ABSTRACT
Traumatic brain injury (TBI) is one of the most common acute neurological illnesses, but the impact of numerous biochemical and genetic indicators on its neurological sequelae has yet to be examined.This work aims to assess the initial circulating neuroglobin (NGB) levels using ELISA assays and the frequency of genetic variants of isocitrate dehydrogenase isoform 1 (IDH1) [c.395 G˃A(R132H)] by restriction fragment length polymorphism using polymerase chain reaction technique (RFLP-PCR) in patients with traumatic brain injury (TBI). The severity of TBI was assessed using the Glasgow coma scale (GSC) and revised trauma scale (RTS). The outcome was evaluated using the Extended Glasgow Outcome Scale and Disability Rating Scale.This research involved 30 patients with TBI comparable with 30 matched healthy controls.The overall results revealed male predominance among TBI. There were significantly higher serum NGB levels among cases compared to the controls, and among patients with severe TBI than those with mild to a moderate degree, p ˂ 0.05 for both. Serum NGB levels were significantly negatively correlated to both GCS (r = −0.498, p = 0.005) and RTS (r = −0.521, p = 0.003). The serum NGB is better in predicting TBI severity (AUC = 0.715) compared to the outcome (AUC = 0.582). The frequencies of both wild and mutant types of IDH1 c.395 G˃A (R132H) showed non-significant differences in TBI patients in terms of severity or outcome, p˃0.05 for both.Serum NGB can be considered a valuable noninvasive marker for predicting the severity and outcome of TBIpatients. There is a lack of association of IDH1 genetic variants with the outcome or severity of TBI in Egyptian patients.
Introduction
Astrocytes are involved in various brain disorders, including Alzheimer’s disease, focal ischemic stroke, and traumatic brain injury (TBI). An imbalance in blood flow and nutrients such as glucose and lactate causes biochemical and molecular changes in the brain, resulting in neuronal death and the loss of cognitive and motor skills [Citation1].
Hemoglobin (Hb) is the most studied member of the globin family of proteins due to its oxygen-binding affinity in blood. Other globin proteins, such as myoglobin (Mb), cytoglobin (CYGB), globin E (GbE), globin Y (GBY), and neuroglobin (NGB), have been found to be expressed in vertebrate erythrocytes in recent investigation [Citation2].NGB, a member of the globin family of proteins, is synthesized and released by astrocytes as a protective molecule for neurons. NGB expression is induced in astrocytes after a brain injury. NGB promotes neuronal survival; hence its enhanced expression in astrocytes following brain damage could be an endogenous neuroprotective mechanism [Citation1,Citation2]. NGB, a member of the globin family of proteins, is synthesized and released by astrocytes as a protective molecule for neurons. NGB expression is induced in astrocytes after a brain injury. NGB increases neuronal survival; hence its enhanced expression in astrocytes following brain damage could be an endogenous neuroprotective mechanism [Citation1].
NGB is a 17-kDa monomeric protein with a characteristic folded shape that contains both ferric and ferrous forms of the heme-hexa-Fe-type HisF8HisE7 [Citation3].According to homology analyses [Citation4], the NGB sequence is highly conserved between species, accounting for around 76% of sequence conservation between humans and amphibians.NGB is found in the neurological system, as well as the eyes, gut, and ovaries; however, it has not been found in the kidney, liver, heart, or skeletal muscle [Citation5,Citation6]. NGB is detected in several brain parts, including the cortex, thalamus, cerebellum, hippocampus, and hypothalamus. These areas are critical for sensation processing, memory, and learning, and they are frequently damaged by hypoxia, ischemic insults, and traumatic injuries [Citation7].
In brain diseases, genome-wide alterations of the cytosolic nicotinamide adenine dinucleotide phosphate (NADP+)-dependent isocitrate dehydrogenase (IDH1) were discovered. Isocitrate dehydrogenase catalyzesisocitrate’s oxidative decarboxylation to -α-ketoglutarate, resulting in NADPH generation. Mutations impact the amino acid arginine at position 132, which is evolutionarily highly conserved and located in the isocitrate binding site. In most cases, histidine replaces wild-type arginine at position 132, [c.395 G˃A(R132H)] [Citation8,Citation9].Under hypoxic situations, such as tumors or trauma, IDH1 mutations have been proven to affect cellular metabolism. It has been reported that IDH-deficient cells are susceptible to mitochondrial metabolism inhibition [Citation10,Citation11].Most studies on neuroglobin’s role in brain diseases have been conducted in vitro using experimental animal models; hence in vivo assessments of neuroglobin in head trauma in people are currently scarce, with a lack of studies regarding the assessment of possible genetic determinants in TBI. Accordingly, the current study set out to determine the serum neuroglobin levels and the frequency of wild and mutant IDH1 genotypes in TBI patients, and to evaluate their likely relationship to the severity and outcome of these patients.
