ABSTRACT
Epidermal growth factor receptor (EGFR) mutations have been reported to be associated with non–small cell lung cancer (NSCLC) and correlated to the responsiveness of tumors to EGFR tyrosine kinase inhibitors (TKIs). EGFR mutations in NSCLC Egyptian patients was investigated in formalin-fixed paraffin-embedded lung tumor tissues of 120 NSCLC patients and 20 control tissues from patients with benign lung tumor using ViennaLab StripAssay. Patients showed higher rates of females (87/120, 72.5%) (P=0.043) and never-smokers (82/120, 68.3%) (P=0.003), than the control group. EGFR mutations were significantly related to NSCLC adenocarcinoma, where 49 (40.8%) of patients were mutant (P=0.013) compared with the control group who have no mutations. EGFR mutations were most found in exons 1821, whereas the most mutated exons were exon 19 (55.1%) and exon 21 (26.5%). While exon 18 (10.2%) and exon 20 (8.2%) were the less mutated exons. Moreover, the most common mutations were; L858R (Leu858Arg) in exon 21 and L747-P753 in exon 19; representing 22.4% and 18.4% of all mutations; respectively. Our findings imply that, somatic EGFR mutations could be helpful for NSCLC diagnosis and can be used in combination with clinical factors to select the patients in Egypt who will respond more effectively to TKIs.
Introduction
Lung cancer is one of the most common malignancies that affect both men and women, it is the leading cause of cancer death in men, and the second cause of cancer death in women, after breast cancer [Citation1,Citation2]. Lung cancer accounts 5–7% of all malignancies in Egypt [Citation3], with incidence rising from 11.9 to 63.3 per 100.000 males and 3.7 to 13.8 per 100.000 women [Citation4,Citation5] during the period 2000 and 2014 [Citation6]. Lung cancers are classified into two major types, small cell lung cancer (SCLC) and non–small cell lung cancer (NSCLC), with the latter consisting of several types, mainly adenocarcinoma; the most commonly diagnosed type [Citation7]; and squamous cell carcinoma [Citation8], and accounts for approximately 85% of all lung cancer cases [Citation9]. In Egypt, NSCLC represents about 40% of total lung cancer patients, with adenocarcinoma accounting for about 63% [Citation10].
Smoking, air pollution, exposure to carcinogenic chemicals, alcohol intake and a family history of lung cancer are all considered risk factors [Citation11]. However, smoking seems to be a less potent oncogenic factor for adenocarcinoma than squamous cell carcinoma and SCLC [Citation12,Citation13]. In addition, genetic factors contribute in the development of sporadic lung cancer and may vary by ethnicity [Citation14]. Previous studies reported that somatic mutations in the epidermal growth factor receptor (EGFR), tumor protein 53 (TP53), Kirsten rat sarcoma viral oncogene (KRAS), proto-oncogene B-Raf (BRAF), c-ROS oncogene 1 (ROS1), anaplastic lymphoma kinase (ALK), and human epidermal growth factor receptor (HER2) are common in lung cancers. These genes are involved in the regulation of gene expression, cell proliferation, differentiation and apoptosis [Citation12,Citation13,Citation15].
The discovery of oncogenic driver mutations in NSCLC patients over the past 20 years has led to new molecular targeted therapies that have dramatically improved treatment efficacy and quality of life [Citation16]. Over-expression of EGFR is associated with angiogenesis and poor prognosis in 50% – 80% NSCLC patients, and that makes it a prime candidate for targeted therapy [Citation17]. EGFR is a transmembrane glycoprotein with cytoplasmic tyrosine kinase activity consisting of an extracellular ligand-binding domain, a transmembrane domain, an intracellular tyrosine kinase (TK) domain and a regulatory region [Citation18]. After ligand binding, specific tyrosine residues of the intracellular domain are autophosphorylated, leading to the initiation of intracellular signaling cascades, including the Ras/Raf/MAPK, JAK/ STAT and PI3K-Akt pathways, this results in a number of consequences including cell proliferation, cell differentiation, angiogenesis, metastasis and anti-apoptosis [Citation19]. Mutations in the EGFR enhance cell proliferation through increasing downstream signaling. The molecular characterization of advanced NSCLC adenocarcinoma in recent years had participated in identifying several driver events, including EGFR constitutive activation [Citation20]. EGFR gene is frequently amplified, and/or mutated in lung adenocarcinoma. In NSCLC cases; EGFR is highly expressed and coding for tyrosine kinase receptor [Citation21]. EGFR mutation analysis is considered as an essential biomarker in the controlling of advanced lung adenocarcinoma, assessment of responsiveness to tyrosine kinase inhibitors (TKIs) [Citation22–24], for prolonged progression free survival (PFS) and overall survival (OS) of EGFR positive NSCLC patients [Citation25]. Therefore, it is essential to examine the basic status of EGFR mutations at diagnosis using the most trustworthy methods [Citation23]. EGFR mutations are frequently found in adenocarcinoma, with Asians having higher rates (38.8%–64.0%) than Caucasians (4.9%–17.4%). Exon 19 deletions and exon 21 leucine to arginine substitutions (L858R) are the most frequent EGFR mutations, accounting for about 90% of all EGFR mutations. Furthermore, exons 18 and 20 also have mutations that are less prevalent [Citation26].
