ABSTRACT
Sirtuin1 (SIRT1) is an epigenetic modulator that belongs to sirtuins family and participates in many physiologic reactions, as genomic stabilization, apoptosis, proliferation, and inflammation. STAT4 (signal transducer and activator of transcription 4) gene is a component of JAK-STAT signaling pathway, which plays key role in activating cellular-mediated immune responses. The present study aimed to investigate the association between SIRT1 rs12778366 SNP, expression level of STAT4 gene and susceptibility to COVID-19 infections as well as their correlation to clinicopathological data.
The present study included 100 ICU patients with severe COVID-19 infection and 100 age- and sex-matched healthy controls. DNA was extracted. Genotyping of SNP in SIRT1 (rs12778366) was performed, Total RNA was extracted from PBMCs. Reverse transcription was done. STAT4 gene expression levels were evaluated with GAPDH as internal control using real-time PCR. We found a significantly higher frequency of ‘C’ allele and C/T genotype in case vs. control. The association was of low strength (φ = 0.105 for alleles, and 0.154 for genotypes). This was associated with higher expression of STAT4 gene (P < 0.001) and increased tendency for lower respiratory complications. Median STAT4 (FC) (Median = 0.18, 95% CI = 0.15–0.27) was lower than normal control value of 1.0. This difference was statistically significant (Hodges-Lehmann location estimator = 0.217, P < 0.001). SIRT1 polymorphism and decreased expression of STAT4 gene are associated with increased susceptibility to COVID-19 infection and correlate to its phenotypic manifestations.
Introduction
Coronavirus disease 2019 (COVID19), is a contagious disease caused by the severe acute respiratory syndrome-coronavirus 2 (SARS-CoV2). It mainly affects the lungs with the severity ranging from mild upper respiratory tract symptoms to severe respiratory distress. However, other organs including the heart, skin, kidneys, and brain may be affected. This indicates a massive proinflammatory response, that comprises cytokines and prothrombotic factors [Citation1,Citation2].
Sirtuins are a family of nicotine adenine dinucleotide (+)-dependent histone deacetylases, working as signaling proteins. The first sirtuin was identified in yeast and named Silent information regulator 2 (SIR2). SIR2 is an antiaging gene which encodes a protein with NAD + dependent histone deacetylase activity [Citation3]. There are seven homologs of SIR2 in mammals [Citation4] deeply involved in gene regulation, genome stability maintenance, apoptosis, autophagy, senescence, proliferation, aging, neuroprotection and tumorigenesis. It also has a key role in the epigenetic regulation of tissue homeostasis and many diseases by deacetylating both histone and non-histone targets [Citation5,Citation6].
SIRT1 is a highly conserved NAD ± dependent deacetylase belonging to the sirtuin family. SIRT1 is a multifunctional protein that plays a central role in various pathways.
During inflammation, It plays an essential role in the regulation of JAK/STAT by controlling HMGB1 translocation from the nucleus to the cytoplasm [Citation7]. However, the role of SIRT1 in COVID-19 has not been clearly defined.
The STAT family involves seven different proteins (STAT1, STAT2, STAT3, STAT4, STAT5A, STAT5B, and STAT6), which play a key role in the regulation of many physiological and pathological processes in humans [Citation8]. Specifically, STAT4 gene is a main component of the JAK-STAT signaling pathway, regulating the cellular-mediated immune responses [Citation9]. STAT4 protein consists of six domains which are stimulated by several cytokines, including interleukin (IL)12, type I interferon (IFN-I), IL23, IL2, IL27, and IL35 [Citation10,Citation11]. The literature regarding STAT4 and COVID-19 is scarce. We hypothesized that patients with COVID19 May have abnormal STAT4 expression, correlating to the clinical severity of the disease.
This work aimed to investigate the association between SIRT1 rs12778366 SNP & the expression level of STAT4 gene and the susceptibility to COVID-19 infections as well as their correlation to the clinicopathological data.
Material and methods
This study has been approved by the Mansoura University ethics committee (Institutional research board) with code (R.20.10.1) all patients gave an informed consent to participate
This study was conducted at the Medical Biochemistry Departments and Mansoura university hospital,Faculty of Medicine, Mansoura University, Egypt over a period of six month.