Materials and methods
The current research has been conducted on 30 patients with traumatic brain injuries recruited from the inpatient Neurosurgery Department who were admitted at South Valley University in Qena, Egypt, during the study period from January 1st, 2020, to December 30th, 2021. They were compared with thirty age and sex-matched healthy volunteers selected as controls. Written informed consent was obtained from each participant or the patient’s legally authorized representative if patients cannot give consent prior to the start of the study and after approval of the local Ethics Committee of the Faculty of Medicine, South Valley University, Egypt. Ethical approval code: SVU-MBC-91-15-4-2019.The study was carried out under the declaration of Helsinki. We adjusted the sample size to attain 80% power and a 5% confidence level of significance (type I error).Patients with other brain pathologies except for TBI were excluded. Patients with chronic systemic diseases or on chronic medications were excluded from the study.
Neurosurgical evaluation and data collection
Personal information, complaint, mode of trauma, the timing of the event, and history of any underlying medical conditions or past surgery were all documented during the history-taking process. The Glasgow coma scale (GCS), and the Revised trauma score (RTS) were used to perform the conscious level and the functional consequences following traumatic brain injury. Every patient had a series of tests, including a complete blood count, kidney function tests, and random blood sugar and serum electrolytes. Radiological examinations include a CT scan of the brain with a bone window. The GCS is a measure of the level of consciousness that is always assessed when a patient arrives. Patients with a GCS of 8 or less were brought to ICU.At the same time, the others were admitted to NeurosurgeryDepartment and monitored during their stay until release. The RTS, which comprises the GCS, systolic blood pressure, and respiratory rate, is employed in practices worldwide and may be obtained instantly. RTS is calculated by the formula below: RTS = 0.7326 * systolic blood pressure+0.2908 * respiratory rate+0.9368 * GCS [Citation12].
These scores were used to categorize the degree of severity of TBI among the patients who were enrolled in the study (mild, moderate, or severe TBI). Patients were evaluated radiologically by CT brain and clinically within 3 months from the moment of trauma. The following scales: The Extended Glasgow Outcome Scale (GOSe) and Disability Rating Scale (DRS) [Citation13], were used to categorize patients’ outcomes as favorable (moderate disability or good recovery) or unfavorable (severe disability or poor recovery or death). Radiological evaluation was conducted by CT brain at the time of hospital admission and within 3 months when indicated.
Biochemical assays and genetic analysis
Blood samples
About 6mLsof venous blood samples was withdrawn from each participant. For the included patients, the blood samples were withdrawn within the first 24 hours after injury. Blood samples were divided into two parts. The first part (3mLs) was evacuated into an EDTA-containing tube and stored at −800C until the genetic assays. While the second part (3 mLs) was placed into a serum gel separator tube and was allowed to be clotted at 37°C for 30 min, then centrifuged at 3500 rpm for 10 min. The separated sera were evacuated into I mL cryotubes and stored at −80°C until the biochemical assays of serum NGB.
Biochemical assays of serum neuroglobin
Serum NGB levels were determined using commercially available sandwich enzyme-linked immunosorbent assay (ELISA) kits [provided by Chongqing biopsies, China, Catalog No: BYEK3249] and a microplate ELISA reader (EMR 500, USA), as directed by the manufacturer [Citation14].
IDHI genetic analysis
Extraction of genomic DNA
Genomic DNA from whole EDTA blood samples was performed using G-spinTM total DNA extraction kit protocol (iNtRON Biotechnology, Inc, Korea) according to the manufacturer’s instructions. The isolated DNA was stored at −80°C for subsequent genetic analysis.
RFLP-PCR analysis of IDH1C.395 G˃A(R132H) mutation
Exon 4 mutations in codon 132 were detected in all participants using polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP). Mismatched primers designed by Meyer et al. [Citation15] were used for amplification. The used primers were designed to detect the presence or absence of IDH1 mutations in codon 132[c.395 G˃A(R132H)]. The forward primer 5’-TGGGTAAAACCT ATC ATCATC GAT-3’. The reverse primer 5′- TGT GTT GAG ATG GAC GCC TA −3′, as per previous protocols [Citation16,Citation17].