Recently, development of TKIs such as gefitinib (2002), erlotinib (2003), bevacizumab (2006), and crizotinib (2011), Rociletinib (2016) Osimertinib (2017); that block EGFR-derived signal transduction are the targeted therapy for EGFR-mutant NSCLC patients [Citation27]. TKIs have a major impact in the treatment of NSCLC [Citation27–29]. Several studies demonstrated the clinical importance of EGFR mutation types in lung cancer adenocarcinoma [Citation26,Citation30,Citation31]. Mutations in exons 19 and 21 have a good prognosis due to their sensitivity to TKIs. While other rare mutations have unknown prognoses [Citation26].
This study aimed primarily to assess the prevalence of the most common EGFR mutations in Egyptian patients diagnosed with NSCLC adenocarcinoma in comparison to the patients with benign lung tumor. In addition, the study aimed to correlate the patients’ characteristics with EGFR mutation status and give an insight on the possible mutations that could be associated with NSCLC in patients from Egypt and thus could help in diagnosis and treatment.
Materials and methods
Patients and tissue samples
Tumor samples were collected from 120 Egyptian patients diagnosed with non-small cell lung cancer. Eligible patients (68 males and 52 females; mean age 62.7 ± 1 years, range 40–86) who were diagnosed as primary NSCLC, adenocarcinoma subtype, smokers, and nonsmokers, stage of tumor from I to IV and with availability of tumor samples (biopsy or surgical specimen) were included in this study. Patients, who were diagnosed with any other type of NSCLC rather than adenocarcinoma, have multiple primaries of malignancies, suffering from any other diseases, received any other treatment with chemo or radiotherapy or pediatric age were excluded from the study. Twenty tissue samples from patients with benign lung tumor were included as control group (10 males,10 females; mean age 58.9 ± 1.5 years, range 40–80). All patients participated in the study after signing consent forms, at Al-Kasr Al-Eini Hospital’s Department of Clinical Oncology and Nuclear Medicine. The study protocol and informed consent were approved by the ethical committee.
Formalin fixed paraffin embedded (FFPE) samples were prepared for subsequent analysis of EGFR mutations. The procedure included three main steps: (1) DNA isolation, (2) PCR amplification using biotinylated primers, and (3) hybridization of the amplified products to a test strip containing allele-specific oligonucleotide probes immobilized as an array of parallel lines using EGFR XL Strip Assay ® kit (ViennaLab Diagnostics GmbH, Austria). Bound biotinylated sequences were detected using streptavidin-alkaline phosphatase and color substrates. The assay covered 30 mutations in the four exons of EGFR gene (18/19/20/21) [Citation32].
DNA extraction
The DNA was extracted from formalin-fixed paraffin-embedded (FFPE) tissue samples using Qiagen kit (QIAamp DNA FFPE tissue kit, UK) according to the manufacturer’s instructions. Paraffin was dissolved in xylene and then tissues were lysed under denaturing conditions with proteinase K and an anionic detergent in the presence of a DNA stabilizer. Then, samples were incubated at 90°C to reverse formalin crosslinking, thus allowing DNA binding to the membrane and contaminants to flow through. Finally, pure, concentrated DNA was eluted and stored at −20°C. DNA concentration was assessed by NanoDrop™ spectrometer device (Nanodrop Technologies, Inc), pure DNA had measured ratio 1.2–8 at wavelength 260/280.
PCR and detection of EGFR mutations
Extracted DNA was amplified by Multiplex PCR amplification of specific DNA sequences using biotinylated primers. Then, the amplification products were analyzed by gel electrophoresis. The amplified products were hybridized to a test strip containing allele-specific oligonucleotide probes immobilized as an array of parallel lines (Precise selective hybridization of specific sequences onto Strip Assay). Malignant tumor and control samples were tested for specific EGFR mutations using ASO strip assay that targets three mutations affecting codon 719 in exon 18 (G719A, G719C, and G719S), grouped deletions within exon 19, insertions within exon 20, and the individual mutations T790M, L858R, L861Q, and S768I. The 30 investigated mutations in the four exons of EGFR gene are listed in .
Table 1. Mutations investigated in the four exons (18, 19, 20, 21) of EGFR gene
Statistical methods
Data management and analysis was performed using the Statistical Package for the Social Sciences (SPSS) version 18.0. Categorical data were summarized as frequencies and percentages. Chi-square (x2) or Fisher’s exact tests were used to compare EGFR mutations’ frequencies between NSCLC patients and control group and to estimate the association between the mutation status and some patient characteristics (gender, age, smoking status, disease status and tumor size, stage, and grade). Student’s t-test was used to compare the age. P-value ≤ 0.05 was considered statistically significant.
Results
Characteristics of patients
The clinical and pathological characteristics of the patients are summarized in . As shown, 120 patients diagnosed with NSCLC adenocarcinoma were eligible for the study. Age was represented by mean ± standard error of mean. The mean age was 62.7 years, 69/120, 57.5% were above 60 years old, 72.5% were female and 68.3% were never-smokers. Significant differences were found in gender and smoking status. Patients showed higher rates of females (87/120, 72.5%) (P = 0.043) and never-smokers (82/120, 68.3%) (P = 0.003), compared with the control group. 74/87 (85%) of females were nonsmokers (P = 0.000).
Table 2. Characteristics of the patients and control groups
Regarding the disease status; 54% of patients had metastases in various regions of their bodies (N = 65). Among them 45/65, (69%), were nonsmokers (P = 0.486). NSCLC spread from the lungs to the bones, liver, brain, pleura, lymph nodes, and other organs. TNM staging was performed according to the Union for International Cancer Control (UICC) guidelines. We found that higher rate of patients included in the study developed stage IV (82/120, 68.3%), grade 2 (96/120, 80%) and size 4 (62/120, 51.7%) of tumor. EGFR mutations were significantly related to NSCLC adenocarcinoma, where 49 (40.8%) of patients were EGFR mutant (P = 0.013) compared with the control group where no mutations were detected (Wild-type).
