ABSTRACT
Methicillin Resistant Staphylococcus aureus (MRSA) is a main cause of severe human infections. The aim of this study was to detect chlorhexidine resistance of MRSA by phenotypic and genotypic methods. Fifty MRSA strains were isolated from different hospitals of Mansoura University over a period of one year from December 2021 till November 2022. The antimicrobial susceptibility test was conducted by the disk diffusion method, except for vancomycin which was detected by minimum inhibitory concentration (MIC) determination using microdilution broth method. The MIC towards chlorhexidine (CHG) was determined by microdilution broth method. Qac genes (qacA/B1, qacA/B2, smr, and norA) were detected by Polymerase Chain Reaction (PCR). Transmission electron microscope was used for determination of cell-wall thickness of chlorhexidine susceptible and resistant strains. The results exhibited that the greatest resistance of the isolates was towards Penicillin (100%) and Ceftazidime (100%) and the greatest susceptibility was towards Vancomycin (72%). Decreased susceptibility to CHG at MIC ≥4 mg/L was in 24 isolates (48%). The highest detected genes were smr (100%), norA (50%), and qacA/B2 (45.8%), with a low prevalence of qacA/B1 (4.16%). It was found that the MRSA strain with high resistance to chlorhexidine had more thickened cell wall than that of the susceptible strain.
Introduction
Staphylococcus aureus strains have spread worldwide and are connected with adverse patient outcomes, longer hospital stays, and a major source of dangerous human infections (nosocomial infections) in both community and healthcare settings [Citation1]. The success of S. aureus as a human pathogen in health care is highly associated with its multidrug resistance [Citation2]. Starting in the 1950s, S. aureus has evolved resistance to mostly all categories of antibiotics. A decade later, this bacterium has been able to resist methicillin because of PBP2a protein [Citation3].
While inactivation and target modification are recognized as antiseptic resistance mechanisms, they are rather infrequent. Resistance frequently results from cellular modifications that reduce the concentrated antibiotics, which include cell wall alteration such as increasing its thickness which limits uptake of biocides or expression of efflux mechanisms [Citation4–7].
The use of biocides (such as antiseptics and disinfectants) clinically plays a major role in limiting nosocomial infections. In hospitals and other healthcare facilities, a wide range of biocides, including quaternary ammonium compounds (QACs) like benzalkonium chloride and divalent cations like chlorhexidine digluconate, are frequently utilized [Citation8].
Up to our knowledge, this is the first time to describe the mode of cell-wall thickening as a possible mechanism of chlorhexidine resistance in MRSA strains isolated from Mansoura University Hospitals.
The activity of multidrug efflux pumps is a known mechanism of resistance to antibiotics and plays a main role in multi-drug resistance (MDR) phenotypes [Citation6]. In S. aureus, various multidrug efflux pumps have been identified and were encoded by genes located on chromosomes (norA, norB, norC, mepA, mdeA, sdrM and sepA) and genes located on plasmids (e.g. qacA/B, smr) [Citation2].
Usually, reduced susceptibility to either antibiotics or biocides can be conferred by chromosomally encoded multidrug efflux pumps, while the plasmid-encoded pumps can contribute to reduced susceptibility to biocides only [Citation6]. The multidrug efflux pump QacA and the closely related QacB are two examples which use the proton-motive force to expel a wide range of monovalent and divalent cationic compounds from MRSA [Citation9].
Long-term use of antiseptics containing QACs may increase prevalence of MRSA isolates which harbor qac genes [Citation10]. This study aimed to detect antiseptic resistance of MRSA using phenotypic and genotypic methods.
Material and method
Sample collection and culture conditions
Staphylococcus aureus strains were collected from Mansoura University Hospitals and grown on Blood agar for blood and sputum samples, Mannitol salt agar (MSA) for nasal swab samples and CLED agar for urine samples and incubated aerobically at 37°C overnight. S. aureus was recognized using traditional techniques (colony morphology, Gram stain, catalase activity, growth on mannitol salt agar, DNase test and tube coagulase) [Citation11]. All media included in this study were prepared according to instructions of manufacturer, and all tests were quality controlled using a positive and a negative control for each test.