This study involved 200 participants (age 45–80 years), assigned into one of two groups; Group (1) included 100 ICU patients with critically ill COVID-19 infection and Group (2) included 100 healthy controls who had no previous history of COVID-19 infection.
Patients who had two consecutive negative PCR tests of COVID19, pregnant women, and patients with mild upper respiratory tract symptoms were excluded from the study.
Samples were collected over the period starting from June 2022 to September 2022 from
Isolation of peripheral blood mononuclear cells
Five ml of whole blood was collected from each participant in an EDTA collection tube. Peripheral mononuclear blood cells (PBMCs) were isolated using Histopaque-1077 (Sigma-Aldrich, USA) Ficoll density-gradient centrifugation as per the manufacturer’s instructions.
RNA extraction& cDNA synthesis
Total RNA was extracted by the TRIzol reagent from PBMCs (Zymo Research, Irvine, CA). NanoDrop2000 (Thermo. Fischer Scientific, Waltham, MA) was used to quantitate RNA. Before processing, the total RNA samples were kept at −80°C. Using SensiFAST cDNA Synthesis Kit [Bioline, Memphis, TN], reverse transcription was performed on the RNA in a final volume of 20 ml reactions.
Real time PCR
The STAT4 gene expression levels were evaluated using GAPDH as an internal control. The Hera plus SYBR Green qPCR kit [Willow fort, Birmingham, UK] was used according to the manufacturer’s protocol. Fold change was calculated using the comparative threshold cycle [2−ΔΔCt] for relative quantification normalized to an endogenous control [Citation12].
The following Primers (purchased from QIAGEN) were used:
DNA extraction
DNA was extracted from whole blood using Qia-amplification DNA extraction kit (Qiagen, USA).
Genotyping of SNP in SIRT-1 (rs12778366)
Real-time polymerase chain reaction with Taq Man allelic discrimination assay (Applied Biosystems,USA) was used. SNP (rs12778366) is detected in the 5′flanking region of the SIRT1 gene. Tagger software (http://www:broadinstitute.org/mpg/tagger/) was used to select tag SNP in the SIRT1 region plus 0.5 Kb downstream, and 30 Kb upstream (NCBI Build 35/UCSC hg17), the minor allele frequency >5% in Caucasian population and r2 > 1 as criteria.
PCR amplification
Predesigned primer/probe sets (C_1340370_10/PN4351379) were used (Applied Biosystems, USA). Probes were synthesized with reporter dye FAM or VIC covalently linked at the 5/and a quencher dye MGB linked to the 3/end of the probe. The rs12778366 (C_1340370_10) was supplied by (Applied Biosystems, USA).
Radiological assessment
All patients underwent non-contrast spiral CT scan on 16 detector GE CT scanner (GE optima 520) with scan area extending from the root of the neck to the level of the upper pole of the kidneys during a single-breath hold using 1 mm slice thickness and 5 mm inter-slice gap. Images were reconstructed in axial, coronal, and sagittal reformats with standard pulmonary filtering window (window width:1000–1200 &window level −500–600). The CT Digital Imaging and Communications in Medicine (DICOM) images of the scanned patients were transferred to picture archiving and communication system (PACS) and blindly correlated to the clinical or laboratory findings. Patients were quantitatively and qualitatively assessed as follows:
Quantitative analysis
All cases were evaluated according to the Radiological Society of North America (RSNA), COVID-19 Reporting and Data System (CORADs), total severity score (TSS), chest x ray score and ground glass percentage evaluation.
Total severity score (TSS)
The TSS is a semi-quantitative scoring system to assess the pulmonary involvement in patients with COVID-19. The total score is determined by summing the five lobe scores [Citation13]. The five lung lobes are assessed for ground-glass opacities, mixed ground-glass opacities, or consolidation. Each lobe is given 0 to 4 points, depending on the percentage of the lobe involvement: 0 (0%), 1 (1–25%), 2 (26–50%), 3 (51–75%), or 4 (76–100%). The TSS cutoff for identifying severe-critical type is 7.5.