Association between EGFR mutation and the clinicopathological characteristics of patients
All EGFR mutations in the four exons; 18, 19, 20, 21; were assessed via allele-specific oligonucleotide Strip Assay. EGFR mutations were detected in 49 NSCLC samples (40.8%), where the occurrence incidence of mutations in each exon was 10.2%, 55.1%, 8.2%, and 26.5% of all the mutations, respectively. EGFR mutation status was correlated to the clinicopathological features of patients as shown in . There was no significant correlation between the age and the positivity to EGFR mutations (P = 0.709). Moreover, females exhibited non-significant higher mutated EGFR (35/49, 71.4%) than males (14/49, 28.6%) (P = 0.827). Although it was observed that nonsmokers showed higher incidence rate of EGFR mutations (30/49, 61.2%) compared to smokers (19/49, 38.8%); however, there is no significant correlation between EGFR mutations and smoking (P = 0.164). On the other hand, EGFR mutation positivity was significantly associated with advanced adenocarcinoma 43 (87.8%) (P = 0.000). Moreover, EGFR positive mutations were significantly associated with stage of tumor where 38 cases (77.6%) were at stage IV (P = 0.036). No significant difference was observed in tumor size between groups ().
Table 3. Clinicopathological characteristics of NSCLC patients in relation to EGFR mutation status, N = 120 patients
Frequencies of EGFR mutated exons and its association with the lung cancer risk:
summarizes the number of participants and their mutations. In addition to the frequency of detected mutations of each EGFR exon in NSCLC patients. Exon 19 was significantly the highest mutated and representing 27 (55.1%) of mutated patients, followed by exon 21 which was mutated in 13 (26.5%) of the mutated patients. On the other hand, exons 18 and 20 were significantly the least mutated, where mutations existed in only 5 (10.2%) and 4 (8.2%) of the mutated patients, respectively. As for the control group, none of these exons were mutated in any of the patients with benign tumor. Regarding the gender of NSCLC patients; there was non-significant difference in the incidence of EGFR mutations in the four exons (18, 19, 20, 21) between male and female patients (P > 0.05) ().
Table 4. The frequencies and percentages of detected mutations in each exon of EGFR gene in NSCLC patients
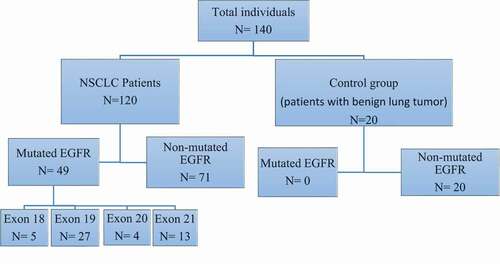
Figure 1. Flow diagram of the study showing the enrolled groups and the detected mutated exons in EGFR gene. As shown; 120 NSCLC patients and 20 control group were enrolled in the study. 49/120 of NSCLC patients have mutated EGFR with high incidence mutations in exon 19, 21,18 and 20. No mutations were detected in the control group. EGFR, epidermal growth factor receptor; NSCLC, non-small-cell lung cancer.
Detected mutations in each exon of EGFR gene:
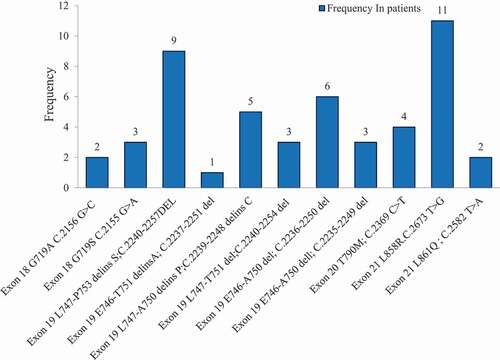
and show the frequency and percentage of each mutation detected in EGFR exons; 18, 19, 20, and 21. The study reported the presence of the following EGFR mutations: two mutations were detected in Exon 18; the first mutation (p. G719A; c. 2156 G > C) was found in 2 patients (4.1%), while the second mutation (p. G719S; c. 2155 G > A) was found in 3 patients (6.1%). Six mutations were detected in Exon 19; the most frequent one was p. L747-P753 delins S; c. 2240–2257 del and was detected in 9 (18.6%) of the mutated patients. Then, p. E746-A750 del; C. 2236–2250 del was found in 6 patients (12.2%), p. L747-A750 delins P; c. 2239–2248 delins C was found in 5 patients (10.2%), p. E746-A750 del; c. 2235–2249 del was found in 3 patients (6.1%), p.L747-T751 del; c. 2240–2254 del was found in 3 patients (6.1%), and the least frequent one was p. E746-T751delinsA; c. 2237–2251 del which was found only in one patient (2%). The only mutation found in Exon 20 was p. T790M; c. 2369 C > T and was detected in 4 (8.1%) of the mutated patients. Two mutations were detected in Exon 21; mutation p. L858R, c. 2573 T > G was detected in 11 (22.5%) of the mutated patients and mutation p. L861Q, c. 2582 T > A was detected only in 2 patients (4.1%). Co-occurrence of two or more EGFR somatic mutations was not reported in the same patient.
