation31]. Interestingly, all H. influenzae identified in NP specimens collected from hospitalised children (EO) belonged to NTHi, with higher prevalence as compared to NU and HA cohorts, and lack of any encapsulated strains that can be explained by older age of EO children and different clinical presentations. Admission to a tertiary paediatric centre could be associated with conditions favouring the presence of NTHi in the upper respiratory tract, e.g. representing nosocomial acquired infection or colonisation [Citation32]. Indeed, NTHi colonisation rates among children with ARTI can considerably vary (from 40% to 85%) and depend on dynamic interactions among H. influenzae, respiratory viruses and other bacterial species in the local microbiota [Citation29,Citation33,Citation34]. Although NTHi can cause serious infections, as documented by an increase in incidence of invasive NTHi disease in Europe and North America over the last two decades [Citation35], encapsulated H. influenzae have superior abilities to resist serum bactericidal activity and phagocytosis that explain their propensity to cause invasive disease [Citation36].
How would 3% Hia detected in NP swabs of NU children relate to high incidence rates of invasive Hia disease? In Northern Ontario, a region with consistent presence of invasive Hia disease, albeit at lower incidence rates as compared to Nunavut, Hia was found in 3% of children with non-invasive disease, such as otitis media, with asymptomatic Hia and NTHi colonisation rates of 9.1% and 51.7%, correspondingly [Citation11,Citation12]. Considering that Hia is an important cause of severe invasive disease in Nunavut children, its presence in 3% of children with ARTI can reflect continuing circulation of the pathogen in the Northern communities that may result in invasive disease outbreaks. Paediatric immunisation with a new Hia conjugate vaccine, which has recently been developed in Canada, can potentially reduce the pathogen transmission among susceptible individuals via decreasing Hia colonisation rates in young children [Citation37]. Indeed, a decrease in H. influenzae detection rates with age in Nunavut children may reflect the maturation of the immune system with age, a natural process leading to an increased resistance to invasive disease; this can be accelerated via immunisation. Our findings corroborate previously published data demonstrating that H. influenzae was less commonly found in children with ARTI > 5 years of age compared to younger children [Citation29,Citation34,Citation38]. As we have not found any Hib out of 436 tested NP specimens, this illustrates the impact of publicly funded paediatric immunisation against this infection. Although invasive Hib disease has not been eradicated, a very low incidence was reported from 2018 to 2021, i.e. 0.03–0.05/100,000 Canadian population [Citation39].
This study has several limitations related to the methodology. We used NP swabs for analysis because they are routinely collected in Canada for virology testing. However, we recognise that there could be differences in detection rates of H. influenzae depending on the specimen collection site. Previous studies have frequently used OP swabs for detecting Hia in Alaska children [Citation10], as well as for study of Hib carriage [Citation22,Citation26,Citation40], although NP swabs have also been used to study H. influenzae carriage in children [Citation6,Citation7]. While some earlier studies found that OP swabs were more efficient than NP for detecting Hib colonisation on older children [Citation22], Rapola et al. (1997) demonstrated higher H. influenzae colonisation rates using NP aspirates or NP swabs as compared to OP swabs [Citation41]. Whether Hia colonisation rates depend on the sampling site and whether Hia presence in a particular niche in the upper respiratory tract can impact the epidemiology and pathogenesis of invasive Hia disease is unknown and this deserves further study.
Because we used only molecular methods, PCR positive results should be considered to indicate the presence of bacterial DNA, but not necessarily viable microorganisms. We could not exclude the possibility that some children had received antibiotics targeting H. influenzae prior to NP swabbing, and that detection of bacterial DNA might have potentially been affected, which could in turn impact the results and comparison between the sites. As concurrent carriage of NTHi and encapsulated H. influenzae could not be ascertained using this methodology, the true proportion of encapsulated strains in the specimens might have been underestimated. For logistic reasons, we were unable to collect clinical data, information on vaccination status or potential confounding factors that could potentially explain the higher prevalence of NTHi in EO compared to NU and HA cohorts. We did not have access to information on specific locations where the NP swabs were taken or determine if Qikiqtani samples were collected from multiple or few communities because of REB restrictions. There was a potential for various biases in specimen collection if a) more sampling in some communities was conducted than in others; b) some clinical presentations could prompt more sampling than others; c) sampling could depend on health care providers; d) frequency of sampling could depend on specific circumstances, such as the local epidemiological situation. Due to the study limitations, the results cannot be generalised to healthy children, or overall Nunavut or Hamilton communities.