Chest X-ray score
According to Borghesi and Maroldi [Citation14], the images have been analyzed in two steps. First: each lung is divided into three upper, middle, and lower zones; the right lung (A, B, C), and the left lung (D, E, F). The upper-level zones above the aortic arch includes zones A and D, the middle level zones include B and E, lying between the aortic arch and the right inferior pulmonary vein, and the lower-level zones (C and F), lying between the right inferior pulmonary vein and the lung bases.
In the second step, we set a score from 0 to 3 points according to the detected lesions as follows: Zero denotes no lung lesions, interstitial pulmonary infiltrates are given a score of one, Interstitial and alveolar infiltrates (more interstitial) are given a score of two, and the highest score of 3 is given for interstitial and alveolar infiltrate (more alveolar). The total score is a sum of the overall points from all lung zones. It ranges between 0 and 18 with a medium of 6.5.
Quantitative analysis of ground glass percentage
All DICOM data of thin cut CT chest were analyzed using Synapse 3D Fujifilm Medical Systems version 3.5 on specific workstations for automated analysis to calculate the ground-glass opacities (GGOs)
The data sets include four groups of density ranges:
Red color representing emphysema: From −1024 to −950 HU.
Yellow color representing normal lung: From −949 to −750 HU.
Blue color representing GGOs: From −749 to −300 HU.
Violet color representing consolidation: From −299 to +40 HU.
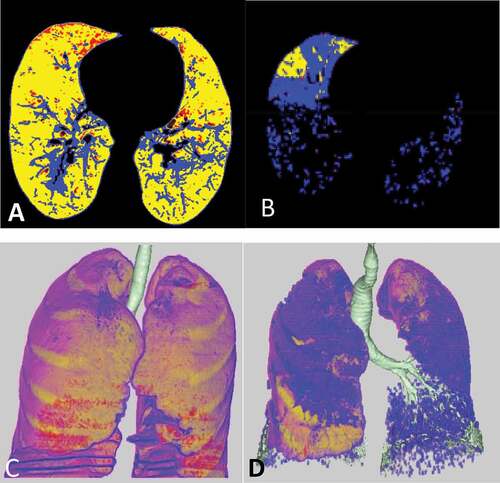
shows the variability between mild and severe cases.
Figure 1. Mild versus marked CT analysis of two cases in our study. A&C) axial colored CT image and 3D image analysis of the lung revealed mild ground glass percentage (20%) in blue color. B&D) axial colored CT image and 3D image analysis of the lung revealed marked ground glass percentage (77%) in blue color.
Qualitative analysis
Additionally, CT was assessed for the presence of other CT findings associated with COVID-19 such as consolidations, crazy paving, subpleural bands, vascular dilatation, and reverse halo sign as well as atypical features of COVID-19 such as pulmonary nodules, mediastinal lymphadenopathy, and pleural effusion with mention of laterality, lobar affection (upper or lower), and distribution pattern (peripheral, central, or diffuse).
Statistical analysis
Data were entered and analyzed using IBM-SPSS software (IBM Corp. Released 2017. IBM SPSS Statistics for Windows, Version 25.0. Armonk, NY: IBM Corp.) and MedCalc® Statistical Software version 20 (MedCalc Software Ltd, Ostend, Belgium; https://www.medcalc.org; 2021). Qualitative data were expressed as N (%) and their association was tested for by Chi-Square test of association (with phi as a measure of the strength of association) and Fisher’s exact test was used if expected counts was smaller than 5. Quantitative data were initially tested for normality using Shapiro-Wilk’s test with data being normally distributed if p > 0.050. The presence of significant outliers (extreme values) was tested for by inspecting box plots. Quantitative data were expressed as mean ± SD or median (Q1-Q3) and compared between two groups using Independent-Samples t-test or Mann-Whitney U test. Binary logistic regression analysis was used to predict the likelihood of occurrence of COVID19. The crude odds ratio was calculated with its 95% CI. One-Sample Wilcoxon’s signed ranks test was used to compare quantitative data vs. a hypothetical value. Spearman’s correlation was used to assess the association direction and strength between non-normally distributed data. For any of the used tests, results were considered as statistically significant if p value ≤ 0.050. Appropriate charts were used to graphically present the results whenever needed.