Identification of MRSA by Cefoxitin Disc diffusion method
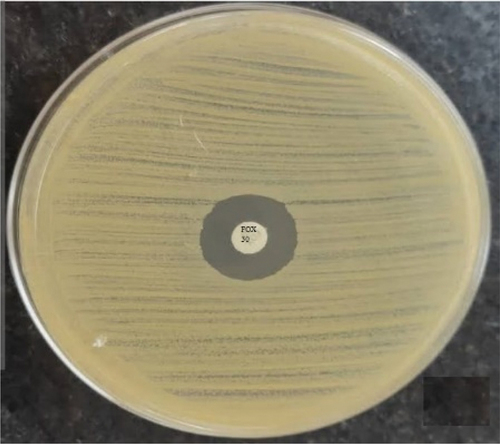
The isolates were confirmed to be methicillin resistant using a cefoxitin disc diffusion test with a 30 µg disc. According to the Clinical and Laboratory Standards Institute, zone sizes were interpreted (resistant if ≤21 mm) [Citation12].
Antibiotic susceptibility testing
The susceptibility of the isolates was determined against nine antibiotic discs by Kirby–Bauer disc diffusion technique using Mueller–Hinton agar [Citation13] with the exception of vancomycin, which was detected by MIC determination using microdilution broth method, according to the Clinical and Laboratory Standards Institute (CLSI) [Citation14].
The antibiotics included: Penicillin, Ceftazidime, Azithromycin, Erythromycin, Tetracycline, Ciprofloxacin, Amikacin and Clindamycin. These antibiotic discs were purchased from Oxoid, England.
Detection of minimum inhibitory concentration (MIC) of the strains toward chlorhexidine by (microdilution broth method)
The MIC of the study strains for chlorhexidine (CHG) (Sigma) was measured using the microdilution broth method, following the recommendations of the Clinical and Laboratory Standards Institute (CLSI) [Citation14]. Microtitre plates were used and each plate had a positive control which did not contain the antiseptic solution (chlorhexidine) and a negative control which did not contain tested bacterial suspension (MRSA).
Detection of quaternary ammonium compounds (QACs) resistance genes in MRSA isolates by multiplex PCR
DNA extraction for PCR
The DNA extraction was carried out using the modified colony direct method [Citation15]. Briefly, one colony on an agar plate was lightly touched with a toothpick and mixed with 100 µl of sterile distilled water and heated for 10 min at 99°C. Centrifugation was done at 30,000 g for 1 min then 3 µl of supernatant were used as PCR templates. The PCR reaction was performed in a volume of 25 μl.
PCR amplification
PCR was used to amplify amplicons which correspond to the QACs resistant genes (qacA/B, smr and norA) with the primers illustrated in . The initial denaturation step was performed at 96°C for 3 min, 25 cycles of 94°C for 20 s, 53°C for 20 s and 72°C for 20 s, and the final extension step at 72°C for 5 min [Citation16–18].
Table 1. Primer sequences and their product size.
PCR products were analyzed on 1.5% (w/v) agarose gels by electrophoresis (Promega). Ethidium bromide (Promega) was added to the agarose gel for staining bands. UV illumination was used to visualize the bands.
Transmission electron microscope examination for determination of cell-wall thickness of chlorhexidine susceptible and resistant strains
MRSA cells were incubated with sub-inhibitory concentration of CHG (32 mg/L for the resistant strain and 1 mg/L for the susceptible strain) overnight at 37°C. Then the cells were collected and washed twice with PBS. For examination with a transmission electron microscope, the cells were centrifuged into a pellet, and their fixation was done with 2.5% of glutaraldehyde, then 1% Osmic acid. Dehydration of the samples was done by passing in an ethanol series (50%, 70%, 90% and 100%), followed by acetone. Then Spurr’s Epon resin was used for embedding of the samples. With the RMC PT-XL Ultra microtome, ultrathin sections were produced, then examined using JEOL JEM-2100EXII transmission electron microscope at 80 kV. The thickness of cell-wall was measured for one chlorhexidine-susceptible strain with MIC of 2 mg/L and one resistant strain with MIC of 64 mg/L because of the high cost of sample examination using transmission electron microscope [Citation19].
Ethical approval
The Institutional Review Board (IRB) of the Medical Research Ethics Committee at Mansoura Faculty of Medicine (University of Mansoura) approved the study protocol (Code Number: MDP.21.11.85) in 22 November 2021.
Results
Collection of samples and isolation of S. aureus isolates
In this study, 50 clinical S. aureus isolates were collected from various hospitals of Mansoura University. The isolates were identified as S. aureus as shown in .
Figure 1. Identification of isolates as S. aureus, A) Microscopic examination of Gram-stained film of S. aureus (100x magnification by oil immersion lens), B) Growth of S. aureus on Blood agar plates, C) Coagulase positive S. aureus with negative control and D) Growth of S. aureus on MSA plate.