After mixing 12.5 μl from the PCR master mix solution (Catalog no. 25,028, iNtRON Biotechnology, Korea) with 1 μl forward primer, 1 μl reverse primer (both primers with a concentration of 10 nmol), 8.5 μl nuclease-free water, and 2 μl of extracted DNA with a total volume 25 μl were used in this test.The following PCR conditions were used: Initial denaturation was carried out at 94°C for 2 min, followed by 35 cycles of 30s each at 94°C, annealing for 30s at 55°C, and extension for 40s at 72°C with a final extension step at 72°C for 5 min, using Biometra thermal cycler (Serial no. 2603204, Biometra, Germany). The PCR products were 261-bp in size, using a 50-bp DNA ladder (Catalog No. 24,072, iNtRON Biotechnology, Korea), and were digested at 37◦C for 3 hours using (FastDigest PvuI supplied by Thermo Fisher Scientific).About 10 μl of the PCR reaction mixture was added to 2 μl of 10X buffer,2 μl of the restriction enzyme mixed with 18 μl nuclease-free water, then loaded in gel electrophoresis (serial no. 283BR11101,Bio-Rad-pac 300, Italy), using 2% agarose gel stained with 5 μL ethidium bromide).
A 50-bp DNA ladder was used to visualize DNA fragments under ultraviolet light (UV Transilluminator 2000, serial no. 642–1045, Bio-Rad, Italy). Detection of a single band (237 bp) indicates a wild genotype variant, while detection of two bands (237 bp and 261 bp) indicates a mutant genotype variant of IDH1gene ()).
Figure 1. Detection of IDH1 c.395 G˃A mutation by RFLP-PCR analysis. Numbers refer to lanes. Lane 1 shows 50 bp DNA ladder. [A] Lane 2 showed PCR amplification product (undigested) with band at 261 bp; Lanes 3–9 showed single band at 237bp corresponding to wild type IDH1. [B] Lane 2–7 showed mutant variant of IDH1 with appearance of two closely placed bands (261 bp and 237 bp).
![Figure 1. Detection of IDH1 c.395 G˃A mutation by RFLP-PCR analysis. Numbers refer to lanes. Lane 1 shows 50 bp DNA ladder. [A] Lane 2 showed PCR amplification product (undigested) with band at 261 bp; Lanes 3–9 showed single band at 237bp corresponding to wild type IDH1. [B] Lane 2–7 showed mutant variant of IDH1 with appearance of two closely placed bands (261 bp and 237 bp).](/cms/asset/4e0a4106-6f40-4b70-a934-43f41fde946b/teba_a_2104073_f0001_oc.jpg)
Statistical methods
For data entry and analysis, SPSS version 19 was utilized (Statistical Package for Social Science). We used numbers, percentages, mean, and standard deviation (SD) for parametric data, and median and inter-quartile range (IQR) for non-parametric data. The Chi-square test and Fisher exact test were employed to compare qualitative variables. Using 95% confidence intervals, the odds ratio (OR) was calculated (CI). The p-value is considered statistically significant when it is less than 0.05. The studied SNP is followed the Hardy Weinberg (HW) equation [Citation18,Citation19].
Results
Demographic and clinical data of the study groups
The present research included 60 participants categorized into 30 patients with various types of TBI (their mean age was 26.3 years ±16.52 SD). A total of 25(83.3%) patients were males, and the remaining 5(16.7%) were females), with male to female ratio was 5:1, and 30 unrelated healthy controls (their mean age was 32.06 years ±10.63 SD, 21(70%) of participants were males, and the remaining 9(30%) were females. There were insignificant differences between the included cases and controls regarding age and sex.
Regarding the severity of TBI, 17 patients with TBI were of a mild degree, 6 patients had moderate severity, and the remaining 7 patients had severe TBI. On follow-up, 26 patients had a favorable outcome, while the remaining 4 had unfavorable outcomes.
Circulating neuroglobin levels in patients with traumatic brain injury
There was a significantly higher median serum NGB value among patients with TBI compared with the healthy controls, with a significantly higher median value of serum NGB among patients with severe TBI compared to those with mild to moderate TBI (p˂0.05 for both).However, serum NGB was higher in TBI patients with the unfavorable outcome than in those with favorable outcomes.This difference was not reachinga significant level (p˃0.05) ().
Table 1. Circulating neuroglobin in patients with traumatic brain injury (TBI) in terms of severity and outcome.