Results
This study involved 200 participants assigned into one of two groups; The first group included 100 ICU patients with confirmed COVID-19 infection compared to 100 healthy participants in the control group.
Association between COVID-19 and SIRT1 SNP
and show higher frequency of ‘T’ allele and T/T genotype in both cases and controls, however, a statistically significant higher frequency of ‘C’ allele and C/T genotype in the case vs. the control group has been found. The association was of low strength (φ = 0.105 for alleles, and 0.154 for genotypes). Participants with ‘C’ allele had 2.56-times higher odds to exhibit COVID-19 compared to participants with ‘T’allele. Participants with C/T genotype had 2.72 times higher odds to exhibit COVID-19 compared to participants with T/T genotype. None of the 200 participants had C/C genotype. SNP exact test for Hardy-Weinberg equilibrium (HWE) was done using SNP Stats web tool (https://www.snpstats.net/start.htm) and it showed that all participants (N = 200), COVID-19 cases (N = 100), and healthy controls (N = 100) were all in a HWE (P = 1.000).
Table 1. Association between COVID-19 and SIRT1 SNP.
Expression levels of STAT4 in COVID-19 patients
One-sample Wilcoxon signed rank test for STAT4 fold change (FC) showed
Median STAT4 (FC) (Median = 0.18, 95% CI = 0.15–0.27) was lower than the normal control value of 1.0. This difference was statistically significant (Hodges-Lehmann location estimator = 0.217, P < 0.001).
Clinico-laboratory, and radiologic differences of the two SIRT1 genotypes
shows a statistically significant higher nodal enlargement, typical RSNA, high/very high CO-RADS, GGO%, and number of lobes affected in T/T vs. C/T genotype, and a significantly higher negative RSNA, very low/low/equivocal CO-RADS, and TSS score in C/T vs. T/T genotype.
Table 2. Clinico-laboratory, and radiologic differences of the two SIRT1 genotypes.
There was a statistically significant difference of RSNA classification, CO-RADS, TSS and percentage of GGO between T/T vs. C/T genotypes with P value < 0.05.
There was a statistically significant difference regarding nodal enlargement and numbers of lobes affected. STAT4 (FC) was lower in T/T vs. C/T genotype though it did not achieve statistical significance.
Correlations between STAT4 (FC) and clinical, laboratory, and radiological parameters
showed a statistically significant positive correlation of medium strength between STAT4 (FC) and body temperature, respiratory rate, and CXR score, and a statistically significant negative correlation of medium strength between STAT4 (FC) and pH and PO2.
Table 3. Correlations between STAT4 (FC) and clinical, laboratory, and radiological parameters.
Diagnostic performance of serum STAT4
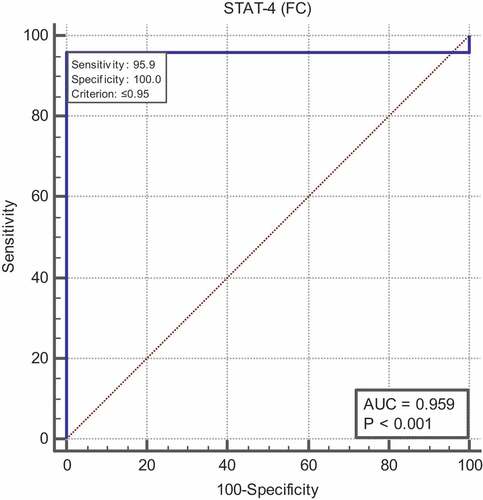
Receiver operating characteristic (ROC) curve analysis revealed A cutoff value of ≤ 0.95 for STAT4 can significantly discriminate COVID-19 from healthy control (Area under the curve (AUC) = 0.959, with 95.9% sensitivity and 100% specificity) ().
Discussion
The present study revealed that SIRT1 polymorphism and decreased expression of STAT4 are associated with increased susceptibility to COVID-19 infection and correlate to its phenotypic manifestations.