Table 5. Frequencies and percentages of different mutations in each mutated exon in EGFR mutation positive patients (N = 49)
Figure 2. The frequencies of each somatic mutation detected in EGFR exons; 18, 19, 20 and 21 in NSCLC patients (N = 49). L858R mutation in exon 21 and L747-P753 mutation in exon 19 showed the highest incidence of all mutations, followed by E746-A750 and L747-A750 mutations in exon 19. The other mutations have low rate of incidence. EGFR, epidermal growth factor receptor; NSCLC, non-small-cell lung cancer.
Discussion
The relationship between the mutational state of EGFR and clinical pathology in NSCLCs was investigated in the present study. The most frequently mutated exons linked to the development of NSCLC were exons 19 and 21 of the TK domain. EGFR gene mutations are the first recognized as targetable driver mutations described in about 17% and 50% of lung adenocarcinoma in Caucasians and Asians; respectively [Citation20,Citation21,Citation33]. In our study, 40.8% of the patients were EGFR mutation positive The incidence differed from published data in the PIONEER Trial which reported lower rate of EGFR mutations among Americans (15%) and higher incidence among Asians (62%) [Citation22]. This prospective, international and epidemiological study correlated the incidence of EGFR mutations to country, gender, ethnicity, smoking status, pack-years, disease stage and histological type. Another study reported that the frequency of EGFR mutations in Middle Eastern and African patients was higher than in white Caucasians, but still lower than in Asian populations [Citation23]. Furthermore, a recent study reported that; the incidence rates based on the population can provide a full picture of the risk of EGFR mutation-positive lung cancer, where the age-specific incidence rates (ASRs) of EGFR mutation-positive NSCLC were around 3.5 times higher for Pacifica and Asians, and two times higher for Maori than New Zealand Europeans [Citation24].
In addition, molecular factors other than EGFR mutations could be attributed to the development of NSCLC including; KRAS, BRAF, c-MET and HER2 mutations or amplification, ALK rearrangement, and ROS1 fusions [Citation25,Citation34]. These molecular factors have impact in targeted therapy, prognosis, and mortality. ALK rearrangements were more common in young, nonsmokers in Asian and Pacific regions [Citation35]. ROS1 mutations were common in advanced adenocarcinomas [Citation36]. Compared to EGFR mutations; KRAS mutations were found to be more common in Western populations (23–33%) than Asian or Australian populations (2–15%) and in smokers (20–44%) than never-smokers (6–10%) [Citation37]. Furthermore, co-occurrence of KRAS with EGFR mutations was more common in Western populations and smokers. Co-occurrence of KRAS with BRAF mutations have been associated with a poor prognosis [Citation38]. While c-MET variations have been linked to a better prognosis [Citation13].
Previous studies specified that histology of adenocarcinoma, status of never smoker, and female gender were features linked with occurrence of EGFR mutations [Citation8,Citation39,Citation40]. In Asians, females had a higher incidence of EGFR mutations than males (61.1% vs. 44), and never smokers had a higher incidence of EGFR mutations than patients who smoked >50 pack-years (60.7% and 31.4%; respectively) [Citation22]. Moreover, in Japan, 30% of NSCLC patients who have mutated-EGFR gene were males or smokers [Citation41]. On the other hand, a lower percentage of women was demonstrated in Egypt [Citation42] and the Middle East [Citation23]. In our findings, 73% of NSCLC patients were females and 68% were nonsmokers. Moreover, higher frequency of EGFR positive patients were females (71%) with 61% were nonsmokers; however, they were not significantly correlated. This could be attributed to the fact that the majority of patients included in the study were female and/or nonsmokers. These results implicate that EGFR mutations might be correlated to gender and factors other than smoking in the Egyptian population. The median age of patients suffering from lung cancer in the developed countries is 60 years [Citation5]. In the current single institutional group of Egyptian patients, the mean age of cancer patients was 48 years. The majority of lung cancer patients in Egypt were in their fifth (55.3%) and sixth (29.3%) decades [Citation43]. That’s why NSCLC patients recruited in our study are over 40 years old (mean age 62.7 years).
IPASS trial (IRESSA Pan-Asia Study) stated that about 96% of patients showed mutations either in exon 19 or exon 21 [Citation44]. Similarly, those mutations were highly detected in our mutated cases: 55.1% and 26.5%; respectively. Other previous studies have shown that the most common EGFR mutations were in exons 19 and 21 representing 85% to 90% of the mutations, where the most frequent mutation in each exon was p. E746-A750 del deletion of exon 19 (45%–50%) and the Leu858Arg (L858R) substitution in exon 21 (40%–45%) [Citation45]. Additional study reported that the most common mutation is exon 19 deletion (45%) followed by exon 21 (L858R) (40%), while other mutations like exon 21 (L861Q) and exon 18 (G719X) were less dominant [Citation39]. G719X mutation might be G719A, G719C, or G719S as a substitution from glycine (G) to alanine (A), cysteine (C), or serine (S); respectively. In the present study, the most common mutations were p. L747-P753 delins in exon 19 and p.L858R in exon 21 which represents 18.4% and 22.4%, respectively. Zaki et al. found exon 19 deletions in 22% of NSCLC cases in a previous Egyptian study of 50 cases, which was similar to our discovered rate and that of Caucasians [Citation46] or East Asians [Citation47], however none of the cases had the L858R point mutation in exon 21 [Citation42]. On the contrary, a higher frequency of the L858R point mutation has been found in East Asians, 16%–46% of NSCLC patients [Citation48].
The T790M mutation at exon 20 is known as a point mutation at codon 790 substituting threonine with methionine in the EGFR gene, it was considered as a rare EGFR mutation [Citation49]. This mutation represents 8.2% in our mutated cases, harmonizing with previous findings, which reported T790M occurrence in 1%–17% of all EGFR mutations, depending on the particular populations, and its presence encourages resistance acquirement and disease progression [Citation50–54].