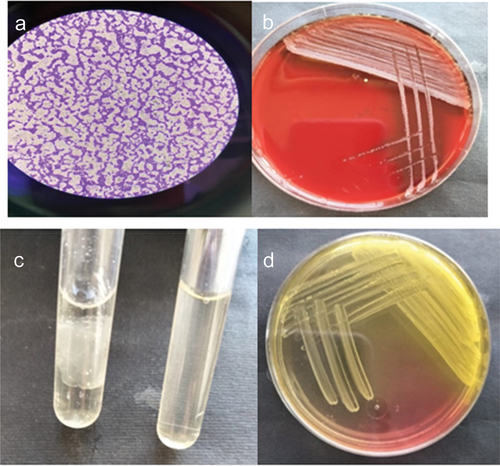
Each sample was cultured in a specific medium. The commonest samples which gave positive bacterial growth were the nasal swab samples as shown in .
Table 2. Number and percentage of positive isolates for S. aureus collected from different samples during the period of the study.
The distribution of S. aureus among Mansoura university hospitals is shown in . It shows that 48% of S. aureus isolates were recovered from the Emergency hospital, 22% from the Pediatric hospital, 12% from the Surgery department in Mansoura University Hospital (MUH), 10% from the Specialized Medical hospital (SMH) and 8% from Oncology hospital. S. aureus were most prevalent in Emergency and least in Oncology.
Table 3. Distribution of S. aureus strains isolated from patients among different Mansoura university hospitals.
Identification of MRSA by Cefoxitin Disc diffusion method
Fifty S. aureus isolates were identified to be methicillin resistant using the cefoxitin disc diffusion test. All the isolates were cefoxitin resistant as shown in .
Antibiotics susceptibility testing
illustrates the antimicrobial susceptibility of 50 MRSA isolates to nine antibiotics from eight different antimicrobial groups. The greatest resistance was against Penicillin (100%) and Ceftazidime (100%) and the greatest susceptibility was exhibited to Vancomycin (72%).
Table 4. Antimicrobial susceptibility test of 50 MRSA isolates.
Determination of minimum inhibitory concentration (MIC) of the strains toward chlorhexidine
Decreased sensitivity to CHG at MIC ≥4 mg/L was found in 48% of isolates (24/50). The prevalence of CHG MICs between the 50 MRSA isolates was summarized in . Reduced susceptibility to CHG was most common with MIC concentration of 16 mg/L.
Table 5. The distribution of CHG MICs among 50 MRSA isolates.
Detection of quaternary ammonium compounds (QACs) resistance genes in MRSA isolates by multiplex PCR
QACs resistance genes were found in the 24 MRSA isolates with decreased susceptibility to chlorhexidine (CHG) at CHG MIC ≥4 mg/L. The most commonly found genes were smr in 24 isolates (100%), norA in 12 isolates (50%) and qacA/B2 in 11 isolates (45.8%), with a low prevalence of qacA/B1 (1 isolate; 4.16%).
The incidence of two genes together was detected in two isolates (4%), which include qacA/B1+ smr in one isolate (4.16%) and norA+ smr in one isolate (4.16%); moreover, 11 isolates (45.8%) had three genes (qacA/B2 + norA + smr) ( and ). There was a moderately positive correlation between the presence of QAC genes and higher CHG MIC associated with qacA/B2 and norA genes ().
Figure 3. Agarose gel electrophoresis of the Multiplex PCR for the qacA/B1, qacA/B2, smr and norA genes for 24 CHG resistant MRSA strains. M is the ladder DNA.
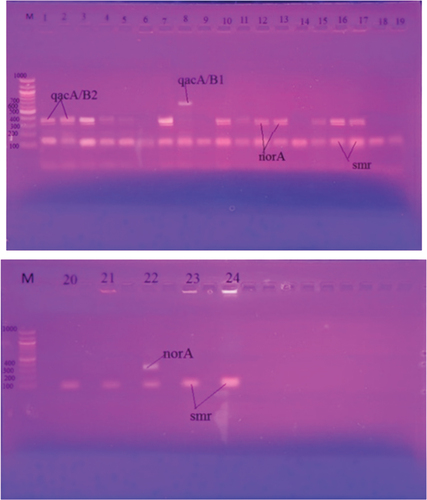
Table 6. Frequency of QACs resistance genes detected by Multiplex PCR among MRSA isolates with MIC ≥4 mg/L (n = 24).