SIRT1 along with the Sirtuin family (SIRT1–7) has been known as a primary defense against DNA and RNA viral pathogens via promoting the down regulation of ADAM 17 (A Disintegrin and Metalloproteinase Domain 17), additionally termed TNF-α converting enzyme (TACE) [Citation14,Citation15]. This consequently decreases the levels of TNF-α, IL-1β and IL-6, which are known to induce an exaggerated inflammatory response as may occur with COVID-19 [Citation14,Citation16]. SIRT1 endorses M2 polarization of the peripheral monocytes to the anti-inflammatory phenotype, an effect mediated by increased STAT6 gene expression [Citation17]. As well, SIRT1 displays a key function in the regulation of JAK/STAT pathway in several inflammatory conditions by regulating the translocation of HMGB1 from the nucleus to the cytoplasm [Citation7]. The JAK/STAT pathway is a fundamental component in the inflammatory process triggered by SARS-CoV-2 infection progressing toward a cytokine storm. STAT signaling represents a potential target to modulate dendritic cell development and function as well as induction of antigen-specific cytotoxic T cell activity [Citation18]. The present study showed that patients with COVID-19 exhibited lower STAT4 gene expression compared to healthy controls. The altered expressions of many genes have been previously reported in COVID-19 patients [Citation19,Citation20]. Viral infections largely contribute to substantial remodeling of the intracellular gene expression due to restricted key gene expression by the active viral proteins as well as populating the cell with viral transcripts [Citation20]. Although patients with COVID-19 react differently to the same viral load, inhibition of STAT phosphorylation and nuclear translocation in SARS-CoV-2-infected cells have been reported in recent studies [Citation21]. The JAK/STAT signaling cascade has been identified to be widely targeted by the SARS-COV-2 virus across cell types [Citation22–24], which suggests that the virus benefits from inhibiting the JAK-STAT pathway [Citation24]. In the same line, STAT1 exhibited lower expression in CD14+ monocytes and plasmablasts in severe cases of COVID-19 compared to mild cases [Citation21]. Taken together, it seems that several viral factors impede STAT function in patients with severe COVID19, as shown in earlier studies [Citation25–27]. Correlating the levels of STAT4 expression to the clinical, laboratory, and radiological data of patients included in the present study, it has been revealed that there is a statistically significant positive correlation between STAT4 and body temperature, respiratory rate, and CXR score.
Several radiological studies considered CXR score a very good predictor for assessing the course and severity of COVID-19 disease [Citation28–30].
On the other hand, a statistically significant negative correlation has been found between STAT4 (FC) and pH and PO2. The above shown clinical profile of altered STAT4 expression with tachypnea, hypoxemia, and PH changes toward acidosis with elevated body temperature reflects the severity of SARS-COV-2 infection. The heightened inflammation in patients with severe COVID-19 causes activation of immune cells and altered gene expression. This may be a trigger to alterations in partial gas pressure of the blood dissolved carbon dioxide (PaCO2) leading to pronounced shifts in the pH level [Citation9,Citation31].
Single nucleotide polymorphisms (SNPs) in SIRT1 has significantly impacted its expression and activity [Citation32]. As regards SIRT 1 SNP, higher frequency of ‘T’ allele and T/T genotype has been noted in both cases and controls, however, a statistically significant higher frequency of ‘C’ allele and C/T genotype in the case vs. the control group has been found. It has also been revealed that participants with ‘C’ allele have 2.56-times higher odds to exhibit COVID-19 compared to participants with ‘T’allele. Furthermore, participants with C/T genotype have 2.72 times higher odds to exhibit COVID-19 compared to participants with T/T genotype. These results may point out the strong association between SIRT-1 gene polymorphism and susceptibility to COVID-19 infection. Hence, it is worth considering the likelihood that specific genes may predispose to the severity of COVID19, and consequently determine the phenotypic presentations and the outcomes. SIRT1 is thought to provide an effective antiviral response mediated by upregulating genes transcribing the Type I IFN (IFNβ) [Citation33], and reduce inflammation in COVID‐19 patients by directly modulating the immune response in macrophages [Citation34]. Previous studies showed significantly altered Sirtuin 1 (SIRT1) expression in COVID-19 patients [Citation35–37]. Infectious diseases, including COVID19, were originally polygenic diseases.