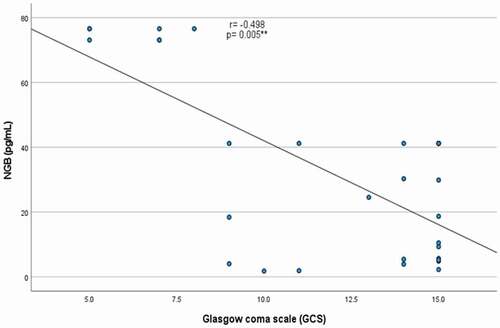
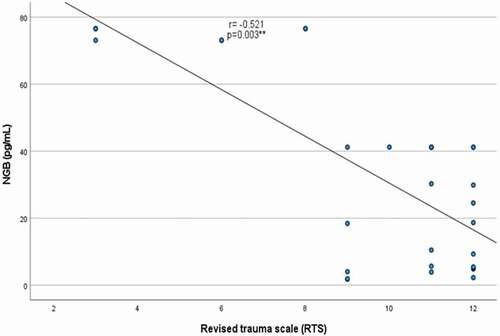
There were significant negative correlations between serum NGB levels with both GCS (r = −0.498, p = 0.005) and RTS (r = −0.521, p = 0.003) [], with a lack of significant correlation between NGB with either GOSe or DRS, p ˃0.05 for both (). The performance characteristics of serum NGB in predicting the severe TBI at cutoff point >41.18 pg/mL showed 53.85% sensitivity, 100% specificity, positive predictive value (PPV) = 100%, negative predictive value (NPP) = 73.9% with AUC = 0.715 (). While its performance in predicting unfavorable outcome at cutoff point >41.18 pg/mL demonstrated 50% sensitivity, 80.77% specificity, positive predictive value (PPV) = 28.6%, negative predictive value (NPP) = 91.3% with AUC = 0.582 ().
Figure 4. ROC Characteristic Curve for serum neuroglobin (pg/mL) in patients with traumatic brain injury. [A] For predicting the severity of TBI. [B] For predicting the outcome of TBI.
![Figure 4. ROC Characteristic Curve for serum neuroglobin (pg/mL) in patients with traumatic brain injury. [A] For predicting the severity of TBI. [B] For predicting the outcome of TBI.](/cms/asset/7dc93733-a702-4ad0-8824-5667f0f0d838/teba_a_2104073_f0004_oc.jpg)
Table 2. Correlation of serum neuroglobin with severity and outcome scores in patients with traumatic brain injury.
Frequency of IDH1 genetic variants among patients with traumatic brain injury in terms of severity and outcome
The frequencies of wild and mutant types of IDH1 [c.395 G˃A(R132H)] in TBI patients with mild to moderate severity were 100% and 69.6%, respectively, while in patients with severe TBI were 0% and 30.4%, respectively. Additionally, the frequencies of both wild and mutant types of IDH1 were 85.7% and 87% in TBI patients with favorable outcomes and 14.3% and 13%, respectively, among cases with unfavorable outcomes. The frequencies of both wild and mutant types of IDH1 showed non-significant differences in patients with TBI in terms of severity or outcome (p˃0.05), as presented in ().
Table 3. Genotypes frequencies of IDH1 gene in patients with traumatic brain injury in terms of severity.
Table 4. Genotypes frequencies of IDH1 gene in patients with traumatic brain injury in terms of outcome.
Discussion
Traumatic brain injury (TBI) is frequently misdiagnosed, misclassified, undertreated (particularly in its longer-term symptoms), and under-researched in comparison to its public health effects [Citation20].TBI is a common source of neurological morbidity worldwide, and neurologic sequelae can arise even in the case of a minor injury. The techniques used to facilitate the diagnostic and prognostic evaluation of TBI patients are currently limited [Citation21].The current study showed male predominance among patients with TBI. Several studies have found sex variations in TBI. They highlighted that the number of male patients is larger than female patients because males have a higher risk of harm [Citation22,Citation23].
Prehospital and point-of-care testing could be made easier with biochemical indicators extracted from serum or whole blood [Citation21].Our findings revealed significantly higher serum NGB levels among patients with TBI compared to the healthy controls with much higher serum levels among severe TBI than those with mild to moderate injury and better characteristic performance of serum NGB in predicting the severity of TBI (AUC = 0.715) than in predicting the outcome (AUC = 0.582).Additionally, there were significant negative correlations between serum NGB with both GCS and RTS among patients with TBI. Chen et al. [Citation24] reported similar findings. After traumatic brain injury, NGB may act as an oxygen sensor, modulating signal pathways, weakening oxidative stress reactions, and maintaining mitochondrial function. Many hazardous chemicals, active free oxygen, and nitrogen radicals accumulate in the brain after hypoxic-ischemic brain injury and NGB can clear up these free radicals [Citation25].Shang et al. [Citation26] used a rat focal brain damage model to find elevated NGB levels at 12 and 36 hours after the lesion.