Conclusions
As current surveillance of H. influenzae infection includes only cases of invasive disease, this does not allow a complete understanding of epidemiology, which is essential for developing preventive measures. Analysis of NP swabs routinely collected from children with respiratory infections for virology testing does not involve any additional invasive procedures and represents a valuable source of data on pathogen circulation in paediatric populations, especially among groups with a high risk of invasive disease. Although analysis of NP specimens collected from children with ARTI does not answer questions on how and when the children acquired H. influenzae, this provides important information that Hia is commonly present in the nasopharynx of sick children in the eastern region of Nunavut. Considering that Hia is an important cause of severe invasive disease in Nunavut children, its 3% presence among children with ARTI can reflect continuing circulation of the pathogen in the Northern communities that may result in invasive disease outbreaks.
Supplemental Material
Download MS Word (15.7 KB)Acknowledgments
We would like to acknowledge the support provided by the Department of Health, Government of Nunavut and thank Dr Amber Miners, Qikiqtani General Hospital, Iqaluit, for helpful discussion and advice on study design. We are grateful to Jennifer Bowes and Chantal Bergeron for organising the collection of specimens in Children’s Hospital of Eastern Ontario, Julia Maciejewski for help with handling specimens at Hamilton Regional Laboratory and Michelle Shuel and Julina Allarie for laboratory analysis of the specimens at the National Microbiology Laboratory.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The datasets used in the current study are not publicly available due to REB restrictions.
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/22423982.2024.2371111.
Additional information
Funding
References
- Murphy TF. Haemophilus species (including H. influenzae and Chancroid). In: Mandell Gerald L, Bennett John E Raphael D, editors. Mandell, Douglas, and Bennett’s Principles and Practice of Infectious Diseases. 7th ed. Philadelphia: Churchill Livingstone Elsevier; 2010. p. 2911–8.
- Slack MPE, Cripps AW, Grimwood K, et al. Invasive Haemophilus influenzae infections after 3 decades of Hib protein conjugate vaccine use. Clin Microbiol Rev. 2021;34(3):e0002821. Epub 2021/06/02. Doi: 10.1128/CMR.00028-21 PubMed PMID: 34076491; PMCID: PMC8262803.
- Degani N, Navarro C, Deeks SL, et al. Invasive bacterial diseases in Northern Canada. Emerg Infect Dis. 2008;14(1):34–40. doi: 10.3201/eid1401.061522
- Ulanova M, Tsang RSW. Haemophilus Influenzae Serotype a as a Cause of Serious Invasive Infections. Lancet Infect Dis. 2014;14(1):70–82. Epub 2013/11/20. Doi: 10.1016/S1473-3099(13)70170-1 PubMed PMID: 24268829.
- Faden H, Duffy L, Williams A, et al. Epidemiology of nasopharyngeal colonization with nontypeable Haemophilus influenzae in the first 2 years of life. J Infect Dis. 1995;172(1):132–135. doi: 10.1093/infdis/172.1.132
- Mackenzie GA, Leach AJ, Carapetis JR, et al. Epidemiology of nasopharyngeal carriage of respiratory bacterial pathogens in children and adults: cross-sectional surveys in a population with high rates of pneumococcal disease. BMC Infect Dis. 2010;10(1):304. doi: 10.1186/1471-2334-10-304
- Jourdain S, Smeesters PR, Denis O, et al. Differences in nasopharyngeal bacterial carriage in preschool children from different socio-economic origins. Clin Microbiol Infect. 2011;17(6):907–914. doi: 10.1111/j.1469-0691.2010.03410.x
- Takala AK, Santosham M, Almeido-Hill J, et al. Vaccination with Haemophilus influenzae type b meningococcal protein conjugate vaccine reduces oropharyngeal carriage of Haemophilus influenzae type b among American Indian children. Pediatr Infect Dis J. 1993;12(7):593–599. doi: 10.1097/00006454-199307000-00010
- Hammitt LL, Hennessy TW, Romero-Steiner S, et al. Assessment of carriage of Haemophilus influenzae type a after a case of invasive disease. Clin Infect Dis. 2006;43(3):386–387. doi: 10.1086/505602
- Nolen LD, Tiffany A, DeByle C, et al. Haemophilus influenzae Serotype a (Hia) carriage in a Small Alaska Community after a cluster of Invasive Hia Disease, 2018. Clin Infect Dis. 2021;73(2):e280–e286. doi: 10.1093/cid/ciaa750
- Ulanova M, Tsang RS, Nix EB, et al. Carriage of Haemophilus influenzae serotype a in children: Canadian Immunization Research Network (CIRN) study. J Assoc Med Microbiol Infect Dis Can. 2024;9(1):20–31. doi: 10.3138/jammi-2023-0020 PMID: 38567364; PMCID: PMC10984318.