Correlating to the radiological data, patients with the T/T genotype showed higher nodal enlargement, typical RSNA, high/very high CO-RADS, GGO%, and number of lobes affected vs. C/T genotype. Previous studies correlate the automated GGO percent with the clinical severity of the COVID-19 patients [Citation38–40]. This suggests that the severity of pulmonary involvement is more pronounced in the T/T genotype. This further confirms that host genetics is a major contributing factor to COVID-19 susceptibility and severity and thus may provide new insights into therapeutic development [Citation41]. In this context, genome-wide association studies may predict the genetic predisposition for COVID-19 in the near future [Citation42].
Upregulation of SIRT1 has been shown to directly inhibit viral replication in addition to its an anti-inflammatory function [Citation43]. On the contrary, SIRT1 depletion provokes viral replication regardless its inhibitory effect on ADAM17 activity [Citation43,Citation44]. Accordingly, the activation of SIRT1 May help prevent the hyperinflammatory response associated with COVID-19 infection. Potent and selective pharmacological Sirt1 activators are available and provides a rational basis for future therapeutic development [Citation45].
In conclusion, SIRT-1 polymorphism together with decreased expression of STAT4 have been found to exhibit a significant association with increased susceptibility to COVID-19 infection and correlate to its phenotypic manifestations.
Ethics
This study has been approved by the Mansoura University Ethics Committee (Institutional Research Board) with code (R.20.10.1) and all patients have signed an informed consent.
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- Furqan MM, Verma BR, Cremer PC, et al. Pericardial diseases in COVID19: a contemporary review. Curr Cardiol Rep. 2021;23(7):1–10. doi: 10.1007/s11886-021-01519-x
- Fauci AS, Lane HC, Redfield RR. Covid-19 — navigating the uncharted. N Engl J Med [Internet]. 2020 Feb 28;382(13):1268–1269. doi: 10.1056/NEJMe2002387
- Shaker OG, Wadie MS, Ali RMM, et al. SIRT1 gene polymorphisms and its protein level in colorectal cancer. Gene Rep. 2017;7:164–168. doi: 10.1016/j.genrep.2017.04.005
- Imai S, Guarente L. Ten Years of NAD-dependent SIR2 family deacetylases: implications for metabolic diseases. Trends Pharmacol Sci. 2010;31(5):212–220. doi: 10.1016/j.tips.2010.02.003
- Alves-Fernandes DK, Jasiulionis MG. The role of SIRT1 on DNA damage response and epigenetic alterations in cancer. Int J Mol Sci. 2019;20(13):3153. doi: 10.3390/ijms20133153
- Michan S, Sinclair D. Sirtuins in mammals: insights into their biological function. Biochem J. 2007;404(1):1–13. doi: 10.1042/BJ20070140
- Kim YM, Park EJ, Kim JH, et al. Ethyl pyruvate inhibits the acetylation and release of HMGB1 via effects on SIRT1/STAT signaling in LPS-activated RAW264. 7 cells and peritoneal macrophages. Int Immunopharmacol. 2016;41:98–105. doi: 10.1016/j.intimp.2016.11.002
- Satarker S, Tom AA, Shaji RA, et al. JAK-STAT pathway inhibition and their implications in COVID-19 therapy. Postgrad Med. 2021;133(5):489–507. doi: 10.1080/00325481.2020.1855921
- Dhont S, Derom E, Van Braeckel E, et al. The pathophysiology of ‘happy’ hypoxemia in COVID-19. Respir Res [Internet];21(1):198. [2020 Jul 28]. Available from: https://pubmed.ncbi.nlm.nih.gov/32723327
- Levy DE, Darnell JE. Stats: transcriptional control and biological impact. Nat Rev Mol Cell Biol. 2002;3(9):651–662. doi: 10.1038/nrm909
- Hoey T, Zhang S, Schmidt N, et al. Distinct requirements for the naturally occurring splice forms Stat4α and Stat4β in IL-12 responses. EMBO J. 2003;22(16):4237–4248. doi: 10.1093/emboj/cdg393
- Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative CT method. Nat Protoc. 2008;3(6):1101–1108. doi: 10.1038/nprot.2008.73
- Li K, Fang Y, Li W, et al. CT image visual quantitative evaluation and clinical classification of coronavirus disease (COVID-19). Eur Radiol. 2020;30(8):4407–4416. doi: 10.1007/s00330-020-06817-6
- Fontani F, Domazetovic V, Marcucci T, et al. MMPs, ADAMs and their natural inhibitors in inflammatory bowel disease: involvement of oxidative stress. J Clin Gastroenterol Treat. 2017;3(39):10–23937. doi: 10.23937/2469-584X/1510039
- Budayeva HG, Rowland EA, Cristea IM, et al. Intricate roles of mammalian sirtuins in defense against viral pathogens. J Virol. 2016;90(1):5–8. doi: 10.1128/JVI.03220-14
- Yoo C-H, Yeom J-H, Heo J-J, et al. Interferon β protects against lethal endotoxic and septic shock through SIRT1 upregulation. Sci Rep. 2014;4(1):1–8. doi: 10.1038/srep04220
- Kolinko L, Shlykova O, Izmailova O, et al. SIRT1 contributes to polarization of peripheral blood monocytes by increasing STAT6 expression in young people with overweight and low-risk obesity. Georgian Med News. 2021 Apr 4;(313):102–112.