A recent study correlated T790M mutation with increased rate of disease progression, mortality, and nonresponse to TKI treatment [Citation55]. As most patients with T790M mutation showed resistance to gefitinib and erlotinib (first-generation TKIs) without inhibiting tumor growth in NSCLC, thus, development of third-generation TKIs; such as Osimertinib (Tagrisso); which focus on overcoming acquired resistance to TKIs have shown promising results for therapeutic responses [Citation49,Citation56,Citation57].
G719X is the most common exon 18 mutation, where Lynch et al. first observed this mutation in EGFR in NSCLC patients in 2004 [Citation58,Citation59]. It was formerly detected in about 2–3% of EGFR-positive NSCLC cases [Citation60]. This mutation is related to responses to TKIs but with less sensitivity than the common mutations (19, 21) in vitro [Citation53,Citation61,Citation62]. In our data, we found only two substitutions at exon 18; substitution from glycine to serine which represents 6.1% (that was reported as the most common mutation of exon 18) and the other one is substitution to alanine represents 4.1%. Those two types; G719A and G719S; collectively represents 10.2% of all mutations. G719X mutation accounts for around 3% of all EGFR mutations in Asian and Caucasian populations [Citation59].
The L861Q mutation is caused by a leucine (L) to glutamine (Q) substitution in exon 21 of the EGFR gene. It was found to be uncommon mutation counting around 2% of all EGFR positive mutations. However, in some preclinical trials, it was associated with poor prognosis and resistance to TKIs, other studies showed moderate response to TKIs with longer PFS [Citation63]. On the other hand, patients with the common mutations of deletions in exon 19 and L858R in exon 21 were luckier showing better response to treatment [Citation53]. In our study, we found that L858R represents 22.4% but L861Q represents only 4.1%.
Exon 19 deletion was detected in 27/49 (55.1%) of our mutated samples, and this goes in agreement with Sharma et al. who reported this mutation in 45% of patients in USA [Citation64]. In-frame deletions and replacements of nucleotides in the region of exon 19 comprising nucleotides 2235–2257 (corresponding to amino acids 746–753) is particularly common in patients diagnosed with lung cancer [Citation65]. Studies have shown that patients with a base-pair deletion at exon 19 (del746-A750) or a point mutation at exon 21 (L858R) had a high response to EGFR-TKI [Citation66]. We found that 2240–2257 del (amino acids L747-P753 delins) is the most common mutation in exon 19 (18.4%) followed by E746-A750 del (18.3%), while the other detected mutations in exon 19 occurred in low frequency. L858R is the most common mutation in exon 21 (22.4%) in our population.
Several types of exon 19 mutations were detected in patients from different populations, such as in Chinese and Korean populations, where the mutation p. E746-A750 del; c. 2235–2249 del is the most common of exon 19 (40.2%) [Citation67] and (16.8%) [Citation68]; respectively. However, this mutation represents only 6.1% in our NSCLC cases. Also, p. L747-P753 delins S; c. 2240–2257 del which is the most common mutation in our population (18.4%) is less common one in the Korean population (2.6%) and in Chinese (8.2%). On the other hand, we detected the mutation p. E746-T751delinsA; c. 2237–2251 del in only 2% of our patients, similar results have been found in Korean (0.5%) and Chinese (3.3%) populations [Citation67,Citation68].
Moreover, the six major in-frame deletions in exon 19: Del (746-A750), del (747–749)ins(S), del (747–751), del (746–752), del (747–749) ins(P) and del (747–753); were found to occupy 90.4% of exon 19 mutations in Chinese NSCLC patients. However, only three of them (del (746-A750), del (747–751) and del (747–753) were detected in our NSCLC cases. On the other hand, other in-frame deletions in exon 19; E746-T751 delinsA, L747-A750 delins P, E746-T751 delinsA may be specific to our population as they were found in our population but were not mentioned to be detected before neither in Chinese nor in Korean NSCLC cases.
NSCLC cases of China reveal that the most prevalent mutation types were L858R in exon 21 and del (746–750) in exon 19, which represented about 38.3% and 37.0% of all EGFR mutations, respectively. In exon 18, G719C was prevailing mutation type (43.9%, 29/66). T790M and insertion mutations represented about 71.8% of all exon 20 mutations. The four main mutations (87.5%, 792/812) in exon 21 were L858R, L858M, L861R, and L861Q. The rate of EGFR-TKI sensitive mutations was 88.5% (1713/1935), comprising deletion of exon 19, L858R, L861Q, G719X (G719C, G719S, and G719A). The rate of EGFR-TKI resistant mutations was 2.6% (76/1935), comprising exon 20 insertion and T790M [Citation69].
Conclusion
EGFR mutations in NSCLC were concentrated in exons 18 to 21, where most mutations were observed in exons 19 and 21. The gender, age, smoking status and EGFR mutation pattern could provide a significant impact on the development of NSCLC in the Egyptian patients. Results of the current single institution study revealed a different EGFR mutation profile in NSCLC Egyptian patients diagnosed with adenocarcinoma compared to published reports in the western countries. Furthermore, mutant EGFR was linked to a higher risk of advanced NSCLC adenocarcinoma with metastases. As a consequence of our research, we believe that selecting patients based on the type of EGFR mutations they have in combination with clinical criteria would aid in identifying the most effective medicine that will result in a good response and increased sensitivity to TKIs in Egypt. It would be desirable to establish a further large-scale prospective study that potentially impact prognosis and patient management that would aid in gaining a deeper understanding of the complexities and clinical relevance of co-existent somatic variants in NSCLC.