Table 7: Relationship between quaternary ammonium compound (QAC) resistance genes and minimum inhibitory concentrations (MICs) of chlorhexidine (CHG) (n = 24).
Transmission electron microscope examination for determination of cell-wall thickness of chlorhexidine susceptible and resistant strains
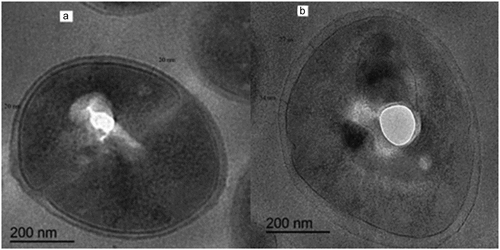
It was found in thin-section micrographs of MRSA cells exposed to sub inhibitory concentration of chlorhexidine (32 mg/L) which showed high resistance to chlorhexidine as they had MIC of chlorhexidine of 64 mg/L had more thickened cell wall (30 nm) than that of the susceptible strain (20 nm) which had MIC of 2 mg/L and exposed to sub inhibitory concentration of chlorhexidine (1 mg/L) as shown in .
Discussion
qac genes, which are antiseptic resistant genes, are so named because of their main resistance action against QACs. In our study, 55% of the MRSA cases were female population. This was similar to the findings of Duza [Citation20].
The antibiotic susceptibility test of MRSA isolates in this study exhibited a high resistance against Penicillin (100%) and Ceftazidime (100%). This is consistent with the results of AlKhazraji et al. [Citation21]. The resistance to Erythromycin, Azithromycin, Tetracycline and Ciprofloxacin were 92%, 86%, 80% and 72%, respectively. These findings were near the findings of Rahimi [Citation22].
The antiseptic resistant genes have been broadly dispersed, particularly in staphylococcal species [Citation23].
According to various geographic regions, the frequency of these genes differs with a vast range varying from 10% up to 80% [Citation23–26]. The position of QACs resistant genes reported in different MRSA clones may impact their incidence rates, as these genes are commonly carried on plasmids causing their quick transfer, while in some MRSA clones, another genes carried on the chromosome control resistance to QACs [Citation27].
In this study, a reduction in susceptibility to chlorhexidine was detected in 48% of isolates detected by CHG MIC ≥4 mg/L. Chlorhexidine resistance genes were found in the 24 MRSA isolates with decreased sensitivity to CHG. The most abundantly observed genes were smr in 24 isolates (100%), norA in 12 isolates (50%) and qacA/B2 in 11 isolates (45.8%), with a low prevalence of qacA/B1 (1 isolate; 4.16%). It was noticed that the 12 MRSA strains with QAC genes (qacA/B2, norA and smr genes) combined had higher CHG MICs like the findings of Lin et al [Citation28].
In our study, smr was the predominant resistance gene, like the findings of Nakipoğlu et al [Citation29] and Vali et al [Citation26]. In contrast to our study, qacA/B was the predominant resistance gene in previous studies [Citation30].
Also the occurrence of qac and smr became more frequently reported than previously [Citation31]. Although qacA/B causes resistance to a wider range of biocides than smr [Citation32].
When the antiseptic susceptibility to a QAC known as acriflavine and gene distributions of MRSA isolates were investigated in Japan in 1992, 71.4% of the MRSA showed resistance to this antiseptic. Also, qacA/B and smr were determined in 10.2% and 20.4% of the isolates, respectively. However, in 1999, qacA/B and smr were found in 47.9% and 3%, respectively [Citation33].
In Europe between 1997 and 1999, qacA/B and smr were detected at 62.6% and 6.4%, respectively, in MRSA strains [Citation34].
These findings strongly imply that antiseptic resistant genes are disseminated rapidly in MRSA. Consequently, monitoring MRSA that is resistant to antiseptics could reveal crucial details about how to prevent nosocomial infection.
It was found in thin-section micrographs of MRSA cells exposed to sub inhibitory concentration of chlorhexidine that the MRSA strain that showed high resistance to chlorhexidine had a more thickened cell wall than that of the susceptible strain. These findings suggest that MRSA’s resistance to chlorhexidine may be caused by increasing the thickness of cell wall. This was like the results of Kawai et al. [Citation19] as they found that cell-wall thickness may be responsible for acriflavine (a quaternary ammonium compound) resistance in MRSA.