- de Haas N, de Koning C, di Blasio S, et al. STAT family protein expression and phosphorylation State during moDC development is altered by Platinum-Based Chemotherapeutics. J Immunol Res [Internet]. 2019 Jun 11;2019:7458238. doi: 10.1155/2019/7458238
- Alobaidy ASH, Elhelaly M, Amer ME, et al. Angiotensin converting enzyme 2 gene expression and markers of oxidative stress are correlated with disease severity in patients with COVID-19. Mol Biol Rep [Internet]. 2023;50(7):5827–5836. doi: 10.1007/s11033-023-08515-0
- Mendez AS, Ly M, González-Sánchez AM, et al. The N-terminal domain of SARS-CoV-2 nsp1 plays key roles in suppression of cellular gene expression and preservation of viral gene expression. Cell Rep [Internet]. 2021/09/30. 2021 Oct 19;37(3):109841. doi: 10.1016/j.celrep.2021.109841
- Rincon-Arevalo H, Aue A, Ritter J, et al. Altered increase in STAT1 expression and phosphorylation in severe COVID-19. Eur J Immunol. 2022 Jan;52(1):138–148. doi: 10.1002/eji.202149575
- Xia H, Cao Z, Xie X, et al. Evasion of type I interferon by SARS-CoV-2. Cell Rep. 2020;33(1):108234. doi: 10.1016/j.celrep.2020.108234
- Miorin L, Kehrer T, Sanchez-Aparicio MT, et al. SARS-CoV-2 Orf6 hijacks Nup98 to block STAT nuclear import and antagonize interferon signaling. Proc Natl Acad Sci. 2020;117(45):28344–28354. doi: 10.1073/pnas.2016650117
- Chen D-Y, Khan N, Close BJ, et al. SARS-CoV-2 Disrupts Proximal Elements in the JAK-STAT pathway. J Virol [Internet]. 2021/09/09. 2021 Sep 9;95(19):e0086221–e0086221. doi: 10.1128/JVI.00862-21
- Sarker B, Rahaman MM, Islam MA, et al. Identification of host genomic biomarkers from multiple transcriptomics datasets for diagnosis and therapies of SARS-CoV-2 infections. PLoS One. 2023;18(3):e0281981. doi: 10.1371/journal.pone.0281981
- Lee JS, Park S, Jeong HW, et al. Immunophenotyping of COVID-19 and influenza highlights the role of type I interferons in development of severe COVID-19. Sci Immunol. 2020;5(49):eabd1554. doi: 10.1126/sciimmunol.abd1554
- Hosseini A, Esmaeili Gouvarchin Ghaleh H, Aghamollaei H, et al. Evaluation of Th1 and Th2 mediated cellular and humoral immunity in patients with COVID-19 following the use of melatonin as an adjunctive treatment. Eur J Pharmacol. 2021 Aug;904:174193.