- Ulanova M, Tsang RSW, Nix EB, et al. Canadian immunization research network investigators (2023) epidemiology of invasive Haemophilus influenzae disease in northwestern Ontario: comparison of invasive and noninvasive H. influenzae clinical isolates. Can J Microbiol. 2023;69(6):219–227. doi: 10.1139/cjm2022-0208 PMID: 36753721.
- Statistics Canada. Census Profile. 2021 Census of Population. Statistics Canada Catalogue no. 98-316-X2021001 Ottawa; 2023 [cited 2023 Nov 15].
- Tsang RSW, Li YA, Mullen A, et al. Laboratory characterization of invasive Haemophilus influenzae isolates from Nunavut, Canada, 2000–2012. International Journal of Circumpolar Health. 2016;75(1):29798. doi: 10.3402/ijch.v75.29798
- Wang X, Mair R, Hatcher C, et al. Detection of bacterial pathogens in Mongolia meningitis surveillance with a new real-time PCR assay to detect Haemophilus influenzae. Int J Med Microbiol. 2011;301(4):303–309. doi: 10.1016/j.ijmm.2010.11.004
- Hare KM, Binks MJ, Grimwood K, et al. Culture and PCR detection of Haemophilus influenzae and Haemophilus haemolyticus in Australian Indigenous children with bronchiectasis. J Clin Microbiol. 2012;50(7):2444–2445. doi: 10.1128/JCM.00566-12
- Falla TJ, Crook DW, Brophy LN, et al. PCR for capsular typing of Haemophilus influenzae. J Clin Microbiol. 1994;32(10):2382–2386. doi: 10.1128/jcm.32.10.2382-2386.1994
- Canadian Notifiable Disease Surveillance System. [cition 2024 Feb 21]. Available from: https://diseases.canada.ca/notifiable/diseases-list
- Huang G, Martin I, Tsang RS, et al. Invasive bacterial diseases in northern Canada, 1999 to 2018. Can Commun Dis Rep. 2021;47(11):491–499. doi: 10.14745/ccdr.v47i11a09 PMID: 34880711; PMCID: PMC8601277.
- McTaggart LR, Cronin K, Seo CY, et al. Increased incidence of invasive Haemophilus influenzae disease driven by non-type B Isolates in Ontario, Canada, 2014 to 2018. Microbiol Spectr. 2021;9(2):e0080321. doi: 10.1128/Spectrum.00803-21 PMID: 34612671; PMCID: PMC8510165.
- Zulz T, Huang G, Rudolph K, et al. Epidemiology of invasive Haemophilus influenzae serotype a disease in the North American Arctic, 2006–2017. International Journal of Circumpolar Health. 2022;81(1):2150382. doi: 10.1080/22423982.2022.2150382 PMID: 36461156; PMCID: PMC9728126.
- Michaels RH, Poziviak CS, Stonebraker FE, et al. Factors affecting pharyngeal Haemophilus influenzae type b colonization rates in children. J Clin Microbiol. 1976;4(5):413–417. doi: 10.1128/jcm.4.5.413-417.1976
- Ward JI, Margolis HS, Lum MK, et al. Haemophilus influenzae disease in Alaskan Eskimos: characteristics of a population with an unusual incidence of invasive disease. Lancet. 1981;1(8233):1281–1285. doi: 10.1016/S0140-6736(81)92458-2
- Hall DB, Lum MK, Knutson LR, et al. Pharyngeal carriage and acquisition of anticapsular antibody to Haemophilus influenzae type b in a high-risk population in southwestern Alaska. Am J Epidemiol. 1987;126(6):1190–1197. doi: 10.1093/oxfordjournals.aje.a114758
- Murphy TV, Pastor P, Medley F, et al. Decreased Haemophilus colonization in children vaccinated with Haemophilus influenzae type b conjugate vaccine. J Pediatr. 1993;122(4):517–523. doi: 10.1016/S0022-3476(05)83529-2
- Singleton R, Bulkow LR, Levine OS, et al. Experience with the prevention of invasive Haemophilus influenzae type b disease by vaccination in Alaska: the impact of persistent oropharyngeal carriage. J Pediatr. 2000;137(3):313–320. doi: 10.1067/mpd.2000.107843
- DeMuri GP, Gern JE, Eickhoff JC, et al. Dynamics of bacterial colonization with Streptococcus pneumoniae, Haemophilus influenzae, and Moraxella catarrhalis during symptomatic and asymptomatic viral upper respiratory tract infection. Clin Infect Dis. 2018;66(7):1045–1053. doi: 10.1093/cid/cix941 PMID: 29121208; PMCID: PMC6019034.