Abbreviations
SCLC: Small Cell Lung Cancer; NSCLC: Non–Small Cell Lung Cancer; EGFR: Epidermal Growth Factor Receptor; FFPE: Formalin-Fixed Paraffin-Embedded; TKIs: Tyrosine Kinase Inhibitors; PFS: Progression Free Survival; OS: Overall Survival.
Ethical approval
This study was approved by the ethical committee of Kasr El-Einy Center of Clinical Oncology and Nuclear Medicine (NEMROCK), Faculty of Medicine, Cairo University, Egypt, and all patients have written an informed consent.
Authors’ contributions
M.Q. performed the experiments, analyzed data and wrote the initial draft of the manuscript. W.H.E. conceived, designed, planned, and supervised the study. H.B. and S.S. planned and supervised the study, analyzed and graphed data, interpreted results, wrote, critically revised, and edited the manuscript. All authors read the final manuscript and approved submission.
Acknowledgments
Not applicable.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Torre LA, Siegel RL, Jemal A. Lung cancer statistics. Lung cancer and personalized medicine . Advances in Experimental Medicine and Biology; 2016. p. 1–19.
- Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021 May;71(3):209–249.
- Ibrahim AS, Khaled HM, Mikhail NN, et al. Cancer incidence in Egypt: results of the national population-based cancer registry program. J Cancer Epidemiol. 2014;437971.
- Larbi A. AMAAC workshop, Algiers November, 19, 2011.
- Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136(5):E359–E386.
- Dubey AK, Gupta U, Jain S. Epidemiology of lung cancer and approaches for its prediction: a systematic review and analysis. Chin J Cancer. 2016;35(1):71.
- Butnor KJ, Beasley MB. Resolving dilemmas in lung cancer staging and histologic typing. Arch Pathol Lab Med. 2007;131(7):1014–1015.
- Tokumo M, Toyooka S, Kiura K, et al. The relationship between epidermal growth factor receptor mutations and clinicopathologic features in non–small cell lung cancers. Clin Cancer Res. 2005;11(3):1167–1173.
- Cufer T, Ovcaricek T, O’Brien ME. Systemic therapy of advanced non-small cell lung cancer: major-developments of the last 5-years. Eur J Cancer. 2013;49(6):1216–1225.
- Abd El-Monaem Mohamed Abd El-Monaem WHE-S, Zekry MSED. Treatment outcome in patients with metastatic non-small cell lung cancer at presentation (Retrospective study) [original article]. Clin Oncol. 2020 January 26;2020:53–60.
- Cao Y, Gao H. Prevalence and causes of air pollution and lung cancer in Xuanwei City and Fuyuan County, Yunnan Province, China. Front Med. 2012;6(1):1–4.
- de Alencar Vtl, Formiga MN, de Lima Vcc. Inherited lung cancer: a review [Review]. Ecancermedicalscience. 2020;14:1008.
- Fois SS, Paliogiannis P, Zinellu A, et al. Molecular epidemiology of the main druggable genetic alterations in non-small cell lung cancer. Int J Mol Sci. 2021;22(2):612.
- Bennett WP, Alavanja MC, Blomeke B, et al. Environmental tobacco smoke, genetic susceptibility, and risk of lung cancer in never-smoking women. J Natl Cancer Inst. 1999;91(23):2009–2014.
- Is OI, Sohara Y. Genes associated with susceptibility to lung adenocarcinoma among never smokers suggest the mechanism of disease. Anticancer Res. 2014;34(10):5229–5240.
- Lee V, Mok T, Goto Y, et al. Differences between the east and the west in managing advanced-stage non-small cell lung cancer. Clin Oncol. 2019;31(8):560–569.
- Belani C, Goss G, Blumenschein G Jr. Recent clinical developments and rationale for combining targeted agents in non-small cell lung cancer (NSCLC). Cancer Treat Rev. 2012;38(3):173–184.
- Du Z, Lovly CM. Mechanisms of receptor tyrosine kinase activation in cancer [Research support, N.I.H., extramural research support, Non-U.S. Gov’t review]. Mol Cancer. 2018 Feb 19; 17(1):58.
- Wee P, Wang Z. Epidermal growth factor receptor cell proliferation signaling pathways [Review]. Cancers (Basel). 2017 May 17; 9(5):52.
- Travis WD, Brambilla E, Nicholson AG, et al., who classification of the pathology and genetics of tumors of the lungThe 2015 World Health Organization Classification of Lung Tumors: Impact of Genetic, Clinical and Radiologic Advances Since the 2004 Classification. Journal of Thoracic Oncology . 2015;10(9):1243-1260.
- Kris MG, Johnson BE, Berry LD, et al. Using multiplexed assays of oncogenic drivers in lung cancers to select targeted drugs. Jama. 2014;311(19):1998–2006.
- Shi Y, Au JS-K, Thongprasert S, et al. A prospective, molecular epidemiology study of EGFR mutations in Asian patients with advanced non–small-cell lung cancer of adenocarcinoma histology (PIONEER). J Thorac Oncol. 2014;9(2):154–162.
- Benbrahim Z, Antonia T, Mellas N. EGFR mutation frequency in Middle East and African non-small cell lung cancer patients: a systematic review and meta-analysis [Meta-analysis systematic review]. BMC Cancer. 2018 Sep 14; 18(1):891.