- Yasin R, Gouda W. Chest X-ray findings monitoring COVID-19 disease course and severity. Egypt J Radiol Nucl Med. 2020;51(1):1–18. doi: 10.1186/s43055-020-00296-x
- Toussie D, Voutsinas N, Finkelstein M, et al. Clinical and chest radiography features determine patient outcomes in young and middle-aged adults with COVID-19. Radiology. 2020;297(1):E197. doi: 10.1148/radiol.2020201754
- Borghesi A, Maroldi R. COVID-19 outbreak in Italy: experimental chest X-ray scoring system for quantifying and monitoring disease progression. Radiol Med. 2020;125(5):509–513. doi: 10.1007/s11547-020-01200-3
- Vaporidi K, Akoumianaki E, Telias I, et al. Respiratory drive in critically ill patients. Pathophysiology and clinical implications. Am J Respir Crit Care Med. 2020;201(1):20–32. doi: 10.1164/rccm.201903-0596SO
- Kurylowicz A. In search of new therapeutic targets in obesity treatment: sirtuins. Int J Mol Sci. 2016;17(4):572. doi: 10.3390/ijms17040572
- Chattree V, Singh K, Singh K, et al. A comprehensive review on modulation of SIRT1 signaling pathways in the immune system of COVID-19 patients by phytotherapeutic melatonin and epigallocatechin-3-gallate. J Food Biochem. 2022 Dec;46(12):e14259. doi: 10.1111/jfbc.14259
- Rajendrasozhan S, Yang S-R, Kinnula VL, et al. SIRT1, an antiinflammatory and antiaging protein, is decreased in lungs of patients with chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2008;177(8):861–870. doi: 10.1164/rccm.200708-1269OC
- Bordoni V, Tartaglia E, Sacchi A, et al. The unbalanced p53/SIRT1 axis may impact lymphocyte homeostasis in COVID-19 patients. Int J Infect Dis IJID Off Publ Int Soc Infect Dis. 2021 Apr;105:49–53.
- Zhou Y, Zhang F, Ding J. As a modulator, multitasking roles of SIRT1 in respiratory diseases. Immune Netw. 2022 Jun;22(3):e21. doi: 10.4110/in.2022.22.e21
- Huarachi Olivera RE, Lazarte Rivera A. Coronavirus disease (COVID-19) and sirtuins. Rev Fac Cien Med Univ Nac Cordoba. 2020 Jun;77(2):117–125. doi: 10.31053/1853.0605.v77.n2.28196
- Bressem K, Adams L, Albrecht J, et al. Is lung density associated with severity of COVID-19? Pol J Radiol. 2020;85(1):600–606. doi: 10.5114/pjr.2020.100788
- Salvatore C, Roberta F, Angela de L, et al. Clinical and laboratory data, radiological structured report findings and quantitative evaluation of lung involvement on baseline chest CT in COVID-19 patients to predict prognosis. Radiol Med. 2021;126(1):29–39. doi: 10.1007/s11547-020-01293-w
- Romanov A, Bach M, Yang S, Franzeck FC, Sommer G, Anastasopoulos C, et al. Automated CT lung density analysis of viral pneumonia and healthy lungs using deep learning-based segmentation, histograms and HU thresholds. Diagnostics. 2021;11(5):738. doi: 10.3390/diagnostics11050738
- Niemi MEK, Karjalainen J, Liao RG, et al. Mapping the human genetic architecture of COVID-19. Nature [Internet]. 2021/07/08. 2021 Dec:600(7889):472–477. doi: 10.1038/s41586-021-03767-x
- Yamamoto N, Nishida N, Yamamoto R, et al. Angiotensin–converting enzyme (ACE) 1 gene polymorphism and phenotypic expression of COVID-19 symptoms. Genes (Basel) [Internet]. 2021 Oct 1;12(10):1572. doi: 10.3390/genes12101572
- Khan H, Patel S, Majumdar A. Role of NRF2 and Sirtuin activators in COVID-19. Clin Immunol [Internet]. 2021/11/16 2021 Dec;233:108879. doi: 10.1016/j.clim.2021.108879
- Miller R, Wentzel AR, Richards GA. COVID-19: NAD+ deficiency may predispose the aged, obese and type2 diabetics to mortality through its effect on SIRT1 activity. Med Hypotheses. 2020;144:110044. doi: 10.1016/j.mehy.2020.110044
- Dai H, Sinclair DA, Ellis JL, et al. Sirtuin activators and inhibitors: promises, achievements, and challenges. Pharmacol Ther. 2018 Aug;188:140–154.