- Ederveen THA, Ferwerda G, Ahout IM, et al. Haemophilus is overrepresented in the nasopharynx of infants hospitalized with RSV infection and associated with increased viral load and enhanced mucosal CXCL8 responses. Microbiome. 2018;6(1):10. doi: 10.1186/s40168-017-0395-y PMID: 29325581; PMCID: PMC5765694.
- Thors V, Christensen H, Morales-Aza B, et al. High-density Bacterial Nasal Carriage in Children is Transient and Associated with Respiratory Viral Infections-Implications for Transmission Dynamics. Pediatr Infect Dis J. 2019;38(5):533–538. doi: 10.1097/INF.0000000000002256 PMID: 30985547.
- Sajjan U, Wang Q, Zhao Y, et al. Rhinovirus disrupts the barrier function of polarized airway epithelial cells. Am J Respir Crit Care Med. 2008;178(12):1271–1281. doi: 10.1164/rccm.200801-136OC PMID: 18787220; PMCID: PMC2599868.
- Langereis JD, de Jonge Mi, de Jonge MI. Unraveling Haemophilus influenzae virulence mechanisms enable discovery of new targets for antimicrobials and vaccines. Curr Opin Infect Dis. 2020;33(3):231–237. doi: 10.1097/QCO.0000000000000645 PubMed PMID: 32304471.
- Miyahara R, Suzuki M, Morimoto K, et al. Nosocomial outbreak of upper respiratory tract infection with β-Lactamase-negative ampicillin-resistant nontypeable Haemophilus influenzae. Infect Control Hosp Epidemiol. 2018;39(6):652–659. doi: 10.1017/ice.2018.56
- Van den Bergh MR, Biesbroek G, Rossen JW, et al. Associations between pathogens in the upper respiratory tract of young children: interplay between viruses and bacteria. PLOS ONE. 2012;7(10):e47711. doi: 10.1371/journal.pone.0047711 PMID: 23082199; PMCID: PMC3474735.
- Giucă MC, Cîlcic C, Mihăescu G, et al. Streptococcus pneumoniae and Haemophilus influenzae nasopharyngeal molecular detection in children with acute respiratory tract infection in SANADOR Hospital, Romania. J Med Microbiol. 2019;68(10):1466–1470. doi: 10.1099/jmm.0.001038 PMID: 31389781.
- Van Eldere J, Slack MP, Ladhani S, et al. Non-typeable Haemophilus influenzae, an under-recognised pathogen. Lancet Infect Dis. 2014;4(12):1281–1292. doi: 10.1016/S1473-3099(14)70734-0
- Moxon ER, Kroll JS. The Role of Bacterial Polysaccharide Capsules as Virulence Factors. Curr Top Microbiol Immunol. 1990:15065–15085. doi: 10.1007/978-3-642-74694-9_4 PubMed PMID: 2404690.
- Barreto L, Cox AD, Ulanova M, et al. The emerging Haemophilus influenzae serotype a infection and a potential vaccine: Implementation science in action. Can Commun Dis Rep. 2017;43(5):85–88. doi: 10.14745/ccdr.v43i05a01
- Navne JE, Koch A, Slotved HC, et al. Effect of the 13-valent pneumococcal conjugate vaccine on nasopharyngeal carriage by respiratory pathogens among Greenlandic children. Int J Circumpolar Health. 2017;76(1):1309504. doi: 10.1080/22423982.2017.1309504
- Government of Canada. Notifiable Diseases Online. [cited 2024 Feb 21]. Available from: https://diseases.canada.ca/notifiable/charts?c=abs
- Millar EV, O’Brien KL, Levine OS, et al. Toward elimination of Haemophilus influenzae type B carriage and disease among high-risk American Indian children. Am J Public Health. 2000;90(10):1550–1554.
- Rapola S, Salo E, Kiiski P, et al. Comparison of four different sampling methods for detecting pharyngeal carriage of Streptococcus pneumoniae and Haemophilus influenzae in children. J Clin Microbiol. 1997;35(5):1077–1079. doi: 10.1128/jcm.35.5.1077-1079.1997