Conclusion
These findings strongly imply that antiseptic resistant genes are disseminated rapidly in MRSA. Consequently, monitoring MRSA that is resistant to antiseptics could reveal crucial details about how to prevent nosocomial infection. Also, the findings of this study highly suggest that increasing cell wall thickness of MRSA may be one of the causes of its resistance to chlorhexidine.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- De Lencastre H, Oliveira D, Tomasz A. Antibiotic resistant Staphylococcus aureus: a paradigm of adaptive power. Curr Opin Microbiol. 2007;10(5):428–435. doi: 10.1016/j.mib.2007.08.003
- Lee J-H, Park J-H, Kim J-A, et al. Low concentrations of honey reduce biofilm formation, quorum sensing, and virulence in Escherichia coli O157: h7. Biofouling. 2011;27(10):1095–1104. doi: 10.1080/08927014.2011.633704
- Cooper R, Jenkins L, Rowlands R. Inhibition of biofilms through the use of manuka honey. Wounds UK. 2011;7(1):24–32.
- Gilbert P, McBain AJ. Potential impact of increased use of biocides in consumer products on prevalence of antibiotic resistance. Clinical Microbiology Reviews. 2003;16(2):189–208. doi: 10.1128/CMR.16.2.189-208.2003
- Piddock LJ. Clinically relevant chromosomally encoded multidrug resistance efflux pumps in bacteria. Clinical Microbiology Reviews. 2006;19(2):382–402. doi: 10.1128/CMR.19.2.382-402.2006
- Poole K. Efflux pumps as antimicrobial resistance mechanisms. Ann Med. 2007;39(3):162–176. doi: 10.1080/07853890701195262
- Russell A. Biocide use and antibiotic resistance: the relevance of laboratory findings to clinical and environmental situations. Lancet Infect Dis. 2003;3(12):794–803. doi: 10.1016/S1473-3099(03)00833-8
- Noguchi N, Suwa J, Narui K, et al. Susceptibilities to antiseptic agents and distribution of antiseptic-resistance genes qacA/B and smr of methicillin-resistant Staphylococcus aureus isolated in Asia during 1998 and 1999. J Med Microbiol. 2005;54(6):557–565. doi: 10.1099/jmm.0.45902-0
- Brown MH, Skurray RA. Staphylococcal multidrug efflux protein QacA. J Mol Microbiol Biotechnol. 2001;3(2):163–170.
- Shi GS, Boost M, Cho P. Prevalence of antiseptic-resistance genes in staphylococci isolated from orthokeratology lens and spectacle wearers in Hong Kong. Investig. Ophthalmol. Vis. Sci. 2015;56(5):3069–3074.
- Forbes BA, Sahm DF, Weissfeld AS. Diagnostic microbiology. Missouri: Mosby St Louis; 2007.
- Humphries R, Bobenchik AM, Hindler JA, et al. Overview of changes to the clinical and laboratory standards institute performance standards for antimicrobial susceptibility testing, M100, 31st edition. J Clin Microbiol. 2021;59(12):e00213–21. doi: 10.1128/JCM.00213-21
- Bauer A. Antibiotic susceptibility testing by a standardized single disc method. Am J Clin Pathol. 1966;45(3):149–158. doi: 10.1093/ajcp/45.4_ts.493
- MA, WIKLER. “Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically: approved standard. Clsi (Nccls).” 2006;26: M7-A7.
- Tsuchizaki N, Ishikawa J, Hotta K. [Colony PCR for rapid detection of antibiotic resistance genes in MRSA and enterococci]. Jpn J Antibiot. 2000;53(6):422–429.
- Huet AA, Raygada JL, Mendiratta K, et al. Multidrug efflux pump overexpression in Staphylococcus aureus after single and multiple in vitro exposures to biocides and dyes. Microbiology. 2008;154(10):3144–3153. doi: 10.1099/mic.0.2008/021188-0
- Zmantar T, Kouidhi B, Miladi H, et al. Detection of macrolide and disinfectant resistance genes in clinical Staphylococcus aureus and coagulase-negative staphylococci. BMC Res Notes. 2011;4(1):1–9. doi: 10.1186/1756-0500-4-453
- Heir E, Sundheim G, Holck A. Resistance to quaternary ammonium compounds in Staphylococcus spp. isolated from the food industry and nucleotide sequence of the resistance plasmid pST827. J Appl Bacteriol. 1995;79(2):149–156. doi: 10.1111/j.1365-2672.1995.tb00928.x
- Kawai M, Yamada S, Ishidoshiro A, et al. Cell-wall thickness: possible mechanism of acriflavine resistance in meticillin-resistant Staphylococcus aureus. J Med Microbiol. 2009;58(3):331–336. doi: 10.1099/jmm.0.004184-0
- Duza, SS. Evaluation of virulence properties of staphylococcus aureus isolated from a tertiary level hospital of Dhaka city. PhD Thesis. University of Dhaka; 2021.