Although predicting the outcome of a TBI is challenging, it is critical for acute management, family counseling, and the provision of rehabilitation services. To predict outcomes after a TBI, serum biomarkers may be effective alone or in combination with clinical factors [Citation27]. Our findings revealed higher serum NGB levels among TBI patients with unfavorable outcomes compared to those with favorable outcomes but did not reach a significant level that could be attributed to the relatively small sample size. Chen et al. [Citation24] reported a strong link between serum NGB and the outcome in individuals with TBI.
Metabolic pathways are essential mechanisms that allow cells to develop and survive. IDH1 is involved in the tricarboxylic acid (TCA) cycle’s biosynthesis of core metabolites as well as the production of cellular NADPH. Because the brain is the primary source of NADPH, IDH1 is extremely crucial. Hypoxia-inducible factor-1α (HIF-1α) levels can also be elevated due to IDH mutations [Citation28–30].Although the prevalence of IDH1 gene mutations has been investigated extensively in numerous ethnic groups worldwide, its role in brain cancers has already been reported [Citation30,Citation31].There was no information about the IDH1 gene’s involvement or role in TBI among Egyptian patients till now. The current study’s findings found no link between IDH1 genetic variations and the severity or outcome of TBI in an Egyptian population sample. Further studies are needed to confirm the current findings, as there were no previous studies on the potential contribution of IDH1 gene mutation in brain cell response to severe head trauma or on using IDH1 mutation as a genetic determinant of prognosis in TBI patients.
Conclusion
Initial serum neuroglobin assay is an essential biomarker to assess the TBI outcome’s severity and extent. The current study reported a lack of a significant role of mutation in IDH1 to determine the severity or predict the outcome in patients with TBI.
Authors’ contributions
Study concept and design: MHH and ARH; Neurosurgical assessments and data collection: ARH; Blood sampling and Biochemical assays: MHH and SAE-S; Genetic assays and analysis: MHH and SAE-S; statistical analysis: MHH, ARH, SAE-S, BEME, THS and OA; Literature research: MHH, BEME, THS, SAE-S, ARH and OA; First manuscript drafting: MHH; all authors approved the final version of the manuscript.
Study’s limitations
The study’s main limitations were the relatively small sample size and lack of serial neuroglobin assays.
Ethics approval and consent to participate
The study was approved by the local Ethics Committee of Medical Research of the Faculty of Medicine, South Valley University, Qena, Egypt (Ethical approval code: SVU-MBC-91-15-4-2019), and was conducted per the Declaration of Helsinki. Informed written consent was obtained from every participant or the patient’s legally authorized representative if patients could not give consent prior to the start of the study.
Availability of data and materials
The datasets used and analyzed in this study are available upon reasonable request.
Acknowledgments
We acknowledged the healthy volunteers involved in this research who were part of our project entitled “ Study of some biochemical and genetic markers in traumatic neuro-injuries and neurological tumors”. The statistical data analysis of the included controls was and will be shared in various publications belonging to this project.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Baez E, Echeverria V, Cabezas R, et al. Protection by neuroglobin expression in brain pathologies. Front Neurol. 2016;7:146.
- Gotting M, Nikinmaa M. More than hemoglobin – the unexpected diversity of globins in vertebrate red blood cells. Physiol Rep. 2015;3:e12284.
- Lee VY, McClintock DS, Santore MT, et al. Hypoxia sensitizes cells to nitric oxide-induced apoptosis. J Biol Chem. 2002;277:16067–16074.
- Hankeln T, Ebner B, Fuchs C, et al. Neuroglobin and cytoglobin in search of their role in the vertebrate globin family. J InorgBiochem. 2005;99. 110–119.
- Fuchs C, Burmester T, Hankeln T. The amphibian globin gene repertoire as revealed by the Xenopus genome. Cytogenet Genome Res. 2006;112:296–306.
- De Marinis E, Fiocchetti M, Acconcia F, et al. Neuroglobin upregulation induced by 17β-estradiol sequesters cytocrome c in the mitochondria preventing H2O2-induced apoptosis of neuroblastoma cells. Cell Death Dis. 2013;4:e508.