- Aye PS, McKeage MJ, Tin Tin S, et al. Population-based incidence rates and increased risk of EGFR mutated non-small cell lung cancer in Maori and Pacifica in New Zealand. PloS One. 2021;16(5):e0251357.
- Wheeler DA, Wang L. From human genome to cancer genome: the first decade [Review]. Genome Res. 2013 Jul;23(7):1054–1062.
- Yoon HY, Ryu JS, Sim YS, et al. Clinical significance of EGFR mutation types in lung adenocarcinoma: a multi-centre Korean study [Multicenter study research support, Non-U.S. Gov’t]. PloS One. 2020 [Jan 26];15(2):e0228925.
- Cagle PT, Chirieac LR. Advances in treatment of lung cancer with targeted therapy. Arch Pathol Lab Med. 2012;136(5):504–509.
- Butterworth S, Cross DA, Finlay MRV, et al. The structure-guided discovery of osimertinib: the first US FDA approved mutant selective inhibitor of EGFR T790M. MedChemComm. 2017;8(5):820–822.
- Passaro A, Guerini-Rocco E, Pochesci A, et al. Targeting EGFR T790M mutation in NSCLC: from biology to evaluation and treatment. Pharmacol Res. 2017;117:406–415.
- Ho HL, Kao HL, Yeh YC, et al. The importance of EGFR mutation testing in squamous cell carcinoma or non-small cell carcinoma favor squamous cell carcinoma diagnosed from small lung biopsies. Diagn Pathol. 2019 Jun 21 14(1):59.
- Hsu WH, Yang JC, Mok TS, et al. Overview of current systemic management of EGFR-mutant NSCLC [Review]. Ann Oncol. 2018 Jan 1 29(suppl_1):i3–i9.
- Popper HH, Gruber-Mösenbacher U, Pall G, et al. The 2020 update of the recommendations of the Austrian working group on lung pathology and oncology for the diagnostic workup of non-small cell lung cancer with focus on predictive biomarkers memo-Magazine of European Medical Oncology. 2020;13:11–26.
- Shi Y, Li J, Zhang S, et al. Molecular epidemiology of EGFR mutations in Asian patients with advanced non-small-cell lung cancer of adenocarcinoma histology–Mainland China subset analysis of the PIONEER study. PloS One. 2015;10(11):e0143515.
- Colombino M, Paliogiannis P, Cossu A, et al. EGFR, KRAS, BRAF, ALK, and cMET genetic alterations in 1440 Sardinian patients with lung adenocarcinoma. BMC Pulm Med. 2019 Nov 11 19(1):209.
- McKeage MJ, Tin Tin S, Khwaounjoo P, et al. Screening for anaplastic lymphoma kinase (ALK) gene rearrangements in non‐small‐cell lung cancer in New Zealand. Intern Med J. 2020;50(6):716–725.
- Chin LP, Soo RA, Soong R, et al. Targeting ROS1 with anaplastic lymphoma kinase inhibitors: a promising therapeutic strategy for a newly defined molecular subset of non-small-cell lung cancer [Review]. J Thorac Oncol. 2012 Nov;7(11):1625–1630.
- Jiang AG, Lu HY. k-RAS mutations in non-small cell lung cancer patients treated with TKIs among smokers and non-smokers: a meta-analysis. Contemp Oncol (Pozn). 2016;20(2):124–129.
- Scheffler M, Ihle MA, Hein R, et al. K-ras Mutation Subtypes in NSCLC and associated co-occuring mutations in other oncogenic pathways [Research support, Non-U.S. Gov’t]. J Thorac Oncol. 2019 Apr;14(4):606–616.
- Shigematsu H, Lin L, Takahashi T, et al. Clinical and biological features associated with epidermal growth factor receptor gene mutations in lung cancers. J Natl Cancer Inst. 2005;97(5):339–346.
- Shafik HE, Al-Shemarri SH, Al-Enezi F, et al. Frequency of epidermal growth factor mutation status and its effect on outcome of patients with adenocarcinoma of the lung. J Cancer Ther. 2014;5(11):1012.
- Toyooka S, Takano T, Kosaka T, et al. Epidermal growth factor receptor mutation, but not sex and smoking, is independently associated with favorable prognosis of gefitinib‐treated patients with lung adenocarcinoma. Cancer Sci. 2008;99(2):303–308.
- Zaki MA, Ramadan RA, Mahmoud MI, et al. Nonenriched PCR versus mutant-enriched PCR in detecting selected epidermal growth factor receptor gene mutations among nonsmall-cell lung cancer patients [Comparative study]. Genet Test Mol Biomarkers. 2015 Aug;19(8):444–449.
- Abo-Elkheir OI, Hafez MR. Characteristics, risk factors and histopathological types of bronchogenic carcinoma among cases presented to Chest Department, Al-Zahraa Hospital, Al-Azhar University. Int J Community Med Public Health. 2018;5(4):1281–1290.
- Fukuoka M, Wu Y-L, Thongprasert S, et al. Biomarker analyses and final overall survival results from a phase III, randomized, open-label, first-line study of gefitinib versus carboplatin/paclitaxel in clinically selected patients with advanced non–small-cell lung cancer in Asia (IPASS). J clin oncol. 2011;29(21):2866–2874.
- Murray S, Dahabreh IJ, Linardou H, et al. Somatic mutations of the tyrosine kinase domain of epidermal growth factor receptor and tyrosine kinase inhibitor response to TKIs in non-small cell lung cancer: an analytical database. J Thorac Oncol. 2008;3(8):832–839.