- AlKhazraji NZ, Al Jubouri AS, Al Ma MF. Detection of antiseptic resistant genes and biofilm formation in multidrug resistant staphylococcus aureus in Baghdad hospitals. Iraqi J Biotechnol. 2020;19(2):21–27.
- Rahimi F. Characterization of resistance to aminoglycosides in methicillin-resistant Staphylococcus aureus strains isolated from a tertiary care hospital in Tehran, Iran. Jundishapur J Microbiol. 2016;9(1). doi: 10.5812/jjm.29237
- Jaglic Z, Cervinkova D. Genetic basis of resistance to quaternary ammonium compounds–the qac genes and their role: a review. Vet Med. 2012;57(6):275–281. doi: 10.17221/6013-VETMED
- Miyazaki NHT, Abreu AO, Marin VA, Rezende CA, Moraes MT, Villas Bôas MH. The presence of qacA/B gene in Brazilian methicillin-resistant Staphylococcus aureus. Memorias do Instituto Oswaldo Cruz. 2007;102(4):539–540. doi: 10.1590/S0074-02762007000400018
- Smith K, Gemmell CG, Hunter IS. The association between biocide tolerance and the presence or absence of qac genes among hospital-acquired and community-acquired MRSA isolates. J Antimicrob Chemother. 2008;61(1):78–84. doi: 10.1093/jac/dkm395
- Vali L, Davies SE, Lai LLG, et al. Frequency of biocide resistance genes, antibiotic resistance and the effect of chlorhexidine exposure on clinical methicillin-resistant Staphylococcus aureus isolates. J Antimicrob Chemother. 2008;61(3):524–532. doi: 10.1093/jac/dkm520
- Pulido RP, Gálvez A. Biocide tolerance in bacteria. Int J Food Microbiol. 2013;162(1):13–25. doi: 10.1016/j.ijfoodmicro.2012.12.028
- Lin K-H, Lin C-Y, Huang C-C, et al. Differentiation of qacA and qacB using high-resolution melt curve analysis, and both qacA and qacB but not qacC or norA types increase chlorhexidine minimal inhibitory concentrations in Staphylococcus aureus isolates. J Microbiol Immunol Infect. 2020;53(6):900–908. doi: 10.1016/j.jmii.2020.09.006
- Nakipoğlu Y, Iğnak S, Gürler N, et al. [The prevalence of antiseptic resistance genes (qacA/B and smr) and antibiotic resistance in clinical Staphylococcus aureus strains]. Mikrobiyoloji bulteni. 2012;46(2):180–189.
- Taheri N, Ardebili A, Amouzandeh-Nobaveh A, et al. Frequency of antiseptic resistance among Staphylococcus aureus and coagulase-negative staphylococci isolated from a university hospital in central Iran. Oman Med J. 2016;31(6):426–432. doi: 10.5001/omj.2016.86
- Ghasemzadeh-Moghaddam H, Azimian A, Bayani G, Dashti V, Nojoomi S, Shirazi N, Solati A, Van Belkum A. High prevalence and expression of antiseptic resistance genes among infectious t037/ST239 methicillin-resistant Staphylococcus aureus (MRSA) strains in North Khorasan Province. Iran. 2022;25(6):775.
- Dashtbani-Roozbehani A, Brown MHJA. Efflux pump mediated antimicrobial resistance by staphylococci in health-related environments: challenges and the quest for inhibition. Antibiotics. 2021;10(12):1502. doi: 10.3390/antibiotics10121502
- Noguchi N, Hase M, Kitta M, et al. Antiseptic susceptibility and distribution of antiseptic-resistance genes in methicillin-resistant Staphylococcus aureus. FEMS Microbiology Letters. 1999;172(2):247–253. doi: 10.1111/j.1574-6968.1999.tb13475.x
- Mayer S, Boos M, Beyer A, Fluit, AC, Schmitz FJ. Distribution of the antiseptic resistance genes qacA, qacB and qacC in 497 methicillin-resistant and-susceptible European isolates of Staphylococcus aureus. J Antimicrob Chemother. 2001;47(6):896–897. doi: 10.1093/jac/47.6.896