- Wystub S, Laufs T, Schmidt M, et al. Localization of neuroglobin protein in the mouse brain. Neurosci Lett. 2003;346:114–116.
- Parsons DW, Jones S, Zhang X, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321(5897):1807–1812.
- Zhao S, Yu Z, Zhao G, et al. Neuroglobin-overexpression reduces traumatic brain lesion size in mice. BMC Neurosci. 2012;13:67.
- Grassian AR, Parker SJ, Davidson SM, et al. IDH1 mutations alter citric acid cycle metabolism and increase dependence on oxidative mitochondrial metabolism. Cancer Res. 2014;74:3317–3331.
- Cuyàs E, Fernández-Arroyo S, Corominas-Faja B, et al. Oncometabolic mutation IDH1 R132H confers a metformin-hypersensitive phenotype. Oncotarget. 2015;6(14):12279–12296.
- Gilpin DA, Nelson PG. Revised trauma score: a triage tool in the accident and emergency department. Injury. 1991;22(1):35–37.
- Scranton J, Fogel ML, Erdman WJ 2nd. Evaluation of functional levels of patients during and following rehabilitation. Arch Phys Med Rehabil. 1970;51:1–21.
- Abuhamdah S, Saleem TH, Elsadek BE, et al. Circulating ubiquitin carboxyl terminal hydrolase L1 and neuroglobin levels in traumatic spinal cord injuries: relation to severity and outcomes. Int J Gen Med. 2022;15:5795–5805.
- Meyer J, Pusch S, Balss J, et al. PCR- and restriction endonuclease-based detection of IDH1 mutations. Brain Pathol. 2010;20(2):298–300.
- Elsayed GM, Nassar HR, Zaher A, et al. Prognostic value of IDH1 mutations identified with PCR-RFLP assay in acute myeloid leukemia patients. J Egypt Natl Canc Inst. 2014;26(1):43–49.
- Mohamed Yusoff AA, Zulfakhar FN, Sul’ain MD, et al. Association of the IDH1 C.395G>A (R132H) mutation with histological type in malay brain tumors. Asian Pac J Cancer Prev. 2016;17(12):5195–5201.
- Hardy GH. Mendelian proportions in a mixed population. Science. 1908;28:49–50.
- Stark AE. A clarification of the Hardy-Weinberg law. Genetics. 2006;174:1695–1697.
- Bowman K, Matney C, Berwick DM. Improving traumatic brain injury care and research: a report from the national academies of sciences, engineering, and medicine. JAMA. 2022;327(5):419–420.
- Sharma R, Laskowitz DT. Biomarkers in traumatic brain injury. Curr Neurol Neurosci Rep. 2012;12:560–569.
- Leitgeb J, Mauritz W, Brazinova A, et al. Effects of gender on outcomes after traumatic brain injury. J Trauma. 2011;71(6):1620–1626.
- Egea-Guerrero JJ, Murillo-Cabezas F, Gordillo-Escobar E, et al. S100B protein may detect brain death development after severe traumatic brain injury. J Neurotrauma. 2013;30(20):1762–1769.
- Chen H, Cao H-L, Chen S-W, et al. Neuroglobin and Nogo-a as biomarkers for the severity and prognosis of traumatic brain injury. Biomarkers. 2015;20(6–7):495–501.
- Ding C, Kang D, Chen P, et al. Early stage neuroglobin level as a predictor of delayed cerebral ischemia in patients with aneurysmal subarachnoid hemorrhage. Brain Behav. 2020;10(3):e01547.
- Shang A, Zhou D, Wang L, et al. Increased neuroglobin levels in the cerebral cortex and serum after ischemia–reperfusion insults. Brain Res. 2006;1078(1):219–226.
- Berger RP. The use of serum biomarkers to predict outcome after traumatic brain injury in adults and children. J Head Trauma Rehabil. 2006;21(4):315–333
- Bleeker FE, Atai NA, Lamba S, et al. The prognostic IDH1(R132) mutation is associated with reduced NADP+-dependent IDH activity in glioblastoma. Acta Neuropathol. 2010;119(4):487–494.
- Huang J, Yu J, Tu L, et al. Isocitrate dehydrogenase mutations in glioma: from basic discovery to therapeutics development. Front Oncol. 2019;9:506.
- Alzial G, Renoult O, Paris F, et al. Wild-type isocitrate dehydrogenase under the spotlight in glioblastoma. Oncogene. 2022;41:613–621.
- Turcan S, Rohle D, Goenka A, et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483:479–483.