- Dufort S, Richard MJ, Lantuejoul S, et al. Pyrosequencing, a method approved to detect the two major EGFR mutations for anti EGFR therapy in NSCLC [Research Support, Non-U.S. Gov’t]. J Exp Clin Cancer Res. 2011 May 16 30(1):57.
- Choi YL, Sun JM, Cho J, et al. EGFR mutation testing in patients with advanced non-small cell lung cancer: a comprehensive evaluation of real-world practice in an East Asian tertiary hospital [Research Support, Non-U.S. Gov’t]. PloS One. 2013;8(2):e56011.
- Kozu Y, Tsuta K, Kohno T, et al. The usefulness of mutation-specific antibodies in detecting epidermal growth factor receptor mutations and in predicting response to tyrosine kinase inhibitor therapy in lung adenocarcinoma [Research support, Non-U.S. Gov’t]. Lung Cancer. 2011 Jul;73(1):45–50.
- Bartholomew C, Eastlake L, Dunn P, et al. EGFR targeted therapy in lung cancer; an evolving story. Respir Med Case Rep. 2017;20:137–140.
- Wu J-Y, Wu S-G, Yang C-H, et al. Lung cancer with epidermal growth factor receptor exon 20 mutations is associated with poor gefitinib treatment response. Clin Cancer Res. 2008;14(15):4877–4882.
- Wu J-Y, Yu C-J, Chang Y-C, et al. Effectiveness of tyrosine kinase inhibitors on “uncommon” epidermal growth factor receptor mutations of unknown clinical significance in non–small cell lung cancer. Clin Cancer Res. 2011;17(11):3812–3821.
- Yasuda H, Kobayashi S, Costa DB. EGFR exon 20 insertion mutations in non-small-cell lung cancer: preclinical data and clinical implications. Lancet Oncol. 2012;13(1):e23–e31.
- Yeh P, Chen H, Andrews J, et al. DNA-Mutation Inventory to Refine and Enhance Cancer Treatment (DIRECT): a catalog of clinically relevant cancer mutations to enable genome-directed anticancer therapy. Clin Cancer Res. 2013;19(7):1894–1901.
- Massarelli E, Johnson FM, Erickson HS, et al. Uncommon epidermal growth factor receptor mutations in non-small cell lung cancer and their mechanisms of EGFR tyrosine kinase inhibitors sensitivity and resistance. Lung Cancer. 2013;80(3):235–241.
- Ye L, Mesbah Ardakani N, Thomas C, et al. Detection of Low-level EGFR c.2369 C > T (p.Thr790Met) resistance mutation in pre-treatment non-small cell lung carcinomas harboring activating EGFR mutations and correlation with clinical outcomes. Pathol Oncol Res. 2020 Oct;26(4):2371–2379.
- Gazdar A. Activating and resistance mutations of EGFR in non-small-cell lung cancer: role in clinical response to EGFR tyrosine kinase inhibitors. Oncogene. 2009;28(S1):S24.
- Yun J, Hong MH, Kim S-Y, et al. YH25448, an irreversible EGFR-TKI with Potent Intracranial Activity in EGFR mutant non-small-cell lung cancer. Clin Cancer Res. 2019;25(8):2575–2587.
- Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non–small-cell lung cancer to gefitinib. N Engl J Med. 2004 [2004 May 20];350(21):2129–2139.
- Li K, Yang M, Liang N, et al. Determining EGFR-TKI sensitivity of G719X and other uncommon EGFR mutations in non-small cell lung cancer: perplexity and solution. Oncol Rep. 2017;37(3):1347–1358.
- Cheng C, Wang R, Li Y, et al. EGFR exon 18 mutations in East Asian patients with lung adenocarcinomas: a comprehensive investigation of prevalence, clinicopathologic characteristics and prognosis. Sci Rep. 2015;5(1):13959.
- Yun C-H, Boggon TJ, Li Y, et al. Structures of lung cancer-derived EGFR mutants and inhibitor complexes: mechanism of activation and insights into differential inhibitor sensitivity. Cancer Cell. 2007;11(3):217–227.
- Kancha RK, Von Bubnoff N, Peschel C, et al. Functional analysis of epidermal growth factor receptor (EGFR) mutations and potential implications for EGFR targeted therapy. Clin Cancer Res. 2009;15(2):460–467.
- Zhang T, Wan B, Zhao Y, et al. Treatment of uncommon EGFR mutations in non-small cell lung cancer: new evidence and treatment [Review]. Transl Lung Cancer Res. 2019 Jun;8(3):302–316.
- Sharma SV, Bell DW, Settleman J, et al. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer. 2007;7(3):169.
- Hoglund B, Jin J, Li Y, et al. Mutation in the epidermal growth factor receptor kinase domain. Google Patents; 2018.
- Pao W, Miller V, Zakowski M, et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Nat Acad Sci. 2004;101(36):13306–13311.
- Tian Y, Zhao J, Ren P, et al. Different subtypes of EGFR exon19 mutation can affect prognosis of patients with non-small cell lung adenocarcinoma. PloS One. 2018;13(11):e0201682.
- Sun P-L, Seol H, Lee HJ, et al. High incidence of EGFR mutations in Korean men smokers with no intratumoral heterogeneity of lung adenocarcinomas: correlation with histologic subtypes, EGFR/TTF-1 expressions, and clinical features. J Thorac Oncol. 2012;7(2):323–330.
- Wang S, Wang Z. EGFR mutations in patients with non-small cell lung cancer from mainland China and their relationships with clinicopathological features: a meta-analysis. Int J Clin Exp Med. 2014;7(8):1